Integrated Analysis of microRNA and mRNA Expression Profiles: An Attempt to Disentangle the Complex Interaction Network in Attention Deficit Hyperactivity Disorder
Abstract
1. Introduction
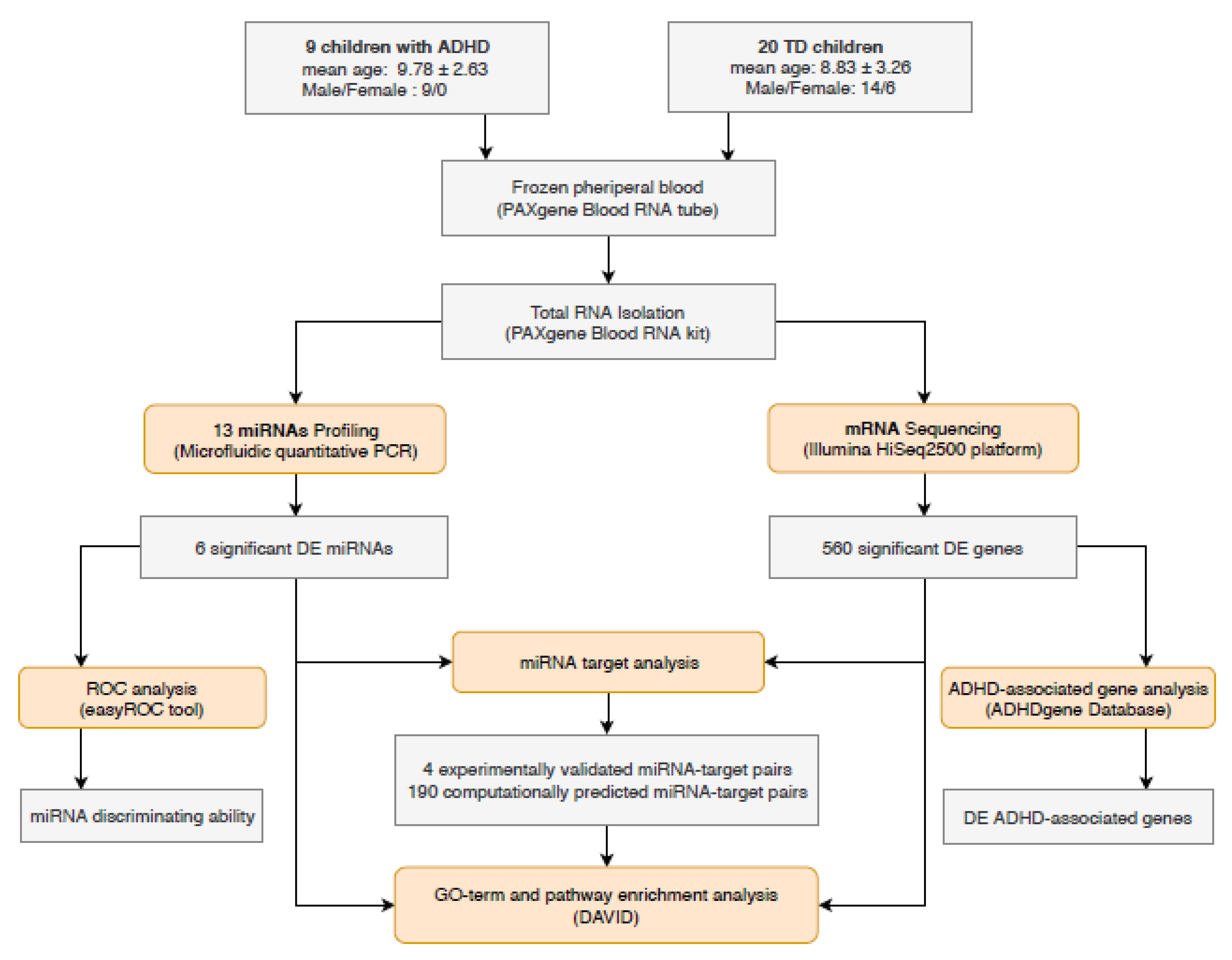
2. Materials and Methods
2.1. Study Population
2.2. Sample Preparation
2.3. miRNA Profiling: Reverse Transcription and Microfluidic qPCR
2.4. mRNA Profiling: HT-NGS
2.5. mRNA Profiling: Bioinformatic Analyses
2.6. miRNA Target Analysis
2.7. Pathway Analysis
3. Results
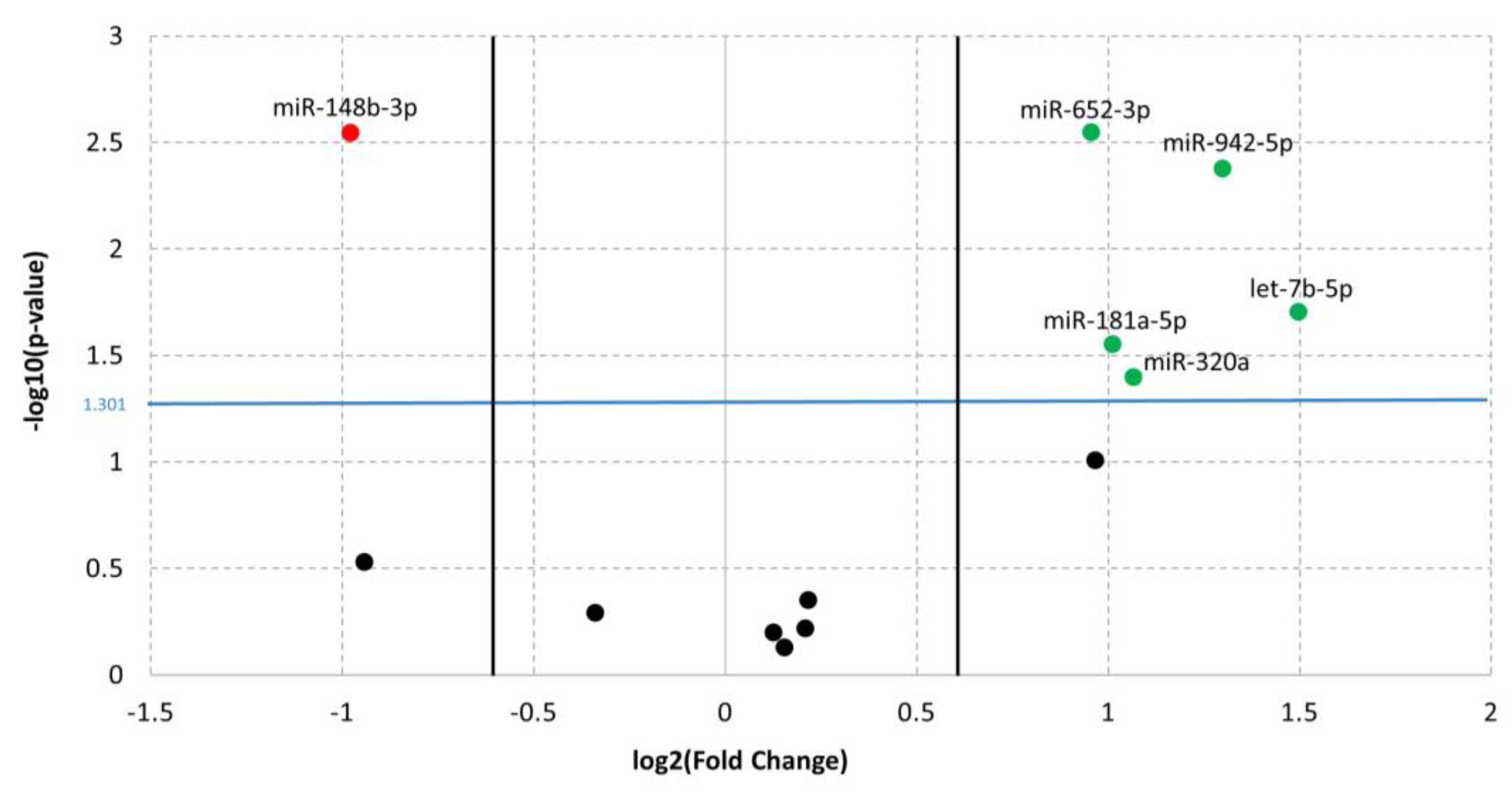
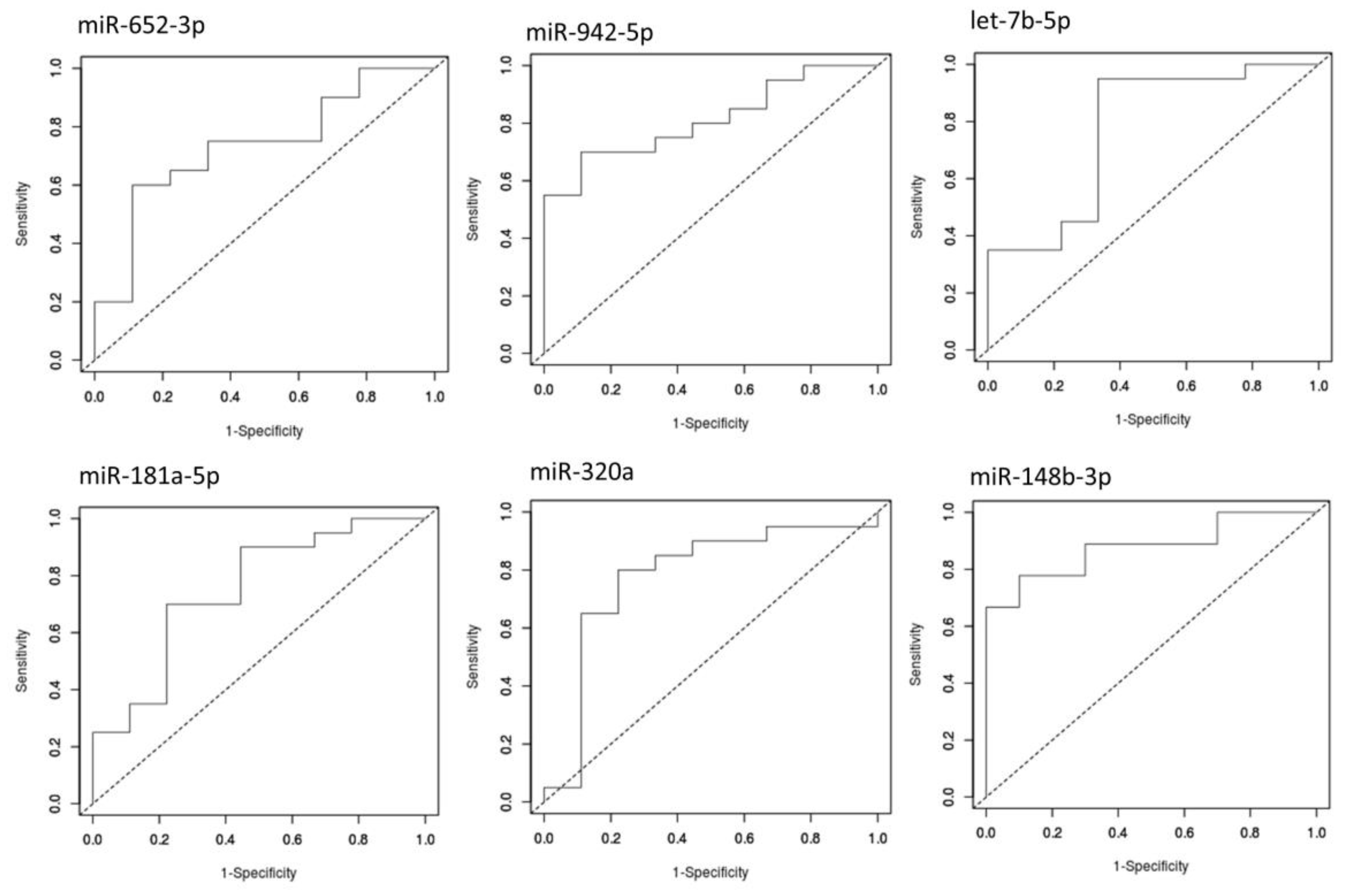
3.1. Differentially Expressed miRNAs in ADHD
3.2. Identification of DE mRNAs
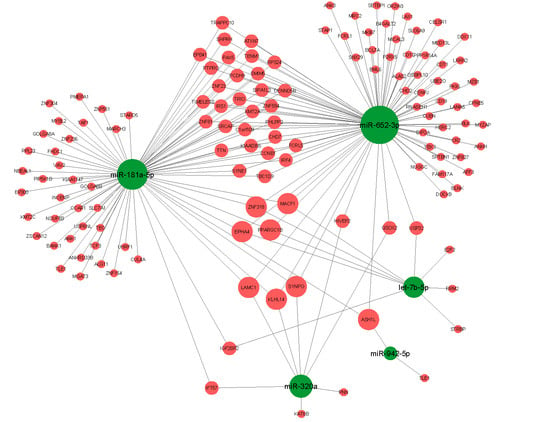
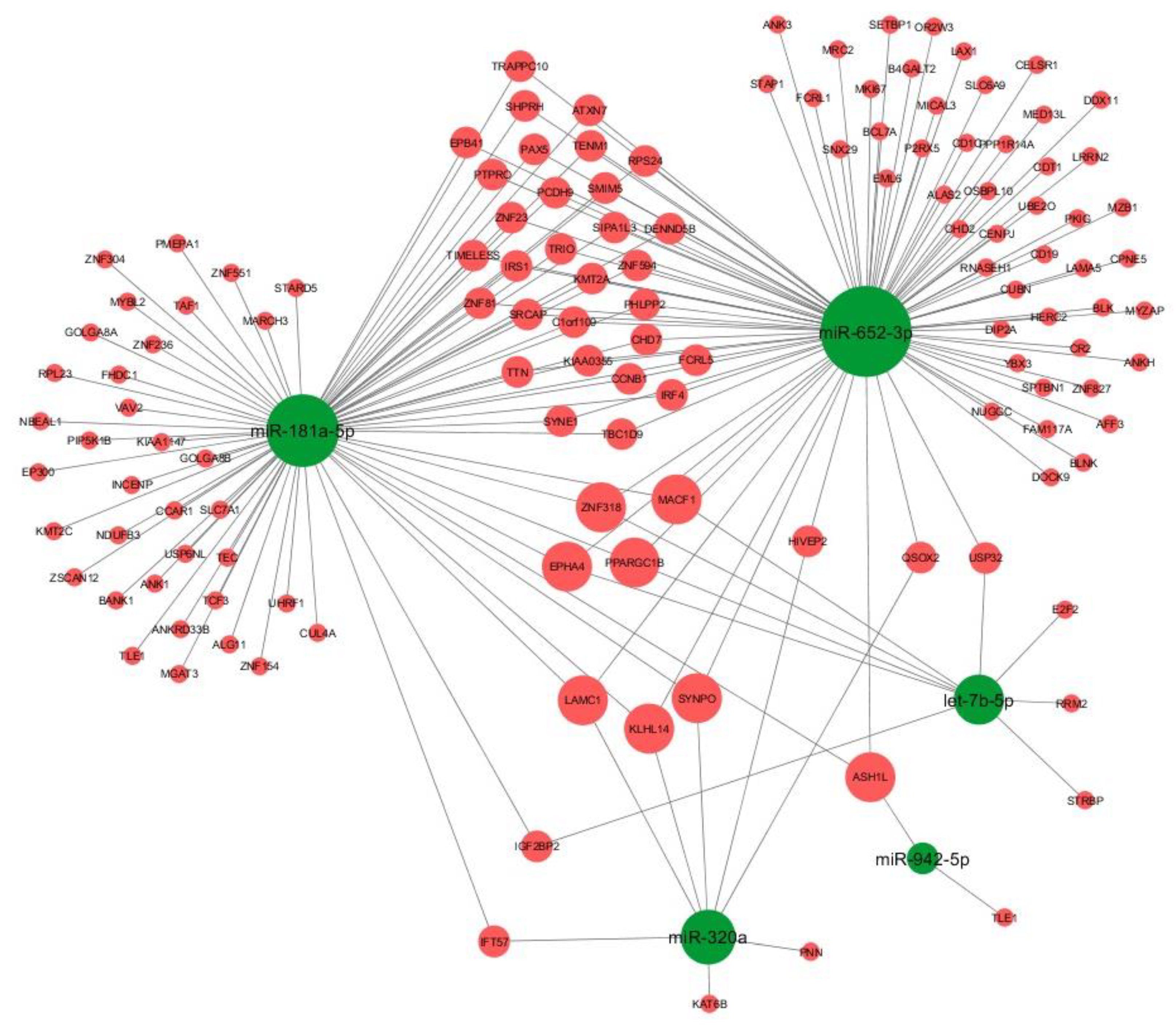
3.3. Target Analysis
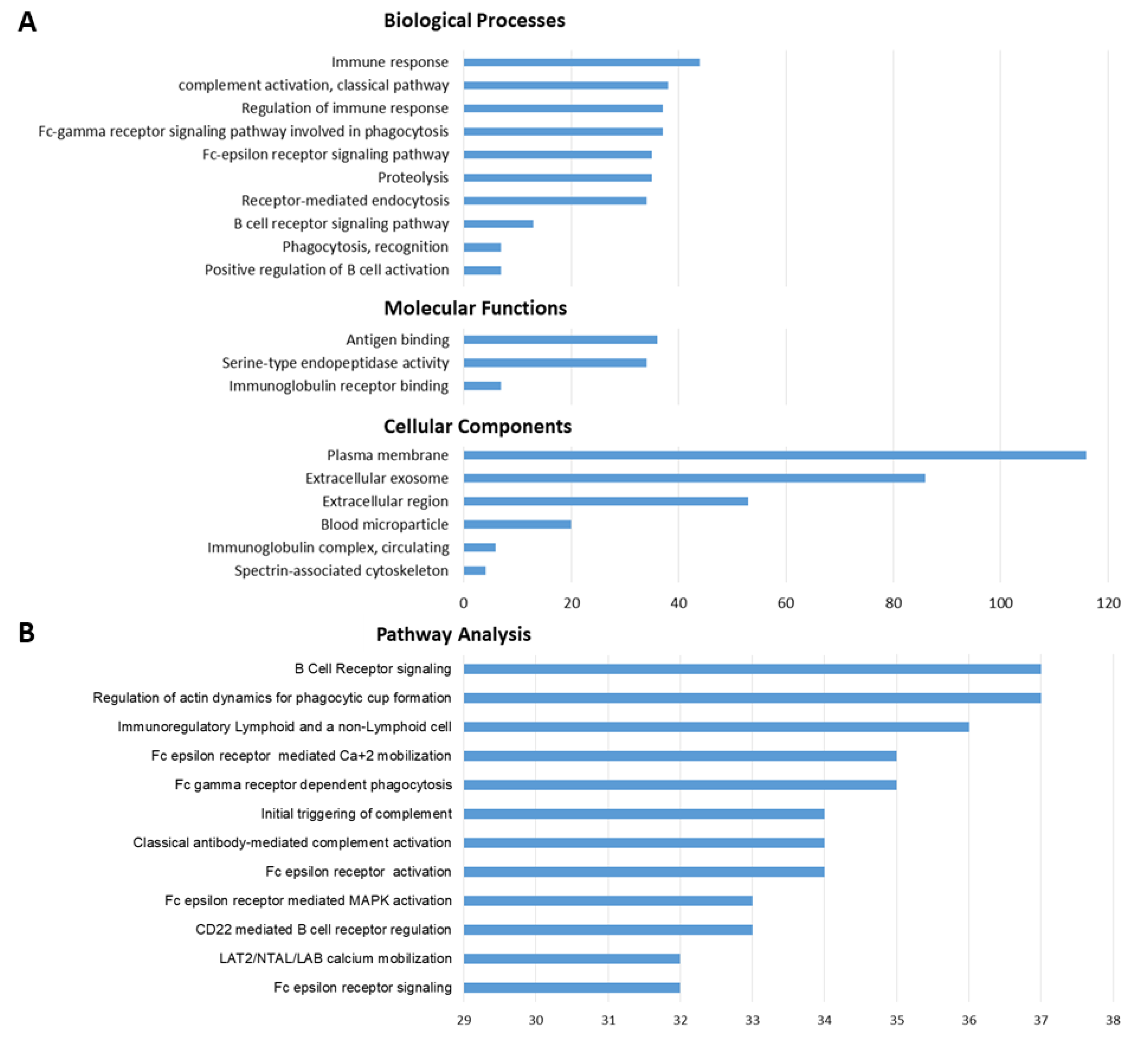
3.4. GO-Term and Pathway Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Thapar, A.; Cooper, M. Attention deficit hyperactivity disorder. Lancet 2016, 387, 1240–1250. [Google Scholar] [CrossRef]
- Hawi, Z.; Cummins, T.D.R.; Tong, J.; Johnson, B.; Lau, R.; Samarrai, W.; Bellgrove, M.A. The molecular genetic architecture of attention deficit hyperactivity disorder. Mol. Psychiatry 2015, 20, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Biederman, J. Attention-Deficit/Hyperactivity Disorder: A Selective Overview. Biol. Psychiatry 2005, 57, 1215–1220. [Google Scholar] [CrossRef] [PubMed]
