Decoding Encoded Cravings: Epigenetic Drivers of Addiction
Abstract
1. Introduction
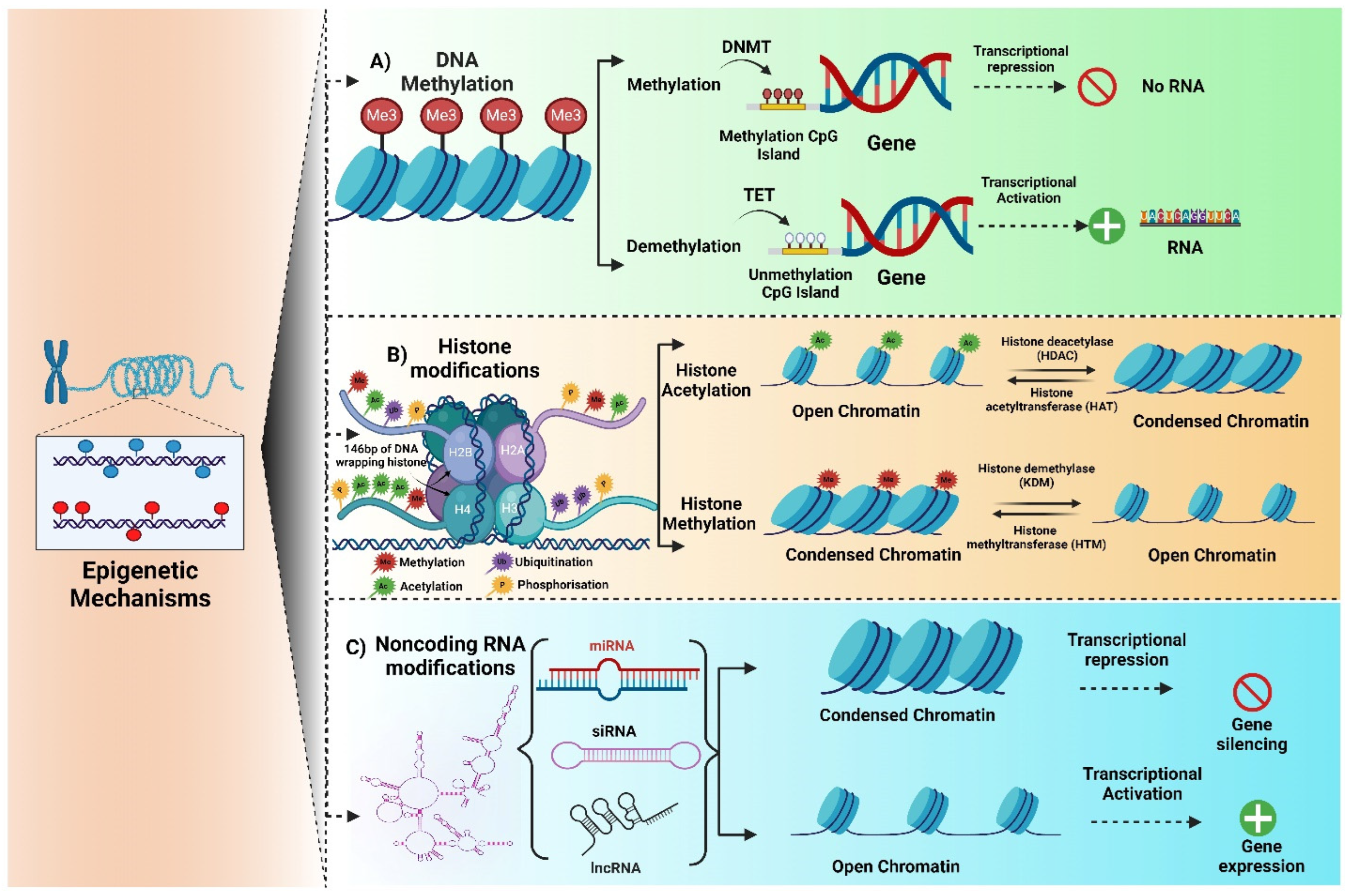
2. Epigenetics
2.1. DNA Methylation
2.2. Histone Modification
2.3. Non-Coding RNA Modification
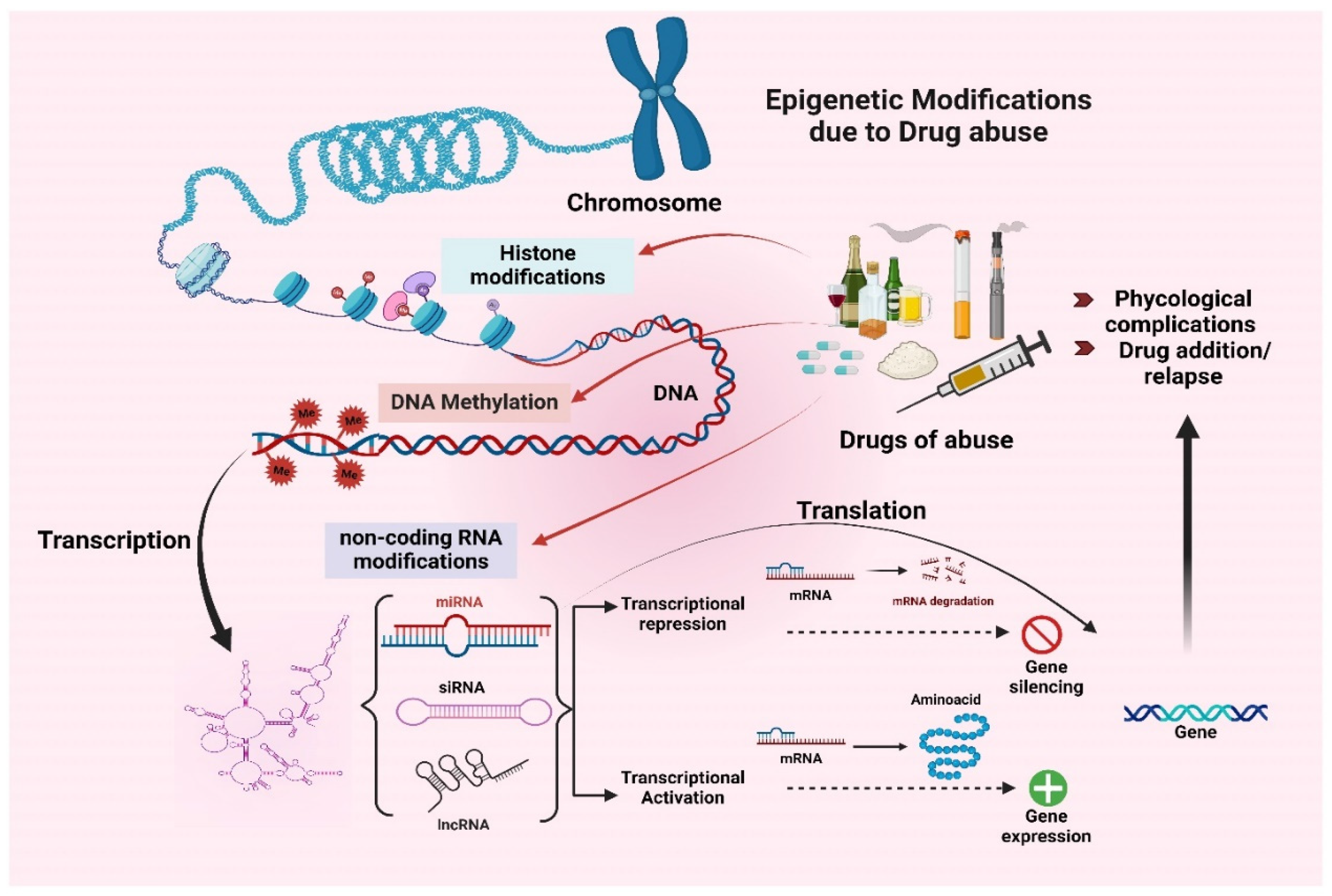
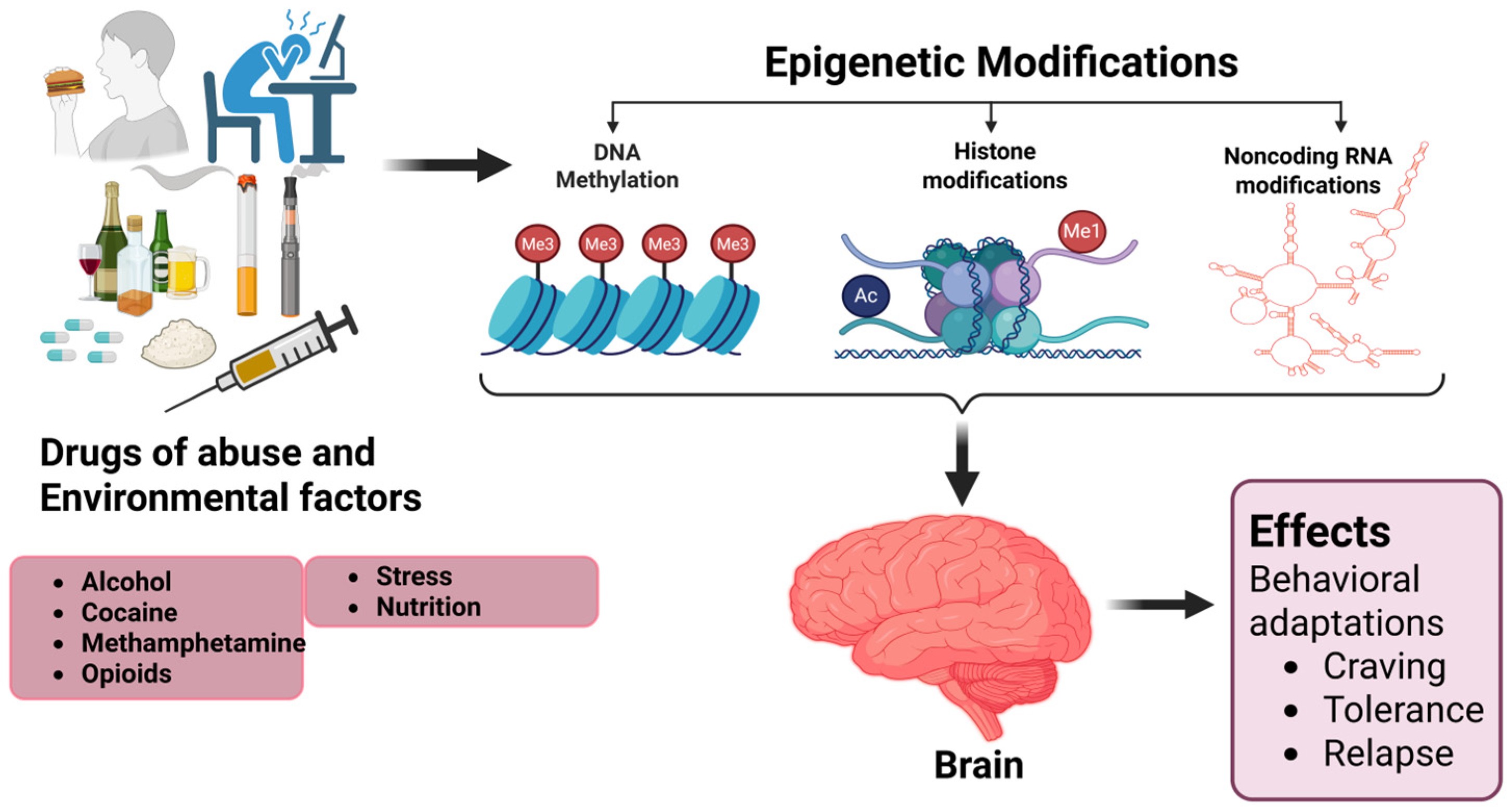
3. Epigenetic Modifications Induced by Drugs Abuse
3.1. Alcohol
3.2. Cocaine
3.3. Methamphetamine (METH)
3.4. Opioids
4. Transgenerational Epigenetic Modification Due to Drug Abuse
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SUD | Substance use disorder |
| AUD | Alcohol use disorder |
| NAc | nucleus accumbens |
| DNA | Deoxyribonucleic acid |
| miRNA | microRNA |
| siRNA | small interfering ncRNA |
| DNMT | DNA methyltransferase |
| HDACs | Histone deacetylase |
| ncRNA | Non-coding RNA |
| RNA | Ribonucleic acid |
| METH | Methamphetamine |
References
- Ignaszewski, M.J. The Epidemiology of Drug Abuse. J. Clin. Pharmacol. 2021, 61, S10–S17. [Google Scholar] [CrossRef]
- Virmani, M.A. Drug Abuse Results in Metabolic and Epigenetic Changes in Body Which May Contribute to Disease Risk: Role of L-Carnitine and Nutrients. Adv. Drug Alcohol Res. 2023, 3, 10901. [Google Scholar] [CrossRef] [PubMed]
- Assistant Secretary for Public Affairs (ASPA). SAMHSA (Substance Abuse and Mental Health Services Administration) Release 2022 National Survey on Drug Use and Health Data. Available online: https://www.hhs.gov/about/agencies/aspa/aspa-organization/index.html (accessed on 9 December 2024).
