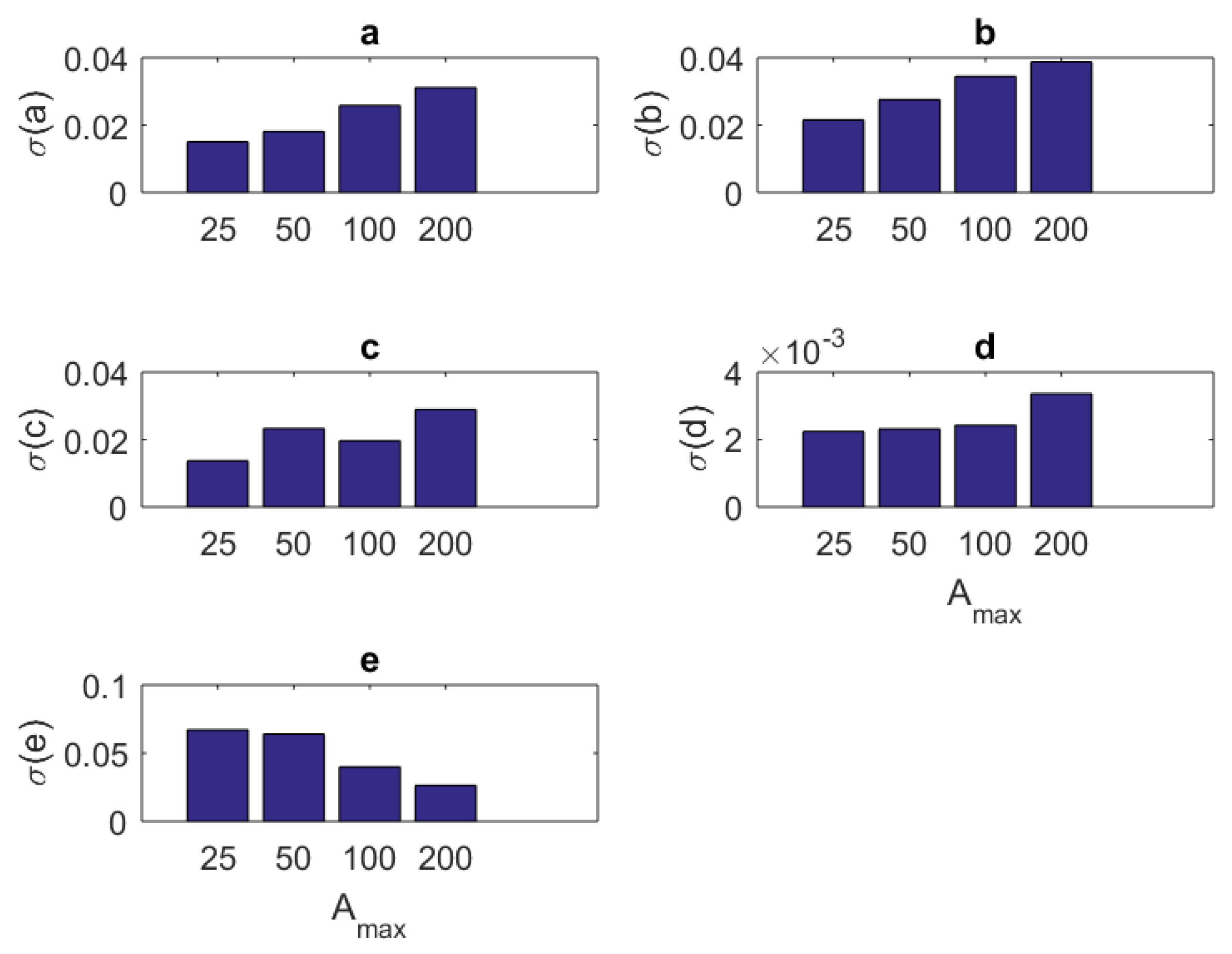
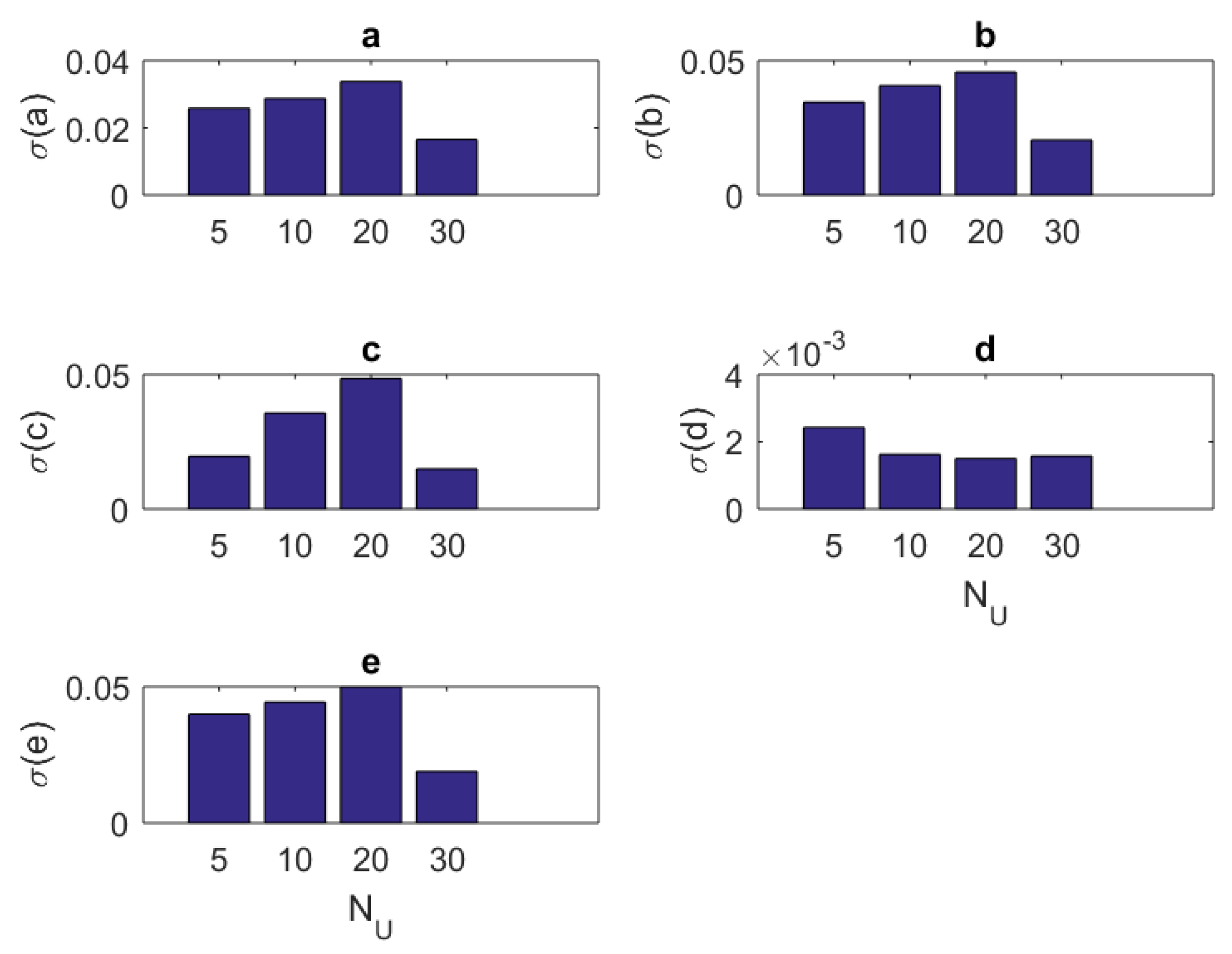
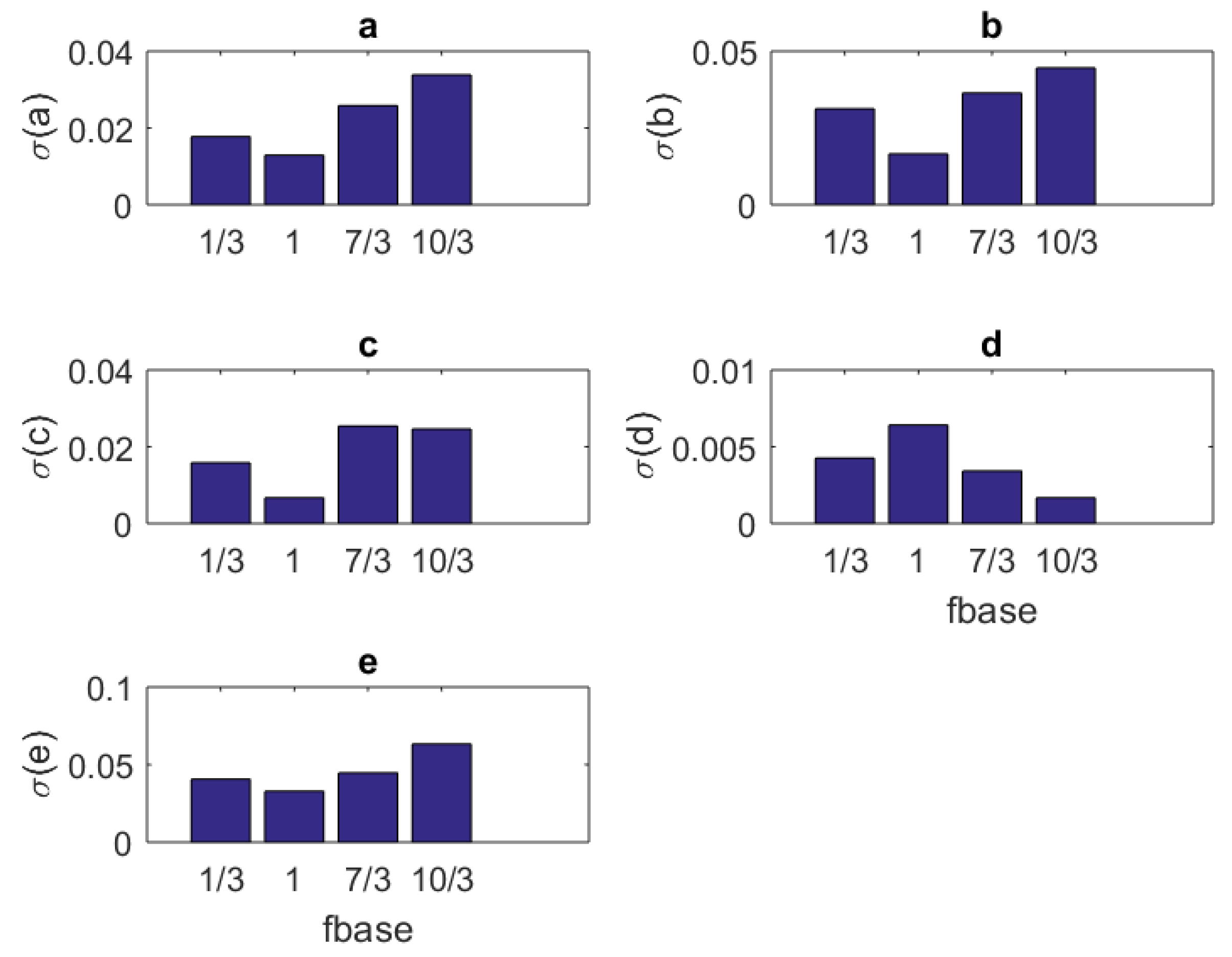
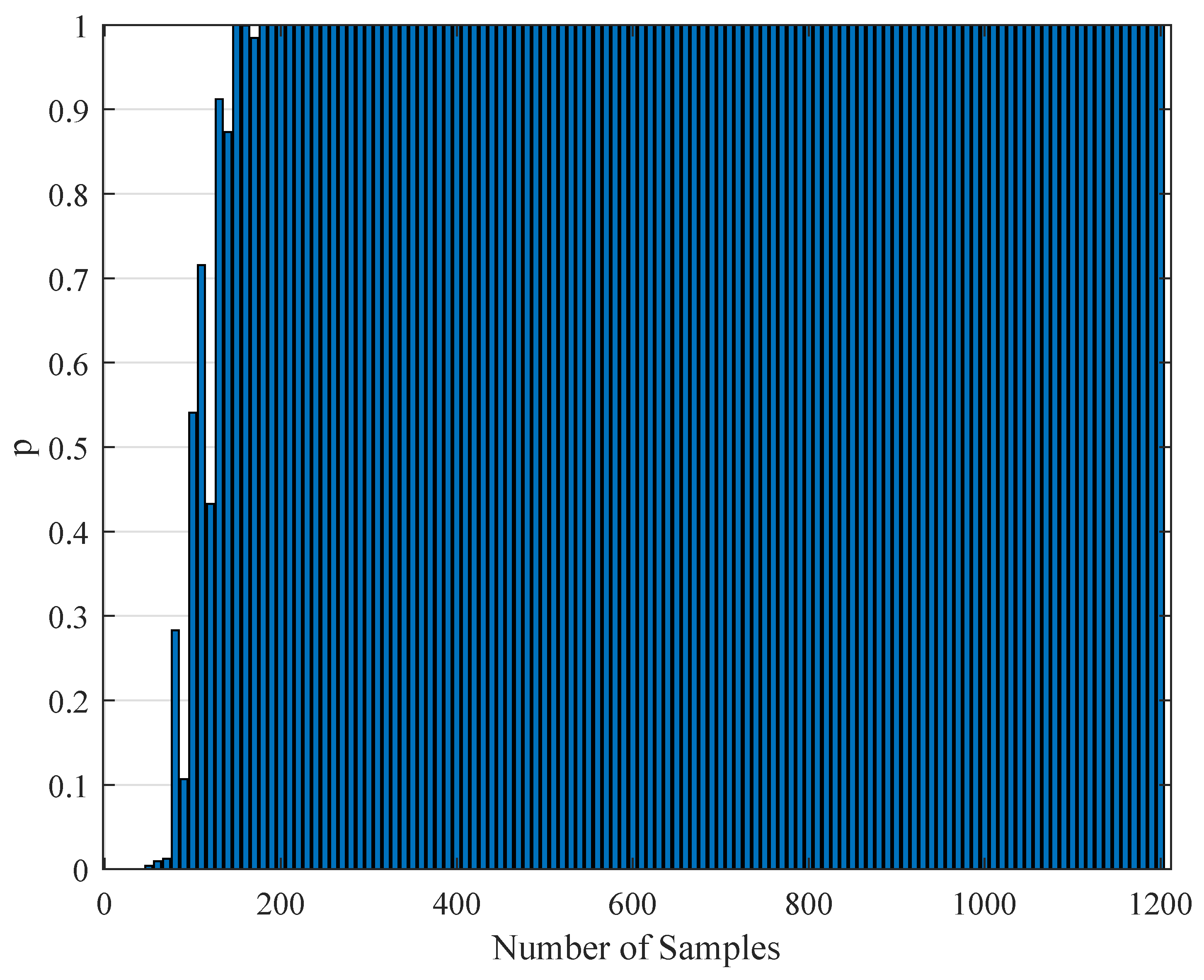
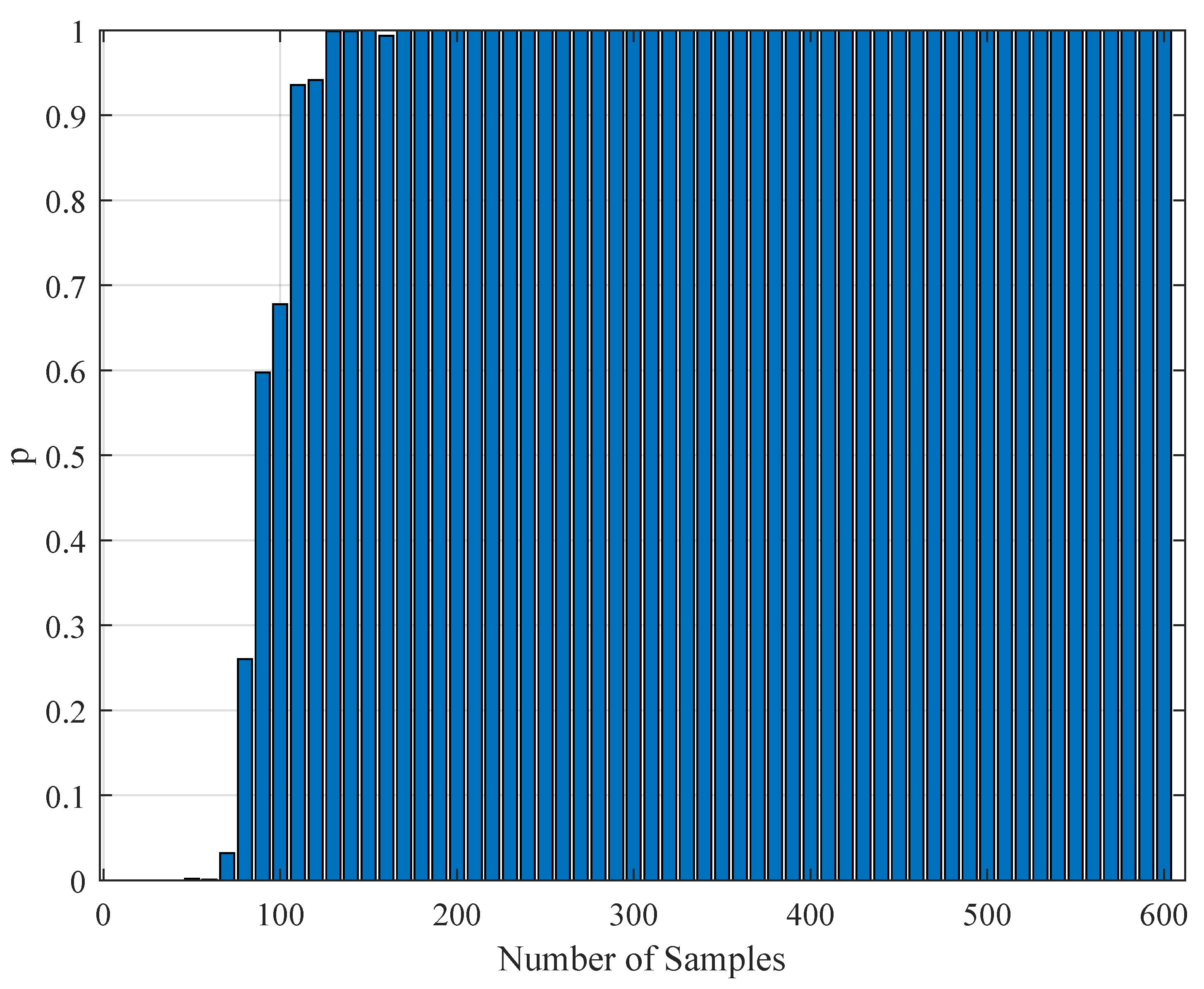
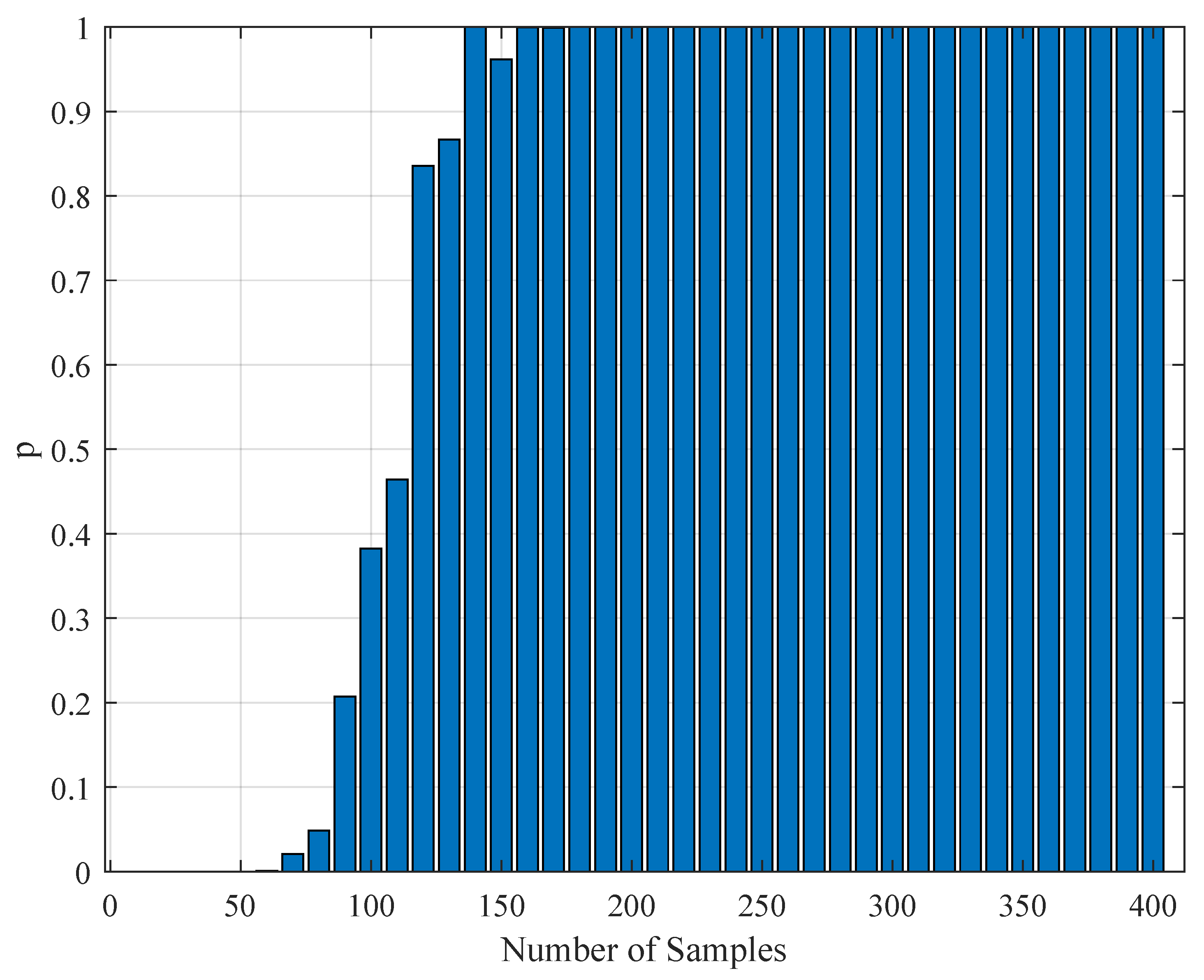
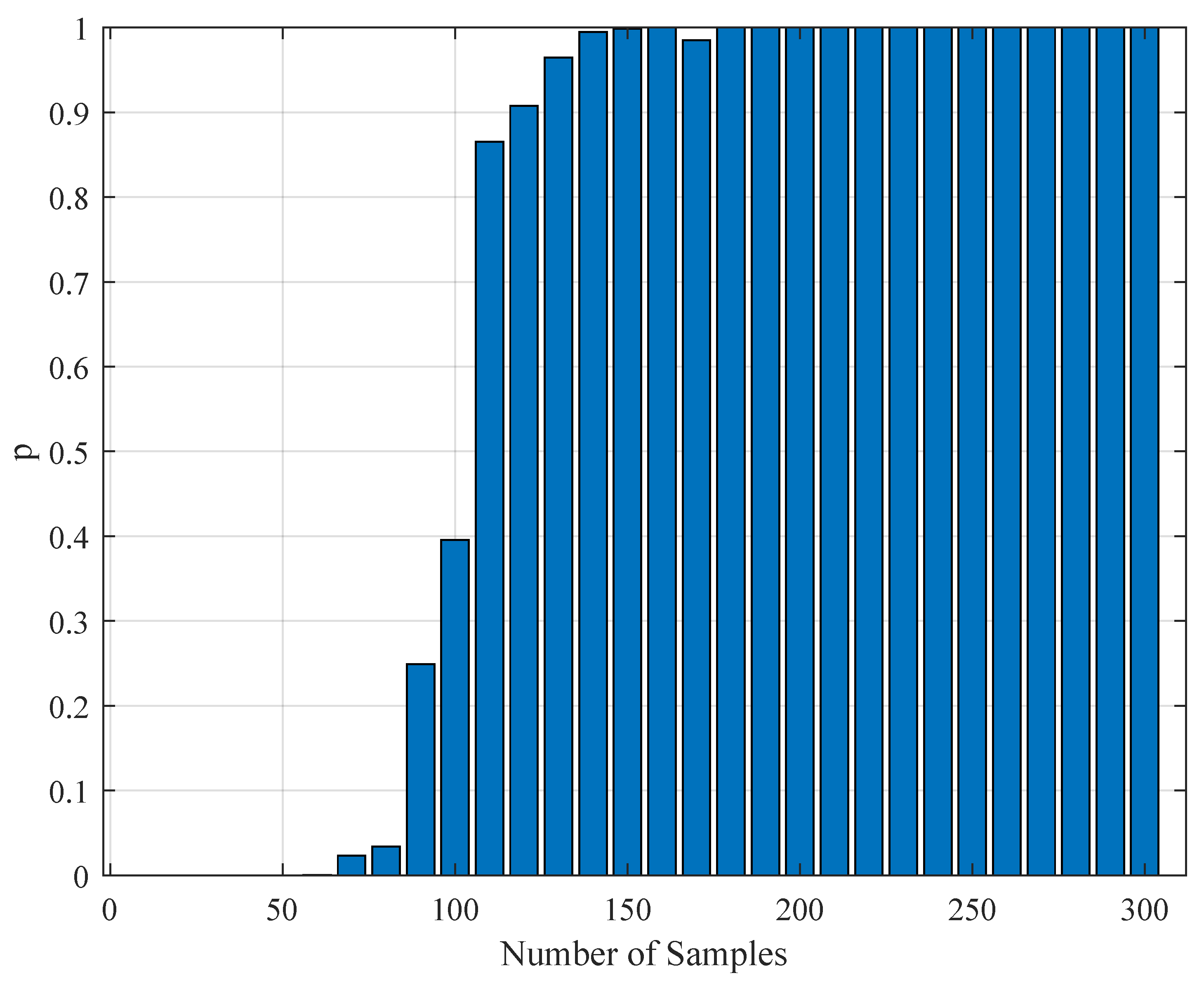
1. Introduction
Application of computational tools in neuroscience is an emerging field of research in the last 50 years. The Hodgkin–Huxley model [
1] is a striking development in the field of theoretical and computational neuroscience. Here, the membrane potential