Probability and Neurodegeneration: Alzheimer’s Disease and Huntington’s Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. The Mechanisms
2.2. Probability in Neurology
2.3. Experimental Verification
3. Results
4. Discussion
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Panegyres, P.K. The clinical spectrum of young onset dementia points to its stochastic origins. J. Alzheimers Dis. Rep. 2021, 5, 663–679. [Google Scholar] [CrossRef]
- Panegyres, P.K. Stochasticity, entropy and neurodegeneration. Brain Sci. 2022, 12, 226. [Google Scholar] [CrossRef]
- Panegyres, P.K. Stochastic considerations into the origins of sporadic adult onset neurodegenerative disorders. J. Alzheimers Dis. Park. 2019, 9, 473. [Google Scholar]
- Hendriks, S.; Peetoom, K.; Bakker, C.; van der Flier, W.M.; Papma, J.M.; Koopmans, R.; Verhey, F.R.J.; de Vugt, M.; Köhler, S.; Young-Onset Dementia Epidemiology Study Group; et al. Global Prevalence of Young-Onset Dementia: A Systematic Review and Meta-analysis. JAMA Neurol. 2021, 78, 1080–1090. [Google Scholar] [CrossRef] [PubMed]
- National Institute of Aging. The NIA Monthly Update. 2 July 2024. Available online: https://www.linkedin.com/pulse/nia-monthly-update-july-2024-national-institute-on-aging-5qxrf?trk=organization_guest_main-feed-card_feed-article-content (accessed on 13 January 2025).
- Livingston, G.; Huntley, J.; Liu, K.Y.; Costafreda, S.G.; Selbaek, G.; Alladi, S.; Ames, D.; Banerjee, S.; Burns, A.; Brayne, C.; et al. Dementia prevention, intervention, and care: 2024 report of the Lancet Sanding Commission. Lancet 2024, 404, 572–628. [Google Scholar] [CrossRef]
- Sato, N. Is organization of living systems explained by probability? Philosophies 2021, 6, 3. [Google Scholar] [CrossRef]
- Nakajima, T. Biological probability: Cognitive processes of generating probabilities of events in biological systems. J. Theor. Biol. 1999, 200, 77–95. [Google Scholar] [CrossRef]
- Nakajima, T. Probability in biology: Overview of a comprehensive theory of probability in living systems. Prog. Biophys. Mol. Biol. 2013, 113, 67–79. [Google Scholar] [CrossRef]
- Nakajima, T. Ecological mechanisms of evolution by natural selection: Causal processes generating density-and-frequency dependent fitness. J. Theor. Biol. 1998, 190, 313–331. [Google Scholar] [CrossRef]
- Nakajima, T. Probabilities of encounters between objects in biological systems: Meta-observer view. J. Theor. Biol. 2001, 211, 347–363. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, T. Probabilities of encounters between objects in biological systems 2: Cognizer view. J. Theor. Biol. 2003, 221, 39–51. [Google Scholar] [CrossRef]
- Root-Bernstein, R.S.; Dillon, P.F. Molecular complementarity I: The complementarity theory of the origin and evolution of life. J. Theor. Biol. 1997, 188, 447–479. [Google Scholar] [CrossRef]
- Vallender, E.J. Genetic correlates of the evolving primate brain. Prog. Brain. Res. 2012, 195, 27–44. [Google Scholar] [CrossRef] [PubMed]
- Enard, W. The Molecular Basis of Human Brain Evolution. Curr. Biol. 2016, 26, R1109–R1117. [Google Scholar] [CrossRef]
- Nakajima, T. Internal Probability Theory and the Evolution of Life. Ann. Jpn. Assoc. Philos. Sci. 2008, 16, 75–94. [Google Scholar] [CrossRef][Green Version]
- Wilson, A.C. The molecular basis of evolution. Sci. Am. 1985, 253, 164–173. [Google Scholar] [CrossRef]
- Ortega, O.O.; Ozen, M.; Wilson, B.A.; Pino, J.C.; Irvin, M.W.; Ildefonso, G.V.; Garbett, S.P.; Lopez, C.F. Signal execution modes emerge in biochemical reaction networks calibrated to experimental data. iScience 2024, 27, 109989. [Google Scholar] [CrossRef]
- Barnes, C.P.; Silk, D.; Sheng, X.; Stumpf, M.P. Bayesian design of synthetic biological systems. Proc. Natl. Acad. Sci. USA 2011, 108, 15190–15195. [Google Scholar] [CrossRef]
- Periwal, V. Bayesian inference of biological systems: The logic of biology. In System Modelling in Cellular Biology: From Concepts to Nuts and Bolts; Szallasi, Z., Stelling, J., Periwal, V., Eds.; MIT Press: Cambridge, MA, USA, 2006. [Google Scholar] [CrossRef]
- Cespedes, M.I.; Fripp, J.; McGree, J.M.; Drovandi, C.C.; Mengersen, K.; Doecke, J.D. Comparisons of neurodegeneration over time between healthy ageing and Alzheimer’s disease cohorts via Bayesian inference. BMJ Open 2017, 7, e012174. [Google Scholar] [CrossRef] [PubMed]