- Volkow, N.D.; Morales, M. The Brain on Drugs: From Reward to Addiction. Cell 2015, 162, 712–725. [Google Scholar] [CrossRef] [PubMed]
- King, N.; Dafny, N. The Ventral Tegmental Area (VTA), the Nucleus Accumbens (NAc), the Caudate Nucleus (CN) and the Prefrontal Cortex (PFC) Role in the Response to Acute and Chronic Methylphenidate. J. Exp. Neurol. 2023, 4, 21–36. [Google Scholar] [CrossRef]
- Cooper, S.; Robison, A.J.; Mazei-Robison, M.S. Reward Circuitry in Addiction. Neurotherapeutics 2017, 14, 687–697. [Google Scholar] [CrossRef]
- Tan, B.; Browne, C.J.; Nöbauer, T.; Vaziri, A.; Friedman, J.M.; Nestler, E.J. Drugs of Abuse Hijack a Mesolimbic Pathway That Processes Homeostatic Need. Science 2024, 384, eadk6742. [Google Scholar] [CrossRef]
- Leshner, A.I. Addiction Is a Brain Disease, and It Matters. Science 1997, 278, 45–47. [Google Scholar] [CrossRef]
- Volkow, N.D.; Koob, G.F.; McLellan, A.T. Neurobiologic Advances from the Brain Disease Model of Addiction. N. Engl. J. Med. 2016, 374, 363–371. [Google Scholar] [CrossRef]
- Greener, M.R.; Storr, S.J. Conflicting Theories on Addiction Aetiology and the Strengths and Limitations of Substance Use Disorder Disease Modelling. Front. Mol. Neurosci. 2023, 16, 1166852. [Google Scholar] [CrossRef]
- Anderson, E.M.; Taniguchi, M. Epigenetic Effects of Addictive Drugs in the Nucleus Accumbens. Front. Mol. Neurosci. 2022, 15, 828055. [Google Scholar] [CrossRef]
- Bogdan, R.; Hatoum, A.S.; Johnson, E.C.; Agrawal, A. The Genetically Informed Neurobiology of Addiction (GINA) Model. Nat. Rev. Neurosci. 2023, 24, 40–57. [Google Scholar] [CrossRef]
- Kendler, K.S.; Schmitt, E.; Aggen, S.H.; Prescott, C.A. Genetic and Environmental Influences on Alcohol, Caffeine, Cannabis, and Nicotine Use From Early Adolescence to Middle Adulthood. Arch. Gen. Psychiatry 2008, 65, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Tsuang, M.T.; Lyons, M.J.; Meyer, J.M.; Doyle, T.; Eisen, S.A.; Goldberg, J.; True, W.; Lin, N.; Toomey, R.; Eaves, L. Co-Occurrence of Abuse of Different Drugs in Men: The Role of Drug-Specific and Shared Vulnerabilities. Arch. Gen. Psychiatry 1998, 55, 967–972. [Google Scholar] [CrossRef] [PubMed]
- Robison, A.J.; Nestler, E.J. Transcriptional and Epigenetic Mechanisms of Addiction. Nat. Rev. Neurosci. 2011, 12, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Al Aboud, N.M.; Tupper, C.; Jialal, I. Genetics, Epigenetic Mechanism. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Handy, D.E.; Castro, R.; Loscalzo, J. Epigenetic Modifications: Basic Mechanisms and Role in Cardiovascular Disease. Circulation 2011, 123, 2145–2156. [Google Scholar] [CrossRef]
- Jiang, S.; Postovit, L.; Cattaneo, A.; Binder, E.B.; Aitchison, K.J. Epigenetic Modifications in Stress Response Genes Associated with Childhood Trauma. Front. Psychiatry 2019, 10, 808. [Google Scholar] [CrossRef]
- Non, A.L. Social Epigenomics: Are We at an Impasse? Epigenomics 2021, 13, 1747–1759. [Google Scholar] [CrossRef]
- Li, Y. Modern Epigenetics Methods in Biological Research. Methods 2021, 187, 104–113. [Google Scholar] [CrossRef]
- Saini, A.; Rawat, Y.; Jain, K.; Mani, I. Chapter Two—State-of-the-Art Techniques to Study Epigenetics. In Progress in Molecular Biology and Translational Science; Singh, V., Mani, I., Eds.; Epigenetics in Health and Disease—Part A; Academic Press: Cambridge, MA, USA, 2023; Volume 197, pp. 23–50. [Google Scholar]
- Koijam, A.S.; Singh, K.D.; Nameirakpam, B.S.; Haobam, R.; Rajashekar, Y. Drug Addiction and Treatment: An Epigenetic Perspective. Biomed. Pharmacother. 2024, 170, 115951. [Google Scholar] [CrossRef]
- Zhang, H.; Gelernter, J. Review: DNA Methylation and Alcohol Use Disorders: Progress and Challenges. Am. J. Addict. 2017, 26, 502–515. [Google Scholar] [CrossRef]
- Mahna, D.; Puri, S.; Sharma, S. DNA Methylation Signatures: Biomarkers of Drug and Alcohol Abuse. Mutat. Res./Rev. Mutat. Res. 2018, 777, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Liu, Y. DNA Methylation in Human Diseases. Genes Dis. 2018, 5, 1–8. [Google Scholar] [CrossRef]
- Zillich, L.; Frank, J.; Streit, F.; Friske, M.M.; Foo, J.C.; Sirignano, L.; Heilmann-Heimbach, S.; Dukal, H.; Degenhardt, F.; Hoffmann, P.; et al. Epigenome-Wide Association Study of Alcohol Use Disorder in Five Brain Regions. Neuropsychopharmacology 2022, 47, 832–839. [Google Scholar] [CrossRef] [PubMed]
- Khodadadi, E.; Fahmideh, L.; Khodadadi, E.; Dao, S.; Yousefi, M.; Taghizadeh, S.; Asgharzadeh, M.; Yousefi, B.; Kafil, H.S. Current Advances in DNA Methylation Analysis Methods. Biomed. Res. Int. 2021, 2021, 8827516. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Pugh, B.F. Nucleosome Positioning and Gene Regulation: Advances through Genomics. Nat. Rev. Genet. 2009, 10, 161–172. [Google Scholar] [CrossRef]