- Fayyad, J.; De Graaf, R.; Kessler, R.; Alonso, J.; Angermeyer, M.; Demyttenaere, K.; De Girolamo, G.; Haro, J.M.; Karam, E.G.; Lara, C.; et al. Cross-national prevalence and correlates of adult attention-deficit hyperactivity disorder. Br. J. Psychiatry 2007, 190, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, J.R. Assessment and monitoring of treatment response in adult ADHD patients: Current perspectives. Neuropsychiatr. Dis. Treat. 2017, 13, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.; Colvin, L.; Hagemann, E.; Bower, C. Environmental Risk Factors by Gender Associated with Attention-Deficit/Hyperactivity Disorder. Pediatrics 2014, 133, e14–e22. [Google Scholar] [CrossRef] [PubMed]
- Hamza, M.; Halayem, S.; Bourgou, S.; Daoud, M.; Charfi, F.; Belhadj, A. Epigenetics and ADHD: Toward an Integrative Approach of the Disorder Pathogenesis. J. Atten. Disord. 2019, 23, 655–664. [Google Scholar] [CrossRef]
- Demontis, D.; Walters, R.K.; Martin, J.; Mattheisen, M.; Als, T.D.; Agerbo, E.; Baldursson, G.; Belliveau, R.; Bybjerg-Grauholm, J.; Bækvad-Hansen, M.; et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 2019, 51, 63. [Google Scholar] [CrossRef]
- Larsson, H.; Lichtenstein, P.; Larsson, J.-O. Genetic Contributions to the Development of ADHD Subtypes from Childhood to Adolescence. J. Am. Acad. Child Adolesc. Psychiatry 2006, 45, 973–981. [Google Scholar] [CrossRef]
