1. Introduction
In previous years, magnetic resonance imaging (MRI) has been efficiently used to study the functionality of the human brain. MRI images are capable of establishing and accomplishing unmatched medical examination to determine whether the brain has any abnormalities. Lately, in addition to scanning adult brains, this imaging technique has also been used as a new non-invasive instrument designed for improving and checking fetal brain development in the uterus. Since around 3 in every 1000 pregnant women carry fetuses with various types of brain abnormalities, fetal brain monitoring is important. Moreover, a series of neuropathological variations can occur, several of which are connected to serious clinical morbidities [
1]. A MRI scan for a fetus provides great details of the soft tissue and the structure of the brain. Therefore, it could be applied for primary identification of brain abnormalities and tumors with no need for medical interferences [
2,
3]. Identifying these defects in a primary phase using machine learning techniques has several advantages. First, it helps doctors to confer an accurate diagnosis, allowing doctors to provide parents with an understanding of the disease in order to prepare them for dealing with the abnormality. Second, it helps in managing the pregnancy and problems that might occur during this period. Third, it improves the quality of diagnosis and aids the decision for the best suitable treatment plan [
4].
Machine learning techniques make use of the ability of computers to train and learn without being explicitly programmed. Machine learning can be used for medical images to assist doctors with making accurate medical diagnoses. Additionally, it can reduce diagnostic errors by clinicians. Moreover, machine learning reduces the time scale and effort made during an examination [
5]. Recently, machine learning techniques have been used on fetal brain MRI images for the early detection of abnormalities, as well as for the identification and classification of these abnormalities [
6]. Most of the previous work that used fetal brain images focused on segmenting fetal brain images to detect abnormalities or separating the fetal brain from the rest of the body. Fewer studies considered the use of machine learning methods to identify defects existing in the fetal brains [
7,
8]. To the best of our knowledge, previous studies that applied classification methods to fetal brain MRI images are limited to References [
9,
10,
11]. Although the first two studies performed classification on fetal MRI brain images, the authors only classified a brain defect in newborns called small for gestational age (SGA). They first assumed that textural patterns of fetal brain MRI images are associated with newborn neurobehavior. Then, they constructed a support vector machine (SVM) classifier for predicting the SGA abnormality after the birth of the fetus (becoming a neonate). These methods have several limitations. First, these approaches only used fetal images of 37 GAs, in contrast to applying a wide range of fetal GAs. Second, their methods only classify one type of abnormality; namely, SGA for newborns. Third, mapping the fetal MRI to its corresponding MRI after birth is an essential hypothesis needed to classify the SGA abnormality. Additionally, the authors used two small datasets which only have 91 and 83 images, respectively. Lastly, the brain of the fetus was clipped in a manual manner, which is a time-wasting process. The authors of Reference [
11] classified several fetus abnormalities, but the highest classification accuracy achieved was limited to 80%.
The literature shows that the majority of work done on detecting and classifying brain abnormalities in early age was limited to infants, neonates, and preterm infants, rather than fetuses. Ball et al. [
12] used the independent component analysis (ICA) technique with a SVM classifier to identify brain defects in preterm infants. Symser et al. [
13] used support vector regression (SVR) to predict the GA of infants. He et al. [
14] proposed a stacked spare auto-encoder (SSAE) framework based on an artificial neural network (ANN) classifier to predict preterm defects. They also used an SVM classifier to detect autism. Similar work was done by Jin et al. [
15] to identify infants with autism.