- Sokolova, V.; Sarkar, S.; Tan, D. Histone Variants and Chromatin Structure, Update of Advances. Comput. Struct. Biotechnol. J. 2022, 21, 299–311. [Google Scholar] [CrossRef]
- Walker, D.M.; Cates, H.M.; Heller, E.A.; Nestler, E.J. Regulation of Chromatin States by Drugs of Abuse. Curr. Opin. Neurobiol. 2015, 30, 112–121. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin Modifications and Their Function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef]
- Liu, R.; Wu, J.; Guo, H.; Yao, W.; Li, S.; Lu, Y.; Jia, Y.; Liang, X.; Tang, J.; Zhang, H. Post-translational Modifications of Histones: Mechanisms, Biological Functions, and Therapeutic Targets. MedComm 2023, 4, e292. [Google Scholar] [CrossRef]
- Renthal, W.; Nestler, E.J. Histone Acetylation in Drug Addiction. Semin. Cell Dev. Biol. 2009, 20, 387–394. [Google Scholar] [CrossRef]
- Romhányi, D.; Szabó, K.; Kemény, L.; Groma, G. Histone and Histone Acetylation-Related Alterations of Gene Expression in Uninvolved Psoriatic Skin and Their Effects on Cell Proliferation, Differentiation, and Immune Responses. Int. J. Mol. Sci. 2023, 24, 14551. [Google Scholar] [CrossRef]
- Bure, I.V.; Nemtsova, M.V.; Kuznetsova, E.B. Histone Modifications and Non-Coding RNAs: Mutual Epigenetic Regulation and Role in Pathogenesis. Int. J. Mol. Sci. 2022, 23, 5801. [Google Scholar] [CrossRef]
- Wei, A.; Wu, H. Mammalian DNA Methylome Dynamics: Mechanisms, Functions and New Frontiers. Development 2022, 149, dev182683. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Shi, Y. Histone Methylation: A Dynamic Mark in Health, Disease and Inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Lan, F.; Matson, C.; Mulligan, P.; Whetstine, J.R.; Cole, P.A.; Casero, R.A.; Shi, Y. Histone Demethylation Mediated by the Nuclear Amine Oxidase Homolog LSD1. Cell 2004, 119, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Van Roy, Z.; Kielian, T. Exploring Epigenetic Reprogramming during Central Nervous System Infection. Immunol. Rev. 2022, 311, 112–129. [Google Scholar] [CrossRef]
- Bernstein, B.E.; Mikkelsen, T.S.; Xie, X.; Kamal, M.; Huebert, D.J.; Cuff, J.; Fry, B.; Meissner, A.; Wernig, M.; Plath, K.; et al. A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell 2006, 125, 315–326. [Google Scholar] [CrossRef]
- Shogren-Knaak, M.; Ishii, H.; Sun, J.-M.; Pazin, M.J.; Davie, J.R.; Peterson, C.L. Histone H4-K16 Acetylation Controls Chromatin Structure and Protein Interactions. Science 2006, 311, 844–847. [Google Scholar] [CrossRef]
- Poliseno, L.; Lanza, M.; Pandolfi, P.P. Coding, or Non-Coding, That Is the Question. Cell Res. 2024, 34, 609–629. [Google Scholar] [CrossRef]
- Inamura, K. Major Tumor Suppressor and Oncogenic Non-Coding RNAs: Clinical Relevance in Lung Cancer. Cells 2017, 6, 12. [Google Scholar] [CrossRef]
- Kaikkonen, M.U.; Lam, M.T.Y.; Glass, C.K. Non-Coding RNAs as Regulators of Gene Expression and Epigenetics. Cardiovasc. Res. 2011, 90, 430–440. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Holoch, D.; Moazed, D. RNA-Mediated Epigenetic Regulation of Gene Expression. Nat. Rev. Genet. 2015, 16, 71–84. [Google Scholar] [CrossRef]
- Yao, H.; Brick, K.; Evrard, Y.; Xiao, T.; Camerini-Otero, R.D.; Felsenfeld, G. Mediation of CTCF Transcriptional Insulation by DEAD-Box RNA-Binding Protein P68 and Steroid Receptor RNA Activator SRA. Genes. Dev. 2010, 24, 2543–2555. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome Regulation by Long Noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Hollander, J.A.; Im, H.-I.; Amelio, A.L.; Kocerha, J.; Bali, P.; Lu, Q.; Willoughby, D.; Wahlestedt, C.; Conkright, M.D.; Kenny, P.J. Striatal microrRNA Controls Cocaine Intake through CREB Signaling. Nature 2010, 466, 197–202. [Google Scholar] [CrossRef]
- Wanet, A.; Tacheny, A.; Arnould, T.; Renard, P. miR-212/132 Expression and Functions: Within and beyond the Neuronal Compartment. Nucleic Acids Res. 2012, 40, 4742–4753. [Google Scholar] [CrossRef]
- Bali, P.; Kenny, P.J. MicroRNAs and Drug Addiction. Front. Genet. 2013, 4, 43. [Google Scholar] [CrossRef]
- Dreyer, J.-L. New Insights into the Roles of microRNAs in Drug Addiction and Neuroplasticity. Genome Med. 2010, 2, 92. [Google Scholar] [CrossRef]
- Wang, Y.; Wei, T.; Zhao, W.; Ren, Z.; Wang, Y.; Zhou, Y.; Song, X.; Zhou, R.; Zhang, X.; Jiao, D. MicroRNA-181a Is Involved in Methamphetamine Addiction Through the ERAD Pathway. Front. Mol. Neurosci. 2021, 14, 667725. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, C.; Sun, G.; Qiu, C.; Fan, J.; Jin, Y.; Liu, K.; Sun, P. Increased Expression of Plasma Mir-9-3p and Let-7b-3p in Methamphetamine Use Disorder and Its Clinical Significance. Sci. Rep. 2024, 14, 31729. [Google Scholar] [CrossRef]