- Faraone, S.V.; Biederman, J.; Weiffenbach, B.; Keith, T.; Chu, M.P.; Weaver, A.; Spencer, T.J.; Wilens, T.E.; Frazier, J.; Cleves, M.; et al. Dopamine D4 Gene 7-Repeat Allele and Attention Deficit Hyperactivity Disorder. AJP 1999, 156, 768–770. [Google Scholar]
- Banerjee, E.; Banerjee, D.; Chatterjee, A.; Sinha, S.; Nandagopal, K. Selective maternal inheritance of risk alleles and genetic interaction between serotonin receptor-1B (5-HTR1B) and serotonin transporter (SLC6A4) in ADHD. Psychiatry Res. 2012, 200, 1083–1085. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Sham, P.C.; Owen, M.J.; He, L. Meta-analysis shows significant association between dopamine system genes and attention deficit hyperactivity disorder (ADHD). Hum. Mol. Genet 2006, 15, 2276–2284. [Google Scholar] [CrossRef] [PubMed]
- Franke, B.; Vasquez, A.A.; Johansson, S.; Hoogman, M.; Romanos, J.; Boreatti-Hümmer, A.; Heine, M.; Jacob, C.P.; Lesch, K.-P.; Casas, M.; et al. Multicenter analysis of the SLC6A3/DAT1 VNTR haplotype in persistent ADHD suggests differential involvement of the gene in childhood and persistent ADHD. Neuropsychopharmacology 2010, 35, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Srivastav, S.; Walitza, S.; Grünblatt, E. Emerging role of miRNA in attention deficit hyperactivity disorder: A systematic review. Adhd Atten. Deficit Hyperact. Disord. 2018, 10, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Kandemir, H.; Erdal, M.E.; Selek, S.; Ay, Ö.