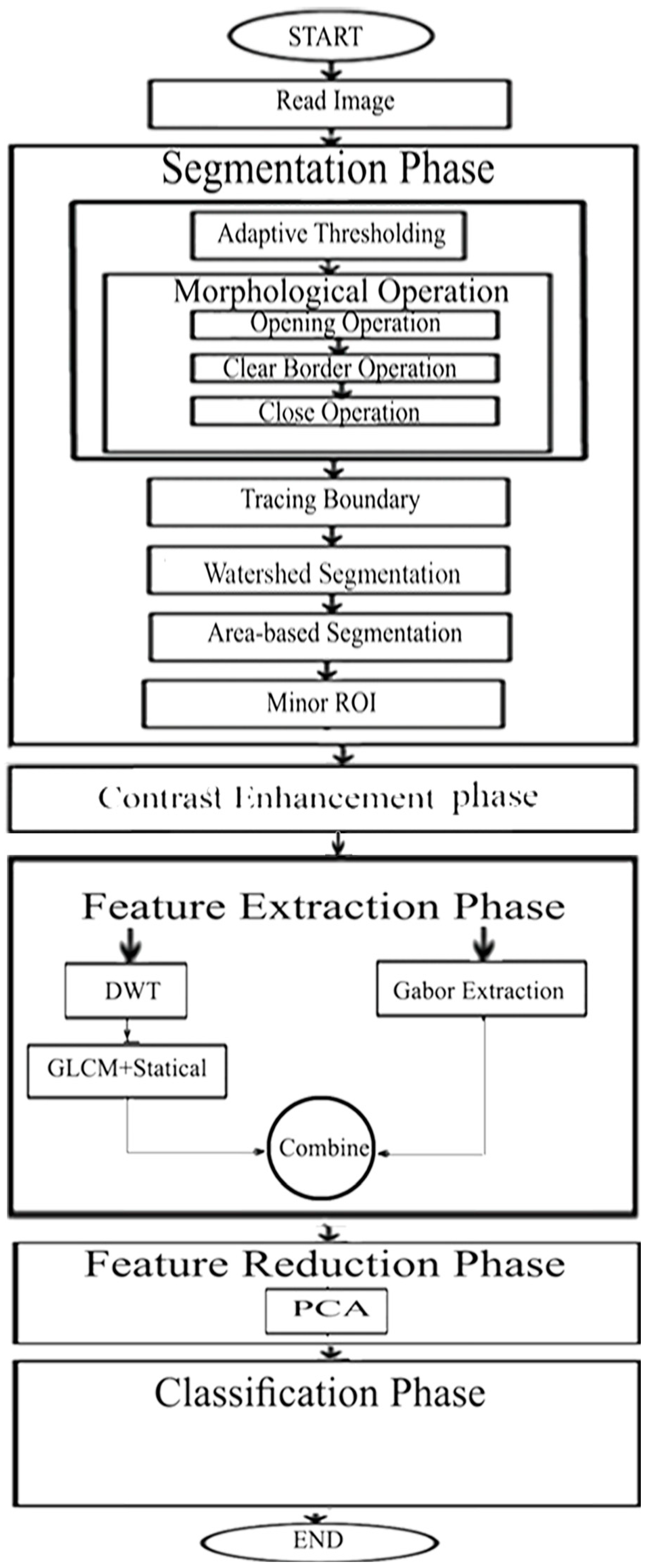
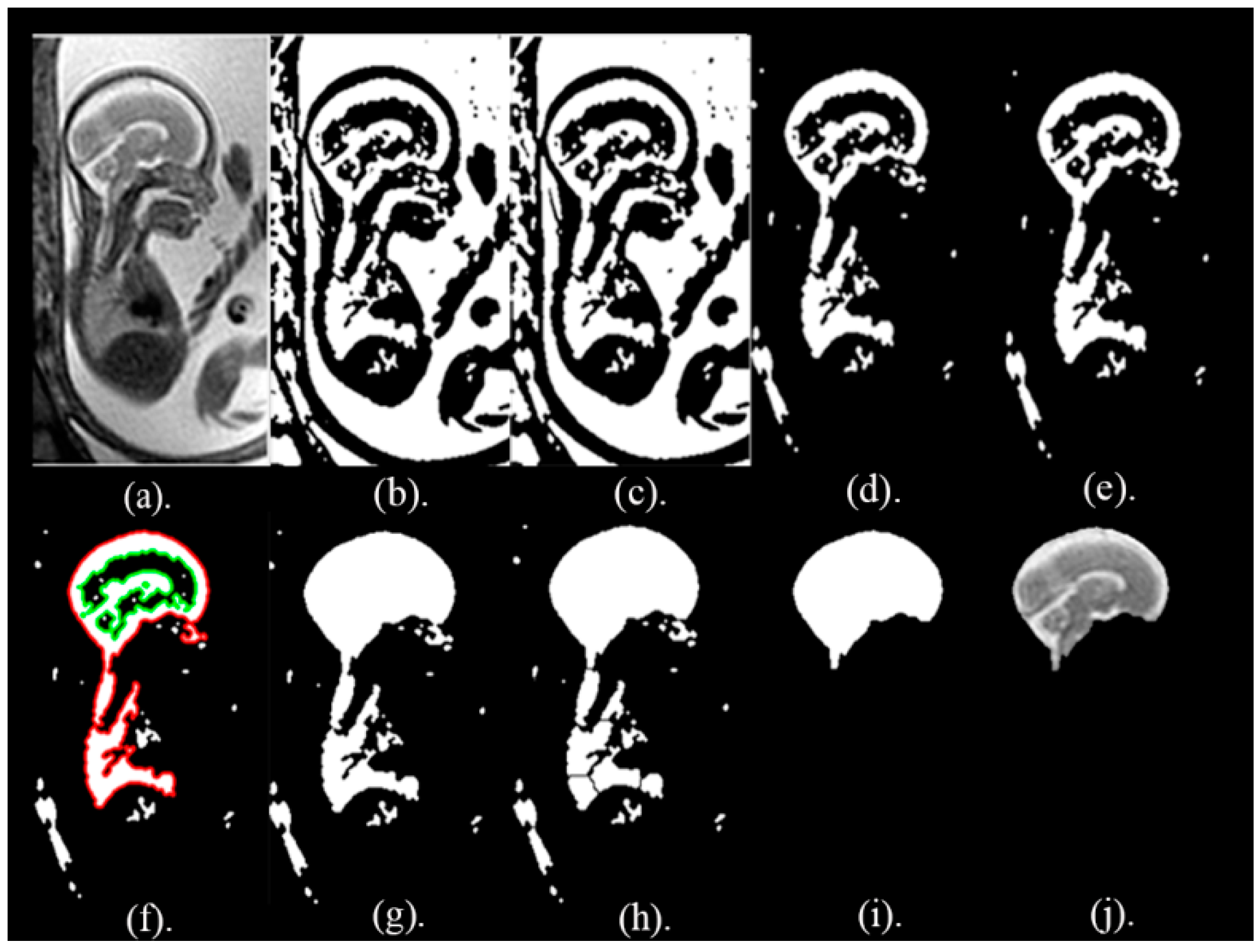
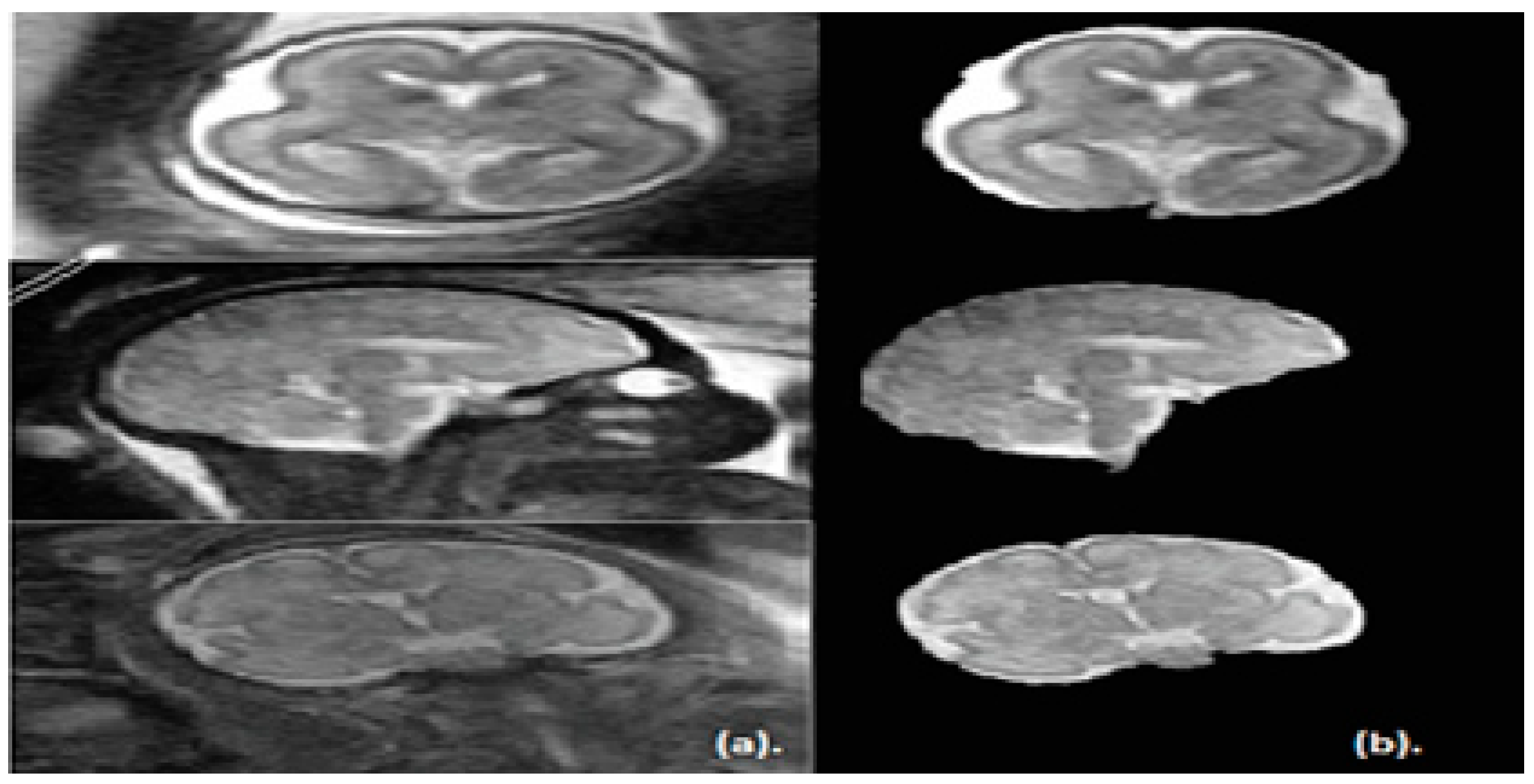
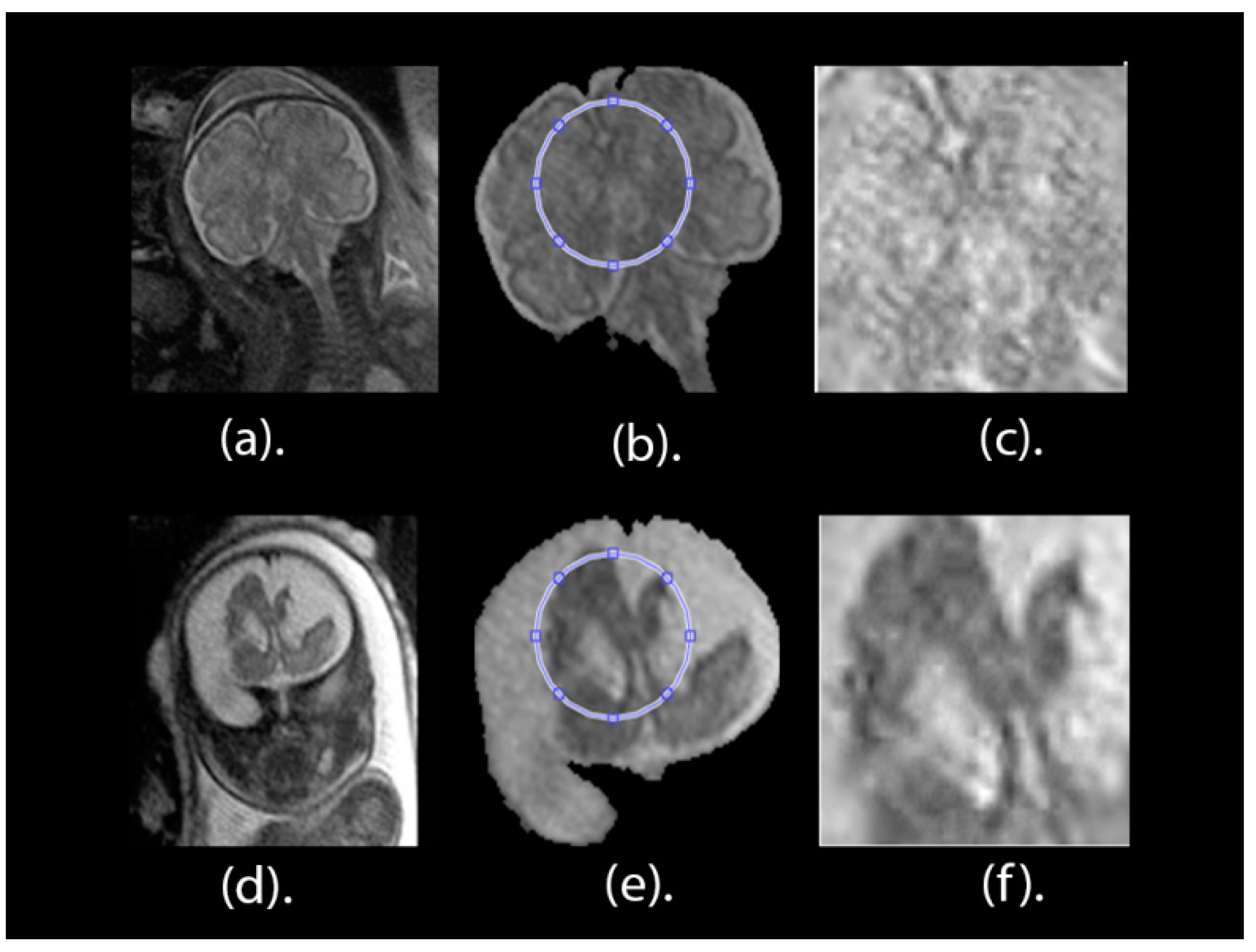
The key objective of this article is classifying between normal (healthy) and abnormal (unhealthy) brains. As it is clear from the previous two paragraphs, the work done in fetal brain classification is very limited. Most of the previous related research dealing with brain defect classification in a primary age have been for infants and preterm neonates. As stated before, the early detection of brain abnormalities is important as it helps in managing the pregnancy and problems that might occur during this period. It could also enhance the diagnosis process and follow up plans. Therefore, this article proposes a novel approach to identify defects in fetal brain scans. This approach is divided into five phases: Segmentation, enhancement, feature extraction, feature reduction, and classification. First is the segmentation, where the brain of the fetus is defined as the region of interest (ROI). It is cropped semi-automatically from other fetal parts. This is performed in three steps. In the first step, the cerebrospinal fluid (CSF), maternal tissues, and strips of the skull are removed from the fetal brain MRI using morphological operations. In the next step, area-based segmentation (ABS) is performed, where the largest area in the image is detected and segmented using region properties and watershed transform methods. In the last step of the segmentation phase, a minor ROI containing the fetal brain abnormality (FBA) is delimited and clipped out of the fetal scan semi-automatically. The following phase is called the enhancement phase. In this phase the minor ROI image is improved using a mixture of filters to increase its contrast. Subsequently comes the feature extraction phase, where six subsets of features are extracted using different textural analysis feature extraction methods. Two of these feature subsets have a high dimensional space; therefore, they are reduced using the principal component analysis (PCA) technique in the feature reduction step. The last phase is the classification phase, where the six subsets of features are used to train and test different classification models, such as random forest decision tree, naïve Bayes, radial basis function (RBF) network, diagonal quadratic discriminates (DQDA), and K-nearest neighbour (K-NN). Several bagging and Adaboosting ensembles are also constructed using naïve Bayes, the RBF network, and random forest classifiers to improve the prediction of their respective individual classifiers.