- Zhang, K.; Jing, X.; Wang, G. MicroRNAs as Regulators of Drug Abuse and Immunity. Cent. Eur. J. Immunol. 2016, 41, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, S.K.; Pachunka, J.M.; Mott, J.L. Role of microRNAs in Alcohol-Induced Multi-Organ Injury. Biomolecules 2015, 5, 3309. [Google Scholar] [CrossRef] [PubMed]
- Upreti, D.; Kumar, R.P.; Chen, J.B.J.; Sonti, S.L.; Bowring, A.V.; Green, S.W.; Miranda, R.C. Noncoding RNA and Alcohol Use Disorder: A Scoping Review of Current Research and Knowledge Gaps. Alcohol Res. 2025, 45, 06. [Google Scholar] [CrossRef] [PubMed]
- Sartor, G.C.; St. Laurent, G.; Wahlestedt, C. The Emerging Role of Non-Coding RNAs in Drug Addiction. Front. Genet. 2012, 3, 106. [Google Scholar] [CrossRef]
- Bannon, M.J.; Savonen, C.L.; Jia, H.; Dachet, F.; Halter, S.D.; Schmidt, C.J.; Lipovich, L.; Kapatos, G. Identification of Long Noncoding RNAs Dysregulated in the Midbrain of Human Cocaine Abusers. J. Neurochem. 2015, 135, 50–59. [Google Scholar] [CrossRef]
- Momen-Heravi, F.; Catalano, D.; Talis, A.; Szabo, G.; Bala, S. Protective Effect of LNA-Anti-miR-132 Therapy on Liver Fibrosis in Mice. Mol. Ther. Nucleic Acids 2021, 25, 155–167. [Google Scholar] [CrossRef]
- Maze, I.; Nestler, E.J. The Epigenetic Landscape of Addiction. Ann. N. Y. Acad. Sci. 2011, 1216, 99–113. [Google Scholar] [CrossRef]
- Cheng, J.; He, Z.; Chen, Q.; Lin, J.; Peng, Y.; Zhang, J.; Yan, X.; Yan, J.; Niu, S. Histone Modifications in Cocaine, Methamphetamine and Opioids. Heliyon 2023, 9, e16407. [Google Scholar] [CrossRef]
- Farris, S.P.; Mayfield, R.D. Epigenetic and Non-Coding Regulation of Alcohol Abuse and Addiction. Int. Rev. Neurobiol. 2021, 156, 63–86. [Google Scholar] [CrossRef]
- Poisel, E.; Zillich, L.; Streit, F.; Frank, J.; Friske, M.M.; Foo, J.C.; Mechawar, N.; Turecki, G.; Hansson, A.C.; Nöthen, M.M.; et al. DNA Methylation in Cocaine Use Disorder-An Epigenome-Wide Approach in the Human Prefrontal Cortex. Front. Psychiatry 2023, 14, 1075250. [Google Scholar] [CrossRef]
- Nestler, E.J. Epigenetic Mechanisms of Drug Addiction. Neuropharmacology 2014, 76, 259–268. [Google Scholar] [CrossRef]
- Kaplan, G.; Xu, H.; Abreu, K.; Feng, J. DNA Epigenetics in Addiction Susceptibility. Front. Genet. 2022, 13, 806685. [Google Scholar] [CrossRef]
- Hamilton, P.J.; Nestler, E.J. Epigenetics and Addiction. Curr. Opin. Neurobiol. 2019, 59, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.N.; Feng, J. Drug Addiction and DNA Modifications. In Neuroepigenomics in Aging and Disease; Delgado-Morales, R., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 105–125. ISBN 978-3-319-53889-1. [Google Scholar]
- Kim, H.-D.; Call, T.; Magazu, S.; Ferguson, D. Drug Addiction and Histone Code Alterations. Adv. Exp. Med. Biol. 2017, 978, 127–143. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.C.Y.; Mill, J.; Fernandes, C. Drugs and Addiction: An Introduction to Epigenetics. Addiction 2011, 106, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Maze, I.; Feng, J.; Wilkinson, M.B.; Sun, H.; Shen, L.; Nestler, E.J. Cocaine Dynamically Regulates Heterochromatin and Repetitive Element Unsilencing in Nucleus Accumbens. Proc. Natl. Acad. Sci. USA 2011, 108, 3035–3040. [Google Scholar] [CrossRef]
- Heinsbroek, J.A.; De Vries, T.J.; Peters, J. Glutamatergic Systems and Memory Mechanisms Underlying Opioid Addiction. Cold Spring Harb. Perspect. Med. 2021, 11, a039602. [Google Scholar] [CrossRef]
- Seyednejad, S.A.; Sartor, G.C. Noncoding RNA Therapeutics for Substance Use Disorder. Adv. Drug Alcohol Res. 2022, 2, 10807. [Google Scholar] [CrossRef]
- Seo, D.; Sinha, R. Neuroplasticity and Predictors of Alcohol Recovery. Alcohol Res. 2015, 37, 143–152. [Google Scholar]
- Nestler, E.J.; Lüscher, C. The Molecular Basis of Drug Addiction: Linking Epigenetic to Synaptic and Circuit Mechanisms. Neuron 2019, 102, 48–59. [Google Scholar] [CrossRef]
- MacKillop, J.; Agabio, R.; Feldstein-Ewing, S.; Heilig, M.; Kelly, J.F.; Leggio, L.; Lingford-Hughes, A.; Palmer, A.; Parry, C.; Ray, L.; et al. Hazardous Drinking and Alcohol Use Disorders. Nat. Rev. Dis. Primers 2022, 8, 80. [Google Scholar] [CrossRef]
- Andrade-Brito, D.E.; Núñez-Ríos, D.L.; Martínez-Magaña, J.J.; Nagamatsu, S.T.; Rompala, G.; Zillich, L.; Witt, S.H.; Clark, S.L.; Lattig, M.C.; Montalvo-Ortiz, J.L. Neuronal-Specific Methylome and Hydroxymethylome Analysis Reveal Significant Loci Associated with Alcohol Use Disorder. Front. Genet. 2024, 15, 1345410. [Google Scholar] [CrossRef]