and its bursting properties are modeled as a fourth-order nonlinear system. In addition to the membrane potential, it describes the behaviors of sodium and potassium ion channels. Its nonlinear properties make some researchers search for possibilities that yield simpler nonlinear differential equations. One such attempt is the second-order Morris–Lecar [
2], which lumps the ion channel activation dynamics into a single recovery variable. It is still a conductance based model. Further simplifications involve complete elimination of physical parameters such as ion conductances. Two major examples are the second-order Fitzhugh–Nagumo [
3,
4] and the third-order Hindmarsh–Rose [
5] models. These can model the pulses and bursts occurring in the membrane potential without the need of physical parameters like ion conductances. In addition, as in the case of Morris–Lecar models, the behaviors of ion channels are lumped into generic variables.
In the case that only the input output (stimulus/response) relationships are important, general neural network models can be a good choice. Some examples from literature are the static feed-forward models [
6,
7] and nonlinear recurrent dynamical neural network models [
8,
9]. The dynamical neural network models can be structured such that one may receive a membrane potential information (bursts can be explicitly recovered) or just the instantaneous firing rate as the output [
10]. In addition, sometimes only the statistical properties of the stimulus/response pair is import and thus statistical black-box models are taken into account [
11,
12].
Regardless of the chosen model, stimulus/response data are required to obtain an accurate relationship. Depending on the experiment, these data may be continuous or discrete in nature. In the case of an in-vitro environment such as a patch clamp experiment, one may record a full time dependent profile of membrane potential. That allows computational biologists to perform an identification (parameter estimation) based on traditional minimum mean square estimation (MMSE) techniques. However, in an in-vivo experiment, it is very difficult to collect continuous data revealing exact (or in an acceptable range at least) membrane potential information. If a membrane potential micro electrode contacts a living neuron membrane, the resistive and capacitive properties of the electrode may alter the operation of the neuron. This is not desired as one will not model a realistically functioning neuron at the end of the identification process.
In [
7,
8], it is suggested that one can record the successive action potential timings if the electrodes are suitably placed in surroundings of the membrane. With that, one is able to form a neural spike train which has the discrete timings of the spikes (or of the action potential bursts). Of course, a spike train cannot have dedicated amplitude information. However, this does not mean that one is hopeless concerning model identification. In [
13], it is suggested that neural spike timings largely obey Inhomogeneous Poisson Point Processes (IPPPs). Being aware of the fact that an IPPP can be approximated by a local Bernoulli process [
14], it would be convenient to derive suitable likelihood functions and apply statistical parameter identification techniques on that.