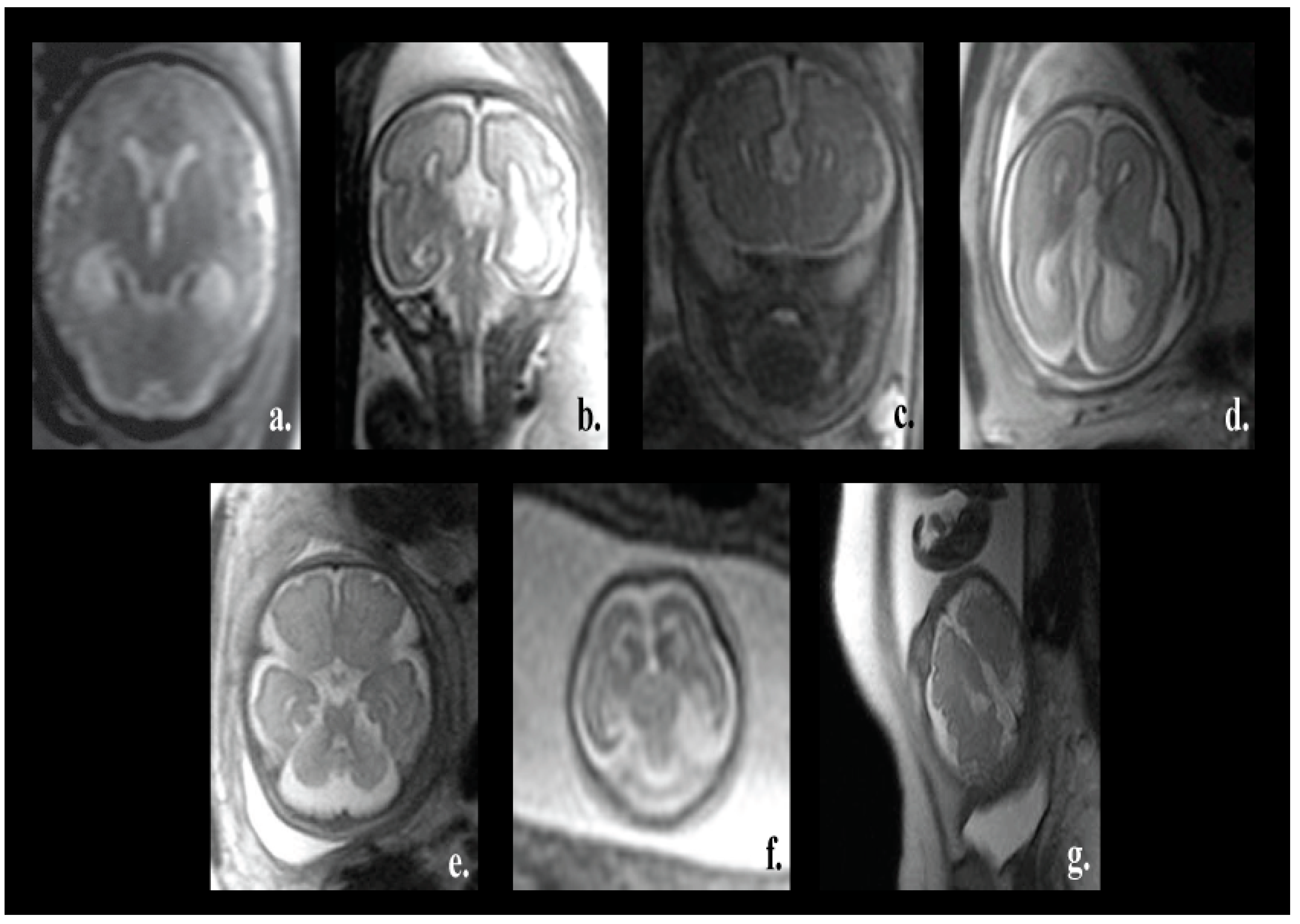
One of the major advantages of our proposed pipeline is its ability to differentiate between normal and various fetal brain abnormalities, including agenesis of the septi pellucidi, Dandy–Walker variant/malformation, colpocephaly, agenesis of the corpus callosum, mega-cisterna magna, cerebellar hypoplasia, and polymicrogyria. All of these abnormalities are shown in
Figure 1.