- Warnault, V.; Darcq, E.; Levine, A.; Barak, S.; Ron, D. Chromatin Remodeling—A Novel Strategy to Control Excessive Alcohol Drinking. Transl. Psychiatry 2013, 3, e231. [Google Scholar] [CrossRef]
- Niinep, K.; Anier, K.; Eteläinen, T.; Piepponen, P.; Kalda, A. Repeated Ethanol Exposure Alters DNA Methylation Status and Dynorphin/Kappa-Opioid Receptor Expression in Nucleus Accumbens of Alcohol-Preferring AA Rats. Front. Genet. 2021, 12, 750142. [Google Scholar] [CrossRef]
- Stouder, C.; Somm, E.; Paoloni-Giacobino, A. Prenatal Exposure to Ethanol: A Specific Effect on the H19 Gene in Sperm. Reprod. Toxicol. 2011, 31, 507–512. [Google Scholar] [CrossRef]
- Monnier, P.; Martinet, C.; Pontis, J.; Stancheva, I.; Ait-Si-Ali, S.; Dandolo, L. H19 lncRNA Controls Gene Expression of the Imprinted Gene Network by Recruiting MBD1. Proc. Natl. Acad. Sci. USA 2013, 110, 20693–20698. [Google Scholar] [CrossRef] [PubMed]
- Roque Bravo, R.; Faria, A.C.; Brito-da-Costa, A.M.; Carmo, H.; Mladěnka, P.; Dias da Silva, D.; Remião, F. Cocaine: An Updated Overview on Chemistry, Detection, Biokinetics, and Pharmacotoxicological Aspects Including Abuse Pattern. Toxins 2022, 14, 278. [Google Scholar] [CrossRef] [PubMed]
- Engmann, O.; Labonte, B.; Mitchell, A.; Bashtrykov, P.; Calipari, E.S.; Rosenbluh, C.; Loh, Y.-H.E.; Walker, D.M.; Burek, D.; Hamilton, P.J.; et al. Cocaine-Induced Chromatin Modifications Associate with Increased Expression and 3D Looping of Auts2. Biol. Psychiatry 2017, 82, 794–805. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Wilkinson, M.; Liu, X.; Purushothaman, I.; Ferguson, D.; Vialou, V.; Maze, I.; Shao, N.; Kennedy, P.; Koo, J.; et al. Chronic Cocaine-Regulated Epigenomic Changes in Mouse Nucleus Accumbens. Genome Biol. 2014, 15, R65. [Google Scholar] [CrossRef]
- Vaillancourt, K.; Yang, J.; Chen, G.G.; Yerko, V.; Théroux, J.-F.; Aouabed, Z.; Lopez, A.; Thibeault, K.C.; Calipari, E.S.; Labonté, B.; et al. Cocaine-Related DNA Methylation in Caudate Neurons Alters 3D Chromatin Structure of the IRXA Gene Cluster. Mol. Psychiatry 2021, 26, 3134–3151. [Google Scholar] [CrossRef]
- González, B.; Pantoja, C.R.G.; Sosa, M.H.; Vitullo, A.D.; Bisagno, V.; González, C.R. Cocaine Alters the Mouse Testicular Epigenome with Direct Impact on Histone Acetylation and DNA Methylation Marks. Reprod. Biomed. Online 2018, 37, 269–278. [Google Scholar] [CrossRef]
- Courtney, K.E.; Ray, L.A. Methamphetamine: An Update on Epidemiology, Pharmacology, Clinical Phenomenology, and Treatment Literature. Drug Alcohol Depend. 2014, 143, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Shirai, T.; Okazaki, S.; Tanifuji, T.; Otsuka, I.; Horai, T.; Mouri, K.; Takemura, Y.; Aso, K.; Yamamoto, N.; Hishimoto, A. Epigenome-wide Association Study on Methamphetamine Dependence. Addict. Biol. 2024, 29, e13383. [Google Scholar] [CrossRef] [PubMed]
- Jayanthi, S.; McCoy, M.T.; Cadet, J.L. Epigenetic Regulatory Dynamics in Models of Methamphetamine-Use Disorder. Genes 2021, 12, 1614. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.; Luo, T.; Dong, H.; Tang, A.; Hao, W. CHN2 Promoter Methylation Change May Be Associated with Methamphetamine Dependence. Shanghai Arch. Psychiatry 2017, 29, 357–364. [Google Scholar] [PubMed Central]
- Jayanthi, S.; McCoy, M.T.; Chen, B.; Britt, J.; Kourrich, S.; Yau, H.-J.; Ladenheim, B.; Krasnova, I.N.; Bonci, A.; Cadet, J.L. Methamphetamine Down-Regulates Striatal Glutamate Receptors via Diverse Epigenetic Mechanisms. Biol. Psychiatry 2014, 76, 47–56. [Google Scholar] [CrossRef]
- Kaboudin, B.; Sohrabi, M. Chemistry and Synthesis of Major Opium Alkaloids: A Comprehensive Review. J. Iran. Chem. Soc. 2021, 18, 3177–3218. [Google Scholar] [CrossRef]
- Liu, J.; Qin, W.; Yuan, K.; Li, J.; Wang, W.; Li, Q.; Wang, Y.; Sun, J.; Deneen, K.M.V.; Liu, Y.; et al. Interaction between Dysfunctional Connectivity at Rest and Heroin Cues-Induced Brain Responses in Male Abstinent Heroin-Dependent Individuals. PLoS ONE 2011, 6, e23098. [Google Scholar] [CrossRef]
- Egervari, G.; Landry, J.; Callens, J.; Fullard, J.F.; Roussos, P.; Keller, E.; Hurd, Y.L. Striatal H3K27 Acetylation Linked to Glutamatergic Gene Dysregulation in Human Heroin Abusers Holds Promise as Therapeutic Target. Biol. Psychiatry 2017, 81, 585–594. [Google Scholar] [CrossRef]
- LaLumiere, R.T.; Kalivas, P.W. Glutamate Release in the Nucleus Accumbens Core Is Necessary for Heroin Seeking. J. Neurosci. 2008, 28, 3170–3177. [Google Scholar] [CrossRef]
- Meyers, J.L.; Salling, M.C.; Almli, L.M.; Ratanatharathorn, A.; Uddin, M.; Galea, S.; Wildman, D.E.; Aiello, A.E.; Bradley, B.; Ressler, K.; et al. Frequency of Alcohol Consumption in Humans; the Role of Metabotropic Glutamate Receptors and Downstream Signaling Pathways. Transl. Psychiatry 2015, 5, e586. [Google Scholar] [CrossRef]