İ.; Karababa, İ.F.; Kandemir, S.B.; Ay, M.E.; Yılmaz, Ş.G.; Bayazıt, H.; Taşdelen, B. Evaluation of several micro RNA (miRNA) levels in children and adolescents with attention deficit hyperactivity disorder. Neurosci. Lett. 2014, 580, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-J.; Li, S.-C.; Lee, M.-J.; Chou, M.-C.; Chou, W.-J.; Lee, S.-Y.; Hsu, C.-W.; Huang, L.-H.; Kuo, H.-C. Blood-Bourne MicroRNA Biomarker Evaluation in Attention-Deficit/Hyperactivity Disorder of Han Chinese Individuals: An Exploratory Study. Front. Psychiatry 2018, 9, 227. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.H.; Peng, M.; Yu, M.; Zhao, Q.L.; Li, C.; Jin, Y.T.; Jiang, Y.; Chen, Z.Y.; Deng, N.H.; Sun, H.; et al. Circulating MicroRNA Let-7d in Attention-Deficit/Hyperactivity Disorder. Neuromol. Med. 2015, 17, 137–146. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Na, Y.-J.; Kim, J.H. Understanding cooperativity of microRNAs via microRNA association networks. BMC Genom. 2013, 14, S17. [Google Scholar] [CrossRef]
- Su, W.-L.; Kleinhanz, R.R.; Schadt, E.E. Characterizing the role of miRNAs within gene regulatory networks using integrative genomics techniques. Mol. Syst. Biol. 2011, 7, 490. [Google Scholar] [CrossRef]
- Zadehbagheri, F.; Hosseini, E.; Bagheri-Hosseinabadi, Z.; Rekabdarkolaee, H.M.; Sadeghi, I. Profiling of miRNAs in serum of children with attention-deficit hyperactivity disorder shows significant alterations. J. Psychiatr. Res. 2019, 109, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Liguori, M.; Nuzziello, N.; Licciulli, F.; Consiglio, A.; Simone, M.; Viterbo, R.G.; Creanza, T.M.; Ancona, N.; Tortorella, C.; Margari, L.; et al. Combined microRNA and mRNA expression analysis in pediatric multiple sclerosis: An integrated approach to uncover novel pathogenic mechanisms of the disease. Hum. Mol. Genet. 2017, 27, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Liguori, M.; Nuzziello, N.; Introna, A.; Consiglio, A.; Licciulli, F.; D’Errico, E.; Scarafino, A.; Distaso, E.; Simone, I.L. Dysregulation of MicroRNAs and Target Genes Networks in Peripheral Blood of Patients with Sporadic Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci. 2018, 11, 288. [Google Scholar] [CrossRef] [PubMed]
- López-Ratón, M.; Rodríguez-Álvarez, M.X.; Cadarso-Suárez, C.; Gude-Sampedro, F. OptimalCutpoints: An R Package for Selecting Optimal Cutpoints in Diagnostic Tests. J. Stat. Softw. 2014, 61, 1–36. [Google Scholar] [CrossRef]
- Babraham Bioinformatics. FastQC a Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 1 October 2019).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Consiglio, A.; Mencar, C.; Grillo, G.; Marzano, F.; Caratozzolo, M.F.; Liuni, S. A fuzzy method for RNA-Seq differential expression analysis in presence of multireads. BMC Bioinform. 2016, 17, 95–110. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- ADHDgene: A Genetic Database for Attention Deficit Hyperactivity Disorder. Available online: http://adhd.psych.ac.cn/ (accessed on 1 October 2019).