The corpus callosum is the curved structure that connects the left and right hemispheres of the brain. Agenesis of the corpus callosum (shown in
Figure 1d) is a brain disorder in which the corpus callosum is partially or completely absent [
16]. The septi pellucidi is a tinny tissue formed in the midline of the fetal brain. Agenesis of the septi pellucidi (shown in
Figure 1a) is a fetal brain defect that affects the development of the septi pellucidi. It commonly occurs in association with other brain disorders [
17,
18]. Dandy–Walker syndrome (shown in
Figure 1b) is a congenital malformation that affects the cerebellum, the third, and the fourth ventricles of the brain. It is characterized by partial or complete agenesis of the cerebellar vermis, cystic dilatation of the fourth ventricle, and the enlargement of the posterior fossa [
19,
20]. Colpocephaly (shown in
Figure 1c) is a fetal brain abnormality in which the posterior of the lateral ventricles of the fetal brain are ballooned. It is characterized by an impaired intellect and microcephaly (an abnormally small head) [
21]. Mega-cisterna magna (
Figure 1e) is another brain defect which is characterized by the enlargement of the cisterna, cerebellar hemispheres, and morphologically intact vermis. The expansion of the cisterna magna measures more than 10 mm from the posterior aspect of the vermis to the internal aspect of the skull arch [
22]. Cerebellar hypoplasia (shown in
Figure 1f) is one more neurological disorder in which the cerebellum is partially developed and its size is small. Polymicrogyria (shown in
Figure 1g) is a complex congenital malformation where the surface of the fetal brain normally has many ridges or folds, called gyri [
23]. Friede et al. [
24] defined polymicrogyria as “an abnormally thick cortex formed by the piling upon each other of many small gyri with a fused surface”.
Another advantage of the proposed approach is that the segmentation phase is a semiautomatic technique, saving the time and effort of manual segmentation. The proposed method was constructed and tested on a dataset consisting of 227 images, which is much larger when compared to the 91 and 45 images that have been used in previous studies [
9,
10]. Most importantly, the proposed approach can be applied for a wider range of fetal GAs and is not limited to a specific age.
The remainder of this paper is organized as follows:
Section 2 introduces the methods and materials. This is followed by
Section 3, which describes the results and comparative analysis of the proposed method.
Section 4 confers the performance of our method and, finally,
Section 5 concludes this article.
3. Results
In this article, 227 (113 healthy and 114 unhealthy) fetal MRI scans the trials were conducted with various GA and positions. The performance of the classification stage depended on the segmentation stage. For this reason, the performance of the segmentation stage for the proposed method was measured using the Dice metric (D) and the segmentation accuracy (SA) were calculated between the ground truth. The results of the segmentation method are shown in
Table 1. It was observed that, out of 50 MRI images, the mean values for Dice (D) and accuracy (AS) were 0.975 and 0.989, respectively. Additionally, the Jaccard Index (JAC), precision (PPV), sensitivity (S), and specificity (Sp) values were 0.951, 0.994, 0.972, and 0.979, correspondingly. These values are close to 1, which shows that our proposed method can successfully segment fetal brain images.
The performance of the new presented approach was calculated using the classification accuracy (ACC) for the five individual classification models for the six subsets of features, as shown in
Table 2. These individual classifiers include DQDA, naïve Bayes, KNN, random forest, and RBF network classifiers. As mentioned before, feature subsets two and six had high dimensional space, therefore the PCA feature reduction method was used to reduce their dimension. It is clear from the table that feature subset six has a higher ACC (91%, 95.6%, 93%, 91.2%, and 90.3% using QDQA, KNN, RBF network, naïve Bayes, and random forest) compared to the other subsets of features. Feature subset six represents features extracted from the Gabor filter feature extraction method, combined with the GLCM and statistical descriptive features extracted from DWT coefficients (fifth subset). The results showed that combining several textural features is better than using one type of feature extraction method. The highest classification accuracy, 95.6%, was achieved using the KNN classifier.