- D’Souza, M.S. Glutamatergic Transmission in Drug Reward: Implications for Drug Addiction. Front. Neurosci. 2015, 9, 404. [Google Scholar] [CrossRef]
- Kim, J.; Ham, S.; Hong, H.; Moon, C.; Im, H.-I. Brain Reward Circuits in Morphine Addiction. Mol. Cells 2016, 39, 645–653. [Google Scholar] [CrossRef]
- Wang, Z.; Yan, P.; Hui, T.; Zhang, J. Epigenetic Upregulation of PSD-95 Contributes to the Rewarding Behavior by Morphine Conditioning. Eur. J. Pharmacol. 2014, 732, 123–129. [Google Scholar] [CrossRef]
- Jia, M.; Wang, X.; Zhang, H.; Wang, X.; Ma, H.; Yang, M.; Li, Y.; Cui, C. MicroRNA-132 Is Involved in Morphine Dependence via Modifying the Structural Plasticity of the Dentate Gyrus Neurons in Rats. Addict. Biol. 2022, 27, e13086. [Google Scholar] [CrossRef] [PubMed]
- Amaral, D.G.; Scharfman, H.E.; Lavenex, P. The Dentate Gyrus: Fundamental Neuroanatomical Organization (Dentate Gyrus for Dummies). Prog. Brain Res. 2007, 163, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Abdulmalek, S.; Hardiman, G. Genetic and Epigenetic Studies of Opioid Abuse Disorder—The Potential for Future Diagnostics. Expert. Rev. Mol. Diagn. 2023, 23, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.E.; Shekarabi, A.; Inan, S. Oxycodone: A Current Perspective on Its Pharmacology, Abuse, and Pharmacotherapeutic Developments. Pharmacol. Rev. 2023, 75, 1062–1118. [Google Scholar] [CrossRef]
- Marie, N.; Noble, F. Oxycodone, an Opioid like the Others? Front. Psychiatry 2023, 14, 1229439. [Google Scholar] [CrossRef]
- Yakout, D.W.; Shree, N.; Mabb, A.M. Effect of Pharmacological Manipulations on Arc Function. Curr. Res. Pharmacol. Drug Discov. 2021, 2, 100013. [Google Scholar] [CrossRef]
- Gerra, M.C.; Dallabona, C.; Arendt-Nielsen, L. Epigenetic Alterations in Prescription Opioid Misuse: New Strategies for Precision Pain Management. Genes 2021, 12, 1226. [Google Scholar] [CrossRef]
- Hong, Q.; Xu, W.; Lin, Z.; Liu, J.; Chen, W.; Zhu, H.; Lai, M.; Zhuang, D.; Xu, Z.; Fu, D.; et al. Role of GABRD Gene Methylation in the Nucleus Accumbens in Heroin-Seeking Behavior in Rats. Front. Pharmacol. 2021, 11, 612200. [Google Scholar] [CrossRef]
- Fan, X.-Y.; Shi, G.; Zhao, P. Methylation in Syn and Psd95 Genes Underlie the Inhibitory Effect of Oxytocin on Oxycodone-Induced Conditioned Place Preference. Eur. Neuropsychopharmacol. 2019, 29, 1464–1475. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.P.; Franklin, L.M.; Johnson, G.S.; Schrott, L.M. Prenatal Oxycodone Exposure Impairs Spatial Learning and/or Memory in Rats. Behav. Brain Res. 2010, 212, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Gilardi, F.; Augsburger, M.; Thomas, A. Will Widespread Synthetic Opioid Consumption Induce Epigenetic Consequences in Future Generations? Front. Pharmacol. 2018, 9, 702. [Google Scholar] [CrossRef] [PubMed]
- Chorbov, V.M.; Todorov, A.A.; Lynskey, M.T.; Cicero, T.J. Elevated Levels of DNA Methylation at the OPRM1 Promoter in Blood and Sperm from Male Opioid Addicts. J. Opioid Manag. 2011, 7, 258–264. [Google Scholar] [CrossRef]
- Mavrikaki, M.; Anastasiadou, E.; Ozdemir, R.A.; Potter, D.; Helmholz, C.; Slack, F.J.; Chartoff, E.H. Overexpression of miR-9 in the Nucleus Accumbens Increases Oxycodone Self-Administration. Int. J. Neuropsychopharmacol. 2019, 22, 383–393. [Google Scholar] [CrossRef]
- Toyama, K.; Kiyosawa, N.; Watanabe, K.; Ishizuka, H. Identification of Circulating miRNAs Differentially Regulated by Opioid Treatment. Int. J. Mol. Sci. 2017, 18, 1991. [Google Scholar] [CrossRef]
- Han, Y.; Yan, W.; Zheng, Y.; Khan, M.Z.; Yuan, K.; Lu, L. The Rising Crisis of Illicit Fentanyl Use, Overdose, and Potential Therapeutic Strategies. Transl. Psychiatry 2019, 9, 282. [Google Scholar] [CrossRef]
- Wu, Q.; Hwang, C.K.; Zheng, H.; Wagley, Y.; Lin, H.-Y.; Kim, D.K.; Law, P.-Y.; Loh, H.H.; Wei, L.-N. MicroRNA 339 Down-Regulates μ-Opioid Receptor at the Post-Transcriptional Level in Response to Opioid Treatment. FASEB J. 2013, 27, 522–535. [Google Scholar] [CrossRef] [PubMed]
- Goodson, R.; Poklis, J.; Elder, H.J.; Walentiny, D.M.; Dewey, W.; Halquist, M. Acute Biodistribution Comparison of Fentanyl and Morphine. Psychoactives 2024, 3, 27. [Google Scholar] [CrossRef]
- Volkow, N.D.; Jones, E.B.; Einstein, E.B.; Wargo, E.M. Prevention and Treatment of Opioid Misuse and Addiction: A Review. JAMA Psychiatry 2019, 76, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Baccarelli, A.; Bollati, V. Epigenetics and Environmental Chemicals. Curr. Opin. Pediatr. 2009, 21, 243–251. [Google Scholar] [CrossRef]