In addition, previous research suggests that the transmitted neural information is not directly coded by the membrane potential level but rather vested in the firing rate [
15], interspiking intervals (ISI) [
16] or individual timings of the spikes [
17]. Thus, training neuron models from discrete and stochastic spiking data is expected to be a beneficial approach to understand the computational features of our nervous system.
Concerning the application of statistical techniques based on point process likelihoods to neural modeling, there are a few research works in the related literature. The authors of [
6,
7] applied maximum a-posteriori estimation (MAP) technique to identification of the weights of a static feed-forward model of the auditory cortex of marmoset monkeys. The authors of [
8,
9] presented a computational study aiming at the estimation of the network parameters and time constants of a dynamical recurrent neural network model using point process maximum likelihood technique. The authors of [
18] applied likelihood techniques to generate models for point process information coding. The authors of [
19] trained a state space model from point process neural spiking data.
In a few research studies, Fitzhugh–Nagumo models are involved in stochastic neural spiking related studies. For example, the authors of [
20] dealt with the interspike interval statistics when the original Fitzhugh–Nagumo model is modified to include noisy inputs. The number of small amplitude oscillations has a random nature and tend to have an asymptotically geometric distribution. Bashkirtseva et al. [
21] studied the effect of stochastic dynamics represented by a standard Wiener process on the limit cycle behavior. In [
22], the authors performed research on the hypoelliptic stochastic properties of Fitzhugh–Nagumo neurons. They studied the effect of those properties on the neural spiking behavior of Fitzhugh–Nagumo models. Finally, Zhang et al. [
23] investigated the stochastic resonance occurring in the Fitzhugh–Nagumo models when trichotomous noise is present in the model. They found that, when the stimulus power is not sufficient to generate firing responses, trichotomous noise itself may trigger the firing.
In this research, we treated a conventional single Fitzhugh–Nagumo equation [
3,
4] as a computational model to form a theoretical stimulus/response relationship. We were interested in the algorithmic details of the modeling. Thus, we modified the original equation to provide firing rate output instead of the membrane potential. Based on the findings in [
8,
9,
10], we mapped the firing rate and membrane potential of the neuron by a gained logistic sigmoid function. Sigmoid functions have a significance in neuron models as they are a feasible way of mapping the ion channel activation dynamics and membrane potential [
1,
2].
Although the output of our model is the neural firing rate, the responses from in vivo neurons are stochastic neural spike timings. To obtain representative data, we simulated the Fitzhugh–Nagumo neurons with a set of true reference parameters and then generated the spikes from the output firing rate by simulating an Inhomogeneous Poisson process on it.
The parameter estimation procedure was based on maximum likelihood method. Similar to that of Eden [
14], the likelihood was derived from the local Bernoulli approximation of the inhomogeneous Poisson process. That depends on the individual spike timings rather than the mean firing rate (which is the case in Poisson distribution’s probability mass function).
The stimulus was modeled as a Fourier series in phased cosine form. This choice was made to investigate the performance of the estimation when the same stimulus as that in [
8,
9] was applied. In the computational framework of this research, the stimulus was applied for a duration 30 ms. This may be observed as a relatively short duration and it is chosen to speed up the computation. In some studies (e.g., [
24,
25,
26]), one can infer that such short duration stimuli may be possible for fast spiking neurons.
In addition, fast spiking responses obtained from a single long random stimulus can be partitioned to segments of short duration such as 30 ms. Thus, the approach in this research can also be utilized in modeling studies that involves longer duration stimuli.
In addition to the computational features of this study, we also investigated the performance of our developments when the training data are taken from a realistic experiment. To achieve this goal, we used the data generated by de Ruyter and Bialek [
27]. The data from this research have a 20 min recording of neural spiking responses obtained from H1 neurons of blowfly vision system against white noise random stimulus. The response was divided into segments of 500 ms and the developed algorithms were applied. Each 500 ms segment can be thought as an independent stimulus and its associated response.