- Predicted MicroRNA Targets & Target Downregulation Scores. Experimentally Observed Expression Patterns. Available online: http://www.microrna.org/microrna/home.do (accessed on 1 October 2019).
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-microT web server v5. 0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef]
- RNA22. Available online: https://cm.jefferson.edu/rna22 (accessed on 1 October 2019).
- mirDIP: microRNA Data Integration Portal. Available online: http://ophid.utoronto.ca/mirDIP (accessed on 1 October 2019).
- TargetScanHuman Prediction of microRNA Targets. Available online: http://www.targetscan.org/vert_71 (accessed on 1 October 2019).
- miRTarBase: The Experimentally Validated microRNA-target Interactions Database. Available online: http://mirtarbase.mbc.nctu.edu.tw (accessed on 1 October 2019).
- Vlachos, I.S.; Paraskevopoulou, M.D.; Karagkouni, D.; Georgakilas, G.; Vergoulis, T.; Kanellos, I.; Anastasopoulos, I.-L.; Maniou, S.; Karathanou, K.; Kalfakakou, D.; et al. DIANA-TarBase v7.0: Indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucl. Acids Res. 2015, 43, D153–D159. [Google Scholar] [CrossRef]
- Simone, M.; Viterbo, R.G.; Margari, L.; Iaffaldano, P. Computer-assisted rehabilitation of attention in pediatric multiple sclerosis and ADHD patients: a pilot trial. BMC Neurol. 2018, 18, 82. [Google Scholar] [CrossRef] [PubMed]
- Bossel Ben-Moshe, N.; Avraham, R.; Kedmi, M.; Zeisel, A.; Yitzhaky, A.; Yarden, Y.; Domany, E. Context-specific microRNA analysis: Identification of functional microRNAs and their mRNA targets. Nucleic Acids Res. 2012, 40, 10614–10627. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chang, S.; Li, Z.; Zhang, K.; Du, Y.; Ott, J.; Wang, J. ADHDgene: A genetic database for attention deficit hyperactivity disorder. Nucleic Acids Res. 2012, 40, D1003–D1009. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Weisbrot, D.; Charvet, L.; Serafin, D.; Milazzo, M.; Preston, T.; Cleary, R.; Moadel, T.; Seibert, M.; Belman, A.; Krupp, L. Psychiatric diagnoses and cognitive impairment in pediatric multiple sclerosis. Mult. Scler. J. 2014, 20, 588–593. [Google Scholar] [CrossRef] [PubMed]
- Tonacci, A.; Bagnato, G.; Pandolfo, G.; Billeci, L.; Sansone, F.; Conte, R.; Gangemi, S. MicroRNA Cross-Involvement in Autism Spectrum Disorders and Atopic Dermatitis: A Literature Review. J. Clin. Med. 2019, 8, 88. [Google Scholar] [CrossRef]
- Mayes, S.D.; Calhoun, S.L.; Mayes, R.D.; Molitoris, S. Autism and ADHD: Overlapping and discriminating symptoms. Res. Autism. Spectr. Disord. 2012, 6, 277–285. [Google Scholar] [CrossRef]
- Taurines, R.; Schwenck, C.; Westerwald, E.; Sachse, M.; Siniatchkin, M.; Freitag, C. ADHD and autism: differential diagnosis or overlapping traits? A selective review. Adhd Atten Def Hyp Disord 2012, 4, 115–139. [Google Scholar] [CrossRef]
- Reiersen, A.M.; Constantino, J.N.; Grimmer, M.; Martin, N.G.; Todd, R.D. Evidence for Shared Genetic Influences on Self-Reported ADHD and Autistic Symptoms in Young Adult Australian Twins. Twin Res. Hum. Genet. 2008, 11, 579–585. [Google Scholar] [CrossRef]
- Ronald, A.; Simonoff, E.; Kuntsi, J.; Asherson, P.; Plomin, R. Evidence for overlapping genetic influences on autistic and ADHD behaviours in a community twin sample. J. Child Psychol. Psychiatry 2008, 49, 535–542. [Google Scholar] [CrossRef]
- Tang, J.; Fan, Y.; Li, H.; Xiang, Q.; Zhang, D.-F.; Li, Z.; He, Y.; Liao, Y.; Wang, Y.; He, F.; et al. Whole-genome sequencing of monozygotic twins discordant for schizophrenia indicates multiple genetic risk factors for schizophrenia. J. Genet. Genom. 2017, 44, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Kos, M.Z.; Carless, M.A.; Peralta, J.; Blackburn, A.; Almeida, M.; Roalf, D.; Pogue-Geile, M.F.; Prasad, K.; Gur, R.C.; Nimgaonkar, V.; et al. Exome Sequence Data from Multigenerational Families Implicate AMPA Receptor Trafficking in Neurocognitive Impairment and Schizophrenia Risk. Schizophr. Bull. 2016, 42, 288–300. [Google Scholar] [CrossRef] [PubMed]