- Knopik, V.S.; Marceau, K.; Bidwell, L.C.; Rolan, E. Prenatal Substance Exposure and Offspring Development: Does DNA Methylation Play a Role? Neurotoxicol. Teratol. 2019, 71, 50–63. [Google Scholar] [CrossRef]
- Viola, T.W.; Danzer, C.; Mardini, V.; Szobot, C.; Chrusciel, J.H.; Stertz, L.; Schmitz, J.M.; Walss-Bass, C.; Fries, G.R.; Grassi-Oliveira, R. Prenatal Cocaine Exposure and Its Influence on Pediatric Epigenetic Clocks and Epigenetic Scores in Humans. Sci. Rep. 2024, 14, 1946. [Google Scholar] [CrossRef]
- Kazemi, M.; Tekieh, E.; Golabi, S.; Sahraei, H. Evaluation of Placental Epigenetic Changes Due to Morphine Consumption. Int. J. Morphol. 2016, 34, 252–261. [Google Scholar] [CrossRef]
- Riyahi, J.; Taslimi, Z.; Gelfo, F.; Petrosini, L.; Haghparast, A. Trans-Generational Effects of Parental Exposure to Drugs of Abuse on Offspring Memory Functions. Neurosci. Biobehav. Rev. 2024, 160, 105644. [Google Scholar] [CrossRef]
- Tomášková, A.; Šlamberová, R.; Černá, M. Influence of Prenatal Methamphetamine Abuse on the Brain. Epigenomes 2020, 4, 14. [Google Scholar] [CrossRef]
- Wanner, N.M.; Colwell, M.L.; Faulk, C. The Epigenetic Legacy of Illicit Drugs: Developmental Exposures and Late-Life Phenotypes. Environ. Epigenetics 2019, 5, dvz022. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of Chromatin by Histone Modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Shibasaki, M.; Mizuno, K.; Kurokawa, K.; Ohkuma, S. L-Type Voltage-Dependent Calcium Channels Facilitate Acetylation of Histone H3 through PKCγ Phosphorylation in Mice with Methamphetamine-Induced Place Preference. J. Neurochem. 2011, 118, 1056–1066. [Google Scholar] [CrossRef] [PubMed]
- Sanjari Moghaddam, H.; Mobarak Abadi, M.; Dolatshahi, M.; Bayani Ershadi, S.; Abbasi-Feijani, F.; Rezaei, S.; Cattarinussi, G.; Aarabi, M.H. Effects of Prenatal Methamphetamine Exposure on the Developing Human Brain: A Systematic Review of Neuroimaging Studies. ACS Chem. Neurosci. 2021, 12, 2729–2748. [Google Scholar] [CrossRef] [PubMed]
- Lo, J.O.; D’Mello, R.J.; Watch, L.; Schust, D.J.; Murphy, S.K. An Epigenetic Synopsis of Parental Substance Use. Epigenomics 2023, 15, 453–473. [Google Scholar] [CrossRef]
- Kundakovic, M.; Jaric, I. The Epigenetic Link between Prenatal Adverse Environments and Neurodevelopmental Disorders. Genes 2017, 8, 104. [Google Scholar] [CrossRef]
- Perera, F.; Herbstman, J. Prenatal Environmental Exposures, Epigenetics, and Disease. Reprod. Toxicol. 2011, 31, 363–373. [Google Scholar] [CrossRef]
- Larsen, E.M.H.; Brock-Nannestad, T.; Skibsted, J.; Reinholdt, A.; Bendix, J. A Homoleptic AgIII Complex Stabilized by Succinimidate Ligands. Chem. Sci. 2024, 15, 18067–18075. [Google Scholar] [CrossRef]
- Knopik, V.S.; Maccani, M.A.; Francazio, S.; McGeary, J.E. The Epigenetics of Maternal Cigarette Smoking During Pregnancy and Effects on Child Development. Dev. Psychopathol. 2012, 24, 1377–1390. [Google Scholar] [CrossRef]
- Geraghty, A.A.; Lindsay, K.L.; Alberdi, G.; McAuliffe, F.M.; Gibney, E.R. Nutrition During Pregnancy Impacts Offspring’s Epigenetic Status—Evidence from Human and Animal Studies. Nutr. Metab. Insights 2016, 8, 41–47. [Google Scholar] [CrossRef]
- DeSocio, J.E. Epigenetics, Maternal Prenatal Psychosocial Stress, and Infant Mental Health. Arch. Psychiatr. Nurs. 2018, 32, 901–906. [Google Scholar] [CrossRef]
- Peña, C.J. Epigenetic Regulation of Brain Development, Plasticity, and Response to Early-Life Stress. Neuropsychopharmacology 2025. [Google Scholar] [CrossRef]
- Baratta, A.M.; Rathod, R.S.; Plasil, S.L.; Seth, A.; Homanics, G.E. Chapter Six—Exposure to Drugs of Abuse Induce Effects That Persist across Generations. In International Review of Neurobiology; Pandey, S.C., Ed.; Epigenetics; Academic Press: Cambridge, MA, USA, 2021; Volume 156, pp. 217–277. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hediyal, T.A.; Shukri, O.; Stone, E.; Foroughi, A.; Samikkannu, T.; Pendyala, G. Decoding Encoded Cravings: Epigenetic Drivers of Addiction. Brain Sci. 2025, 15, 927. https://doi.org/10.3390/brainsci15090927
Hediyal TA, Shukri O, Stone E, Foroughi A, Samikkannu T, Pendyala G. Decoding Encoded Cravings: Epigenetic Drivers of Addiction. Brain Sciences. 2025; 15(9):927. https://doi.org/10.3390/brainsci15090927
Chicago/Turabian StyleHediyal, Tousif Ahmed, Omar Shukri, Elizabeth Stone, Amin Foroughi, Thangavel Samikkannu, and Gurudutt Pendyala. 2025. "Decoding Encoded Cravings: Epigenetic Drivers of Addiction" Brain Sciences 15, no. 9: 927. https://doi.org/10.3390/brainsci15090927
APA StyleHediyal, T. A., Shukri, O., Stone, E., Foroughi, A., Samikkannu, T., & Pendyala, G. (2025). Decoding Encoded Cravings: Epigenetic Drivers of Addiction. Brain Sciences, 15(9), 927. https://doi.org/10.3390/brainsci15090927