The sensitivity (
S), precision (
P), area under ROC (
AUC) were also calculated for the feature subset six (the results are shown in
Table 3). This features set has achieved the highest accuracy compared to other feature subsets (see
Table 1). The results of
Table 3 show that the classifiers have an acceptable performance in classifying the abnormalities. The
AUC of KNN was 99%, which was greater than the other classifiers (98.5%, 93%, 97.6%, and 96.3% of the DQDA, RBF network, naïve Bayes, and random forest classifiers, respectively). Additionally, the DQDA classifiers had a
P of 99%, which was higher than the other classifiers (97%, 94%, 91.2%, and 91% of the K-NN, RBF network, naïve Bayes, and random forest classifiers, respectively). Finally, the
S of the DQDA, K-NN, RBF network, naïve Bayes, and random forest classifiers were 83%, 94%, 93%, 92%, and 90.3%, respectively.
Ensemble classifiers combine the power of individual classification models available in the ensemble. This prevents the possibility of poor results that could be produced from a certain unsuitably individual model. Therefore, bagging and Adaboosting ensembles were constructed using naïve Bayes, random forest, and RBF networks. Their performances were compared with each other and the individual version of these classifiers. These results are presented in
Table 4.
Table 4 shows that the performance of bagging and Adaboosting ensembles constructed using naïve Bayes, RBF neural network, and random forest classifiers outperformed the performance of their individual models. This is because the ACC of the bagged and Adaboosted naïve Bayes were 92.1% and 92.5%, which are greater than the 91.63% of the individual naïve Bayes. In addition, the ACC of the bagged and Adaboosted random forest were 92.5% and 91.2%, which are greater than the 90.3% of the individual random forest classifier. In addition, the ACC of the Adaboosted RBF network is 93.4% which is greater than that of the individual RBF network classifier. However, the ACC of the bagged, RBF network was 93%, which was the same as the individual RBF Network classifier. The highest
P, of 93.6%, was achieved by the RBF network, the bagged RBF network, and bagged naïve Bayes classifiers. The highest
S of 93.4% was achieved by the Adaboosted RBF network.
To test and validate the statistical significance of the results, a one-way analysis of variance test was performed on the results obtained from the repeated fivefold cross-validation process. The null hypothesis
Ho for this experiment was that the mean accuracies of all the classifiers were the same. This test was first performed on the accuracy results of individual classifiers to test the statistical significance between them. The results are shown in
Table 5. Then the test was done on the results of the ensemble classifiers to test the statistical significance between them. The results of the test are shown in
Table 6. Finally, the test was applied on the results of both individual classifiers and their ensemble. The results of the test are shown in
Table 7. It can be observed from
Table 5,
Table 6 and
Table 7 that the
p-values achieved were lower than α, where α = 0.05. Therefore, it can be concluded that there was a statistically significant difference between the accuracies of the classifiers.
As it is commonly known, in order to construct an efficient classification model that achieves high classification accuracy, the number of images for each class label used to construct the model should be large enough. However, the dataset used in the proposed approach contained several types of abnormalities with a limited number of images for each abnormality. Therefore, we gathered all images of several abnormalities, of not only one type, along with normal images, and classified them to either normal or abnormal brains. Note that the datasets that are freely available online contain images of normal and not abnormal, which is why were constrained to classify between normal and abnormal only.
The main aim of this article is to classify fetal brain abnormalities. Several constraints arose for reaching this aim. First, we were restricted with the limited number of related research articles discussing this topic (3 articles, as stated in the introduction). The majority of the previous research has been limited to studies classifying brain deficiencies in early age for the preterm and neonatal stages, but not at the fetus stage. The second constraint was that datasets that contain images of fetal brains, as well as those from neonates and preterm infants are not available online. Finally, datasets that are available online for the fetal stage contain only images of normal brains, so we were not able to use them as our main target was to classify fetal brain images. Therefore, we compared our results with the performance of such approaches for fetal, neonates, and preterm infants to validate and indicate that the performance of our method is competitive. The results are displayed in
Table 8.
The proposed method has an
ACC that ranges between 90.3% and 95.6%, which is higher than the
ACC of the previous methods proposed for preterm and newborn infants [
12,
13,
14,
15] and which achieved 80%, 84%, 71%, and 76%, respectively. The AUC of the proposed method is also higher than the 80% achieved by the method used for classifying fetal abnormalities [
10]. The new approach presented in this article identified different brain deficiencies, unlike [
12,
13,
14,
15] methods that were limited to the classification of only one type of defect for preterm infants or newborns. Lastly, our new approach employed a larger number of MRI images compared to other approaches. Identifying fetal brain defects is a complicated task compared to scanning of newborns and preterm infants. Since scanning of fetal brains displays the head of the fetus along with the maternal parts of the mother and other surrounding fluids [
7], our approach, therefore, has several strengths over other methods reported in the previous related work. First, it is considered to be simple and reliable. Second, the results showed that it works well on T2 weighted scans. Third, it is capable of classifying different GA and several brain deficiencies. Finally, our approach classifies the defect before the fetus is born.