- Kripke, D.F.; Nievergelt, C.M.; Tranah, G.J.; Murray, S.S.; Rex, K.M.; Grizas, A.P.; Hahn, E.K.; Lee, H.-J.; Kelsoe, J.R.; Kline, L.E. FMR1, circadian genes and depression: Suggestive associations or false discovery? J. Circadian Rhythm. 2013, 11, 3. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kripke, D.F.; Nievergelt, C.M.; Joo, E.; Shekhtman, T.; Kelsoe, J.R. Circadian polymorphisms associated with affective disorders. J. Circadian Rhythm. 2009, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Melhuish Beaupre, L.; Brown, G.M.; Kennedy, J.L. Circadian genes in major depressive disorder. World J. Biol. Psychiatry 2018, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Peter-Ross, E.M. Molecular hypotheses to explain the shared pathways and underlying pathobiological causes in catatonia and in catatonic presentations in neuropsychiatric disorders. Med. Hypotheses 2018, 113, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Sadybekov, A.; Tian, C.; Arnesano, C.; Katritch, V.; Herring, B.E. An autism spectrum disorder-related de novo mutation hotspot discovered in the GEF1 domain of Trio. Nat. Commun. 2017, 8, 601. [Google Scholar] [CrossRef]
- Schimmelmann, B.G.; Hinney, A.; Scherag, A.; Pütter, C.; Pechlivanis, S.; Cichon, S.; Jöckel, K.-H.; Schreiber, S.; Wichmann, H.E.; Albayrak, Ö.; et al. Bipolar disorder risk alleles in children with ADHD. J. Neural. Transm. (Vienna) 2013, 120, 1611–1617. [Google Scholar] [CrossRef]
- Landaas, E.T.; Johansson, S.; Halmøy, A.; Oedegaard, K.J.; Fasmer, O.B.; Haavik, J. Bipolar disorder risk alleles in adult ADHD patients. Genes Brain Behav. 2011, 10, 418–423. [Google Scholar] [CrossRef]
- Stergiakouli, E.; Hamshere, M.; Holmans, P.; Langley, K.; Zaharieva, I.; Hawi, Z.; Kent, L.; Gill, M.; Williams, N.; Owen, M.J.; et al. Investigating the Contribution of Common Genetic Variants to the Risk and Pathogenesis of ADHD. Am. J. Psychiatry 2012, 169, 186–194. [Google Scholar] [CrossRef]
- Dela Peña, I.; Kim, H.J.; Sohn, A.; Kim, B.-N.; Han, D.H.; Ryu, J.H.; Shin, C.Y.; Noh, M.; Cheong, J.H. Prefrontal cortical and striatal transcriptional responses to the reinforcing effect of repeated methylphenidate treatment in the spontaneously hypertensive rat, animal model of attention-deficit/hyperactivity disorder (ADHD). Behav. Brain Funct. 2014, 10, 17. [Google Scholar] [CrossRef] [PubMed]
- Lotan, A.; Fenckova, M.; Bralten, J.; Alttoa, A.; Dixson, L.; Williams, R.W.; van der Voet, M. Neuroinformatic analyses of common and distinct genetic components associated with major neuropsychiatric disorders. Front. Neurosci. 2014, 8, 331. [Google Scholar] [CrossRef] [PubMed]
- Dastidar, S.G.; Narayanan, S.; Stifani, S.; D’Mello, S.R. Transducin-like Enhancer of Split-1 (TLE1) Combines with Forkhead Box Protein G1 (FoxG1) to Promote Neuronal Survival. J. Biol. Chem. 2012, 287, 14749–14759. [Google Scholar] [CrossRef]
- Hess, J.L.; Akutagava-Martins, G.C.; Patak, J.D.; Glatt, S.J.; Faraone, S.V. Why is there selective subcortical vulnerability in ADHD? Clues from postmortem brain gene expression data. Mol. Psychiatry 2018, 23, 1787. [Google Scholar] [CrossRef]
- Iqbal, Z.; Vandeweyer, G.; van der Voet, M.; Waryah, A.M.; Zahoor, M.Y.; Besseling, J.A.; Roca, L.T.; Vulto-van Silfhout, A.T.; Nijhof, B.; Kramer, J.M.; et al. Homozygous and heterozygous disruptions of ANK3: At the crossroads of neurodevelopmental and psychiatric disorders. Hum. Mol. Genet. 2013, 22, 1960–1970. [Google Scholar] [CrossRef] [PubMed]
- Zong, W.; Liu, S.; Wang, X.; Zhang, J.; Zhang, T.; Liu, Z.; Wang, D.; Zhang, A.; Zhu, M.; Gao, J. Trio gene is required for mouse learning ability. Brain Res. 2015, 1608, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Steinfeld, H.; Cho, M.T.; Retterer, K.; Person, R.; Schaefer, G.B.; Danylchuk, N.; Malik, S.; Wechsler, S.B.; Wheeler, P.G.; van Gassen, K.L.I.; et al. Mutations in HIVEP2 are associated with developmental delay, intellectual disability and dysmorphic features. Neurogenetics 2016, 17, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Nieto-Estevez, V.; Hsieh, J. CHD2: One Gene, Many Roles. Neuron 2018, 100, 1014–1016. [Google Scholar] [CrossRef]
- Zhang, J.; Weinrich, J.A.P.; Russ, J.B.; Comer, J.D.; Bommareddy, P.K.; DiCasoli, R.J.; Wright, C.V.E.; Li, Y.; van Roessel, P.J.; Kaltschmidt, J.A. A Role for Dystonia-Associated Genes in Spinal GABAergic Interneuron Circuitry. Cell Rep. 2017, 21, 666–678. [Google Scholar] [CrossRef]
- Rathje, M.; Waxman, H.; Benoit, M.; Tammineni, P.; Leu, C.; Loebrich, S.; Nedivi, E. Genetic variants in the bipolar disorder risk locus SYNE1 that affect CPG2 expression and protein function. Mol. Psychiatry 2019. [Google Scholar] [CrossRef]
- Naaijen, J.; Bralten, J.; Poelmans, G.; Glennon, J.C.; Franke, B.; Buitelaar, J.K. Glutamatergic and GABAergic gene sets in attention-deficit/hyperactivity disorder: association to overlapping traits in ADHD and autism. Transl. Psychiatry 2017, 7, e999. [Google Scholar] [CrossRef] [PubMed]
- Edden, R.A.E.; Crocetti, D.; Zhu, H.; Gilbert, D.L.; Mostofsky, S.H. Reduced GABA Concentration in Attention-Deficit/Hyperactivity Disorder. Arch. Gen. Psychiatry 2012, 69, 750–753. [Google Scholar] [CrossRef] [PubMed]
- Goetz, M.; Veselá, M.; Ptá\vcek, R. Notes on the Role of the Cerebellum in ADHD. Austin J. Psychiatry Behav. Sci. 2014, 1, 1013. [Google Scholar]
- Ferrin, M.; Vance, A. Examination of neurological subtle signs in ADHD as a clinical tool for the diagnosis and their relationship to spatial working memory. J. Child Psychol. Psychiatry 2012, 53, 390–400. [Google Scholar] [CrossRef]
- Goetz, M.; Schwabova, J.; Hlavka, Z.; Ptacek, R.; Zumrova, A.; Hort, V.; Doyle, R. Cerebellar Symptoms Are Associated with Omission Errors and Variability of Response Time in Children With ADHD. J. Atten. Disord. 2017, 21, 190–199. [Google Scholar] [CrossRef]
- Verlaet, A.A.J.; Noriega, D.B.; Hermans, N.; Savelkoul, H.F.J. Nutrition, immunological mechanisms and dietary immunomodulation in ADHD. Eur. Child Adolesc. Psychiatry 2014, 23, 519–529. [Google Scholar] [CrossRef]
- Schmitt, J.; Romanos, M.; Schmitt, N.M.; Meurer, M.; Kirch, W. Atopic Eczema and Attention-Deficit/Hyperactivity Disorder in a Population-Based Sample of Children and Adolescents. JAMA 2009, 301, 724–726. [Google Scholar] [CrossRef]
- Fasmer, O.B.; Halmøy, A.; Eagan, T.M.; Oedegaard, K.J.; Haavik, J. Adult attention deficit hyperactivity disorder is associated with asthma. BMC Psychiatry 2011, 11, 128. [Google Scholar] [CrossRef]
- Buske-Kirschbaum, A.; Schmitt, J.; Plessow, F.; Romanos, M.; Weidinger, S.; Roessner, V. Psychoendocrine and psychoneuroimmunological mechanisms in the comorbidity of atopic eczema and attention deficit/hyperactivity disorder. Psychoneuroendocrinology 2013, 38, 12–23. [Google Scholar] [CrossRef]
| miRNA | Regulation | qPCR | ROC | Target | ADHD-Associated Target Genes | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| log2FC | p-Value | adj. p-Value | AUC | p-Value | |||||||
| miR-652-3p | up | 0.95594 | 2.84 × 10-3 | 1.83 × 10-2 | 0.733 | 2.33 × 10-2 | 89 | B4GALT2, ANK3, SLC6A9 | |||
| miR-148b-3p | down | −0.97755 | 2.85 × 10-3 | 1.83 × 10-2 | 0.878 | 5.46 × 10-6 | 8 | ||||
| miR-942-5p | up | 1.29942 | 4.22 × 10-3 | 1.83 × 10-2 | 0.811 | 1 × 10-4 | 2 | TLE1 | |||
| let-7b-5p | up | 1.49735 | 1.98 × 10-2 | 6.45 × 10-2 | 0.772 | 1.16 × 10-2 | 9 | ||||
| miR-181a-5p | up | 1.01275 | 2.82 × 10-2 | 7.33 × 10-2 | 0.75 | 1.82 × 10-2 | 75 | TAF1, TRIO, SYNE1 | |||
| miR-320a | up | 1.06673 | 4.02 × 10-2 | 8.72 × 10-2 | 0.778 | 1.16 × 10-2 | 8 | ||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nuzziello, N.; Craig, F.; Simone, M.; Consiglio, A.; Licciulli, F.; Margari, L.; Grillo, G.; Liuni, S.; Liguori, M. Integrated Analysis of microRNA and mRNA Expression Profiles: An Attempt to Disentangle the Complex Interaction Network in Attention Deficit Hyperactivity Disorder. Brain Sci. 2019, 9, 288. https://doi.org/10.3390/brainsci9100288
Nuzziello N, Craig F, Simone M, Consiglio A, Licciulli F, Margari L, Grillo G, Liuni S, Liguori M. Integrated Analysis of microRNA and mRNA Expression Profiles: An Attempt to Disentangle the Complex Interaction Network in Attention Deficit Hyperactivity Disorder. Brain Sciences. 2019; 9(10):288. https://doi.org/10.3390/brainsci9100288
Chicago/Turabian StyleNuzziello, Nicoletta, Francesco Craig, Marta Simone, Arianna Consiglio, Flavio Licciulli, Lucia Margari, Giorgio Grillo, Sabino Liuni, and Maria Liguori. 2019. "Integrated Analysis of microRNA and mRNA Expression Profiles: An Attempt to Disentangle the Complex Interaction Network in Attention Deficit Hyperactivity Disorder" Brain Sciences 9, no. 10: 288. https://doi.org/10.3390/brainsci9100288
APA StyleNuzziello, N., Craig, F., Simone, M., Consiglio, A., Licciulli, F., Margari, L., Grillo, G., Liuni, S., & Liguori, M. (2019). Integrated Analysis of microRNA and mRNA Expression Profiles: An Attempt to Disentangle the Complex Interaction Network in Attention Deficit Hyperactivity Disorder. Brain Sciences, 9(10), 288. https://doi.org/10.3390/brainsci9100288