4. Discussion
The proposed framework is for the classification FBA. This framework uses several types of machine learning techniques. Most of the work done for brain MRI detection and classification in a primary age is for newborns and preterm infants. To the best of our own knowledge, the work done for FBC is limited to References [
9,
10,
11]. In the articles [
9,
10], the authors identified a brain deficiency called SGA in neonates, as opposed to fetuses. Their approach was applied on a very small number of images. These images were only limited to 37 weeks of fetal GA. Reference [
11] classified several fetal abnormalities, but the highest classification accuracy achieved was limited to 80%, which is insufficient for medical applications. Therefore, in this article, a new approach was proposed to classify brain defects from fetal images at various GAs.
The proposed framework consists of five steps, as follows: Segmentation, enhancing the minor ROI image contrast, feature extraction, feature reduction, and classification. In the segmentation phase, the minor ROI was cropped semi-automatically from the remaining parts of the fetal images. Afterwards, the minor ROI images were improved by a mixture of contrast stretching methods. Next, in the feature extraction phase, six subsets of features were extracted. The first subset represents the horizontal, vertical, approximate, and diagonal coefficients of the 4th decomposing stage of the DWT. The second subset feature was extracted using Gabor filters. The third subset of features was extracted using the gray-level co-occurrence matrix (GLCM) and statistical features. The forth subset is a combination of features extracted from DWT, GLCM, and statistical features. The fifth subset is the GLCM and statistical features extracted from the fourth level 2D-DWT decomposition of the minor ROI. The sixth subset represents features extracted using a Gabor filter combined with the GLCM and statistical descriptive features extracted from the DWT coefficients (fifth subset); that is to say that the sixth subset is a combination of the Gabor filter feature vector with the fifth subset. Note that feature subsets two and six are of high dimension; therefore, principal component analysis (PCA) was applied to them to reduce their dimension. Finally, the six subsets of features were used to train and validate different classification models, such as naïve Bayes, random forest decision tree, RBF network, DQDA, and (K-NN) classifiers. In addition, bagging and Adaboosting ensembles were constructed using naïve Bayes, random forest decision tree, and RBF network classifiers to see if they could enhance predictions of their individual models. The proposed pipeline reached a classification accuracy of 95.6%, which outperformed other methods shown in the literature. Therefore, it can be considered as a powerful tool that can help physicians in the early detection and diagnosis of fetal brain abnormalities.
Proper comparison with other previous work is mandatory and this comparison is more appropriate when it is based on the same dataset. However, as we explained before, our key goal was to classify fetal brain abnormalities. We faced a problem when searching for related research papers discussing this topic. We found that there is a limited number of papers which have explored fetal brain classification (only three relevant papers, as mentioned before). The authors of these papers used a machine learning classifier for either predicting one type of abnormality after the birth of the infant (becoming a neonate), which is not similar to our case (we want to differentiate between several types of abnormalities and normal brains and we strive to classify fetal abnormality before the fetus is born), or to classify several abnormalities and achieve a classification accuracy limited to 80% (which is not very high, especially in medical applications). The datasets are not freely available online. Moreover, all the fetal brain datasets that are available online only include images of normal brains. The dataset used in our work is the only dataset that is free online and contains abnormal images. In addition, the literature showed that the majority of work done on detecting and classifying brain abnormalities in early age was limited to infants, neonates, and preterm infants, rather than fetuses. We thought that the previous work done for infants, neonates, and preterm infants would be the closest work we could use for comparison. Therefore, we compared these approaches (examples [
12,
13,
14,
15]), but their data was not freely available online. We also compared with other papers that used fetal images (examples [
9,
11]).
As it is clear from the comparison with recent related work, most of them used the SVM classifier to construct their models. To make our study novel, we did not employ SVM classifier. SVM is a popular classifier; however, it would not be highly efficient if the number of features is large [
59]. In our case, several features sets were extracted. Most of these features had a high number of features. Moreover, SVM would produce insufficient results when the datasets are partitioned into subsets and used in parallel training, where each subset is used to train a classification model and then the knowledge is combined together [
60]. In our case, the dataset was split using a bootstrap aggregating resampling technique. These subsets were then used to train the bagging ensemble models. In addition, it is difficult to choose the parameters as well as the best kernel of SVM [
61]. For these reasons, SVM was not adopted in our article.
MATLAB R2017b and Waikato Environment for Knowledge Analysis (Weka) software [
62] were used for the implementation of the proposed framework.