- Cobigo, Y.; Goh, M.S.; Wolf, A.; Staffaroni, A.M.; Kornak, J.; Miller, B.L.; Rabinovici, G.D.; Seeley, W.W.; Spina, S.; Boxer, A.L.; et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI) and the Dominantly Inherited Alzheimer’s Network (DIAN). Detection of emerging neurodegeneration using Bayesian linear mixed-effect modeling. Neuroimage Clin. 2022, 36, 103144. [Google Scholar] [CrossRef] [PubMed]
- Khrennikov, A. Open Systems, Quantum Probability, and Logic for Quantum-like Modeling in Biology, Cognition, and Decision-Making. Entropy 2023, 25, 886. [Google Scholar] [CrossRef]
- Arndt, M.; Juffmann, T.; Vedral, V. Quantum physics meets biology. HFSP J. 2009, 3, 386–400. [Google Scholar] [CrossRef] [PubMed]
- Maury, E.A.; Walsh, C.A.; Kahle, K.T. Neurosurgery elucidates somatic mutations. Science 2023, 382, 1360–1362. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E. Revolutionary cryo-EM is taking over structural biology. Nature 2020, 578, 201. [Google Scholar] [CrossRef]
- McMullan, G.; Naydenova, K.; Mihaylov, D.; Yamashita, K.; Peet, M.J.; Wilson, H.; Dickerson, J.L.; Chen, S.; Cannone, G.; Lee, Y.; et al. Structure determination by cryoEM at 100 keV. Proc. Natl. Acad. Sci. USA 2023, 120, e2312905120. [Google Scholar] [CrossRef]
- Evrony, G.D.; Cai, X.; Lee, E.; Hills, L.B.; Elhosary, P.C.; Lehmann, H.S.; Parker, J.J.; Atabay, K.D.; Gilmore, E.C.; Poduri, A.; et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell 2012, 151, 483–496. [Google Scholar] [CrossRef]
- Evrony, G.D.; Lee, E.; Mehta, B.K.; Benjamini, Y.; Johnson, R.M.; Cai, X.; Yang, L.; Haseley, P.; Lehmann, H.S.; Park, P.J.; et al. Cell lineage analysis in human brain using endogenous retroelements. Neuron 2015, 85, 49–59. [Google Scholar] [CrossRef]
- Lodato, M.A.; Woodworth, M.B.; Lee, S.; Evrony, G.D.; Mehta, B.K.; Karger, A.; Lee, S.; Chittenden, T.W.; D’gAma, A.M.; Cai, X.; et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science 2015, 350, 94–98. [Google Scholar] [CrossRef]
- Lodato, M.A.; Rodin, R.E.; Bohrson, C.L.; Coulter, M.E.; Barton, A.R.; Kwon, M.; Sherman, M.A.; Vitzthum, C.M.; Luquette, L.J.; Yandava, C.N.; et al. Aging and neurodegeneration are associated with increased mutations in single human neurons. Science 2018, 359, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Poduri, A.; Evrony, G.D.; Cai, X.; Walsh, C.A. Somatic mutation, genomic variation, and neurological disease. Science 2013, 341, 1237758. [Google Scholar] [CrossRef]
- Keogh, M.J.; Wei, W.; Aryaman, J.; Walker, L.; Ameele, J.v.D.; Coxhead, J.; Wilson, I.; Bashton, M.; Beck, J.; West, J.; et al. High prevalence of focal and multi-focal somatic genetic variants in the human brain. Nat. Commun. 2018, 9, 4257. [Google Scholar] [CrossRef] [PubMed]
- Bae, T.; Fasching, L.; Wang, Y.; Shin, J.H.; Suvakov, M.; Jang, Y.; Norton, S.; Dias, C.; Mariani, J.; Jourdon, A.; et al. Analysis of somatic mutations in 131 human brains reveals aging-associated hypermutability. Science 2022, 377, 511–517. [Google Scholar] [CrossRef]
- Bin Kim, I.; Kim, M.-H.; Jung, S.; Kim, W.K.; Lee, J.; Ju, Y.S.; Webster, M.J.; Kim, S.; Kim, J.H.; Kim, H.J.; et al. Low-level brain somatic mutations in exonic regions are collectively implicated in autism with germline mutations in autism risk genes. Exp. Mol. Med. 2024, 56, 1750–1762. [Google Scholar] [CrossRef]
- Evrony, G.D. One brain, many genomes. Science 2016, 354, 557–558. [Google Scholar] [CrossRef]
- Cai, X.; Evrony, G.D.; Lehmann, H.S.; Elhosary, P.C.; Mehta, B.K.; Poduri, A.; Walsh, C.A. Single-cell, genome-wide sequencing identifies clonal somatic copy-number variation in the human brain. Cell Rep. 2014, 8, 1280–1289. [Google Scholar] [CrossRef] [PubMed]
- Handsaker, R.E.; Kashin, S.; Reed, N.M.; Tan, S.; Lee, W.S.; McDonald, T.M.; Morris, K.; Kamitaki, N.; Mullally, C.D.; Morakabati, N.R.; et al. Long somatic DNA-repeat expansion drives neurodegeneration in Huntington’s disease. Cell 2025, 188, 623–639. [Google Scholar] [CrossRef]
- Scahill, R.I.; Farag, M.; Murphy, M.J.; Hobbs, N.Z.; Leocadi, M.; Langley, C.; Knights, H.; Ciosi, M.; Fayer, K.; Nakajima, M.; et al. Somatic CAG repeat expansion in blood associates with biomarkers of neurodegeneration in Huntington’s disease decades before clinical motor diagnosis. Nat. Med. 2025, 31, 807–818. [Google Scholar] [CrossRef]
- Leija-Salazar, M.; Piette, C.L.; Proukakis, C. Review: Somatic mutations in neurodegeneration. Neuropathol. Appl. Neurobiol. 2018, 44, 267–285. [Google Scholar] [CrossRef]
- Proukakis, C. Somatic mutations in neurodegeneration: An update. Neurobiol. Dis. 2020, 144, 105021. [Google Scholar] [CrossRef]
- Frank, S.A. Evolution in health and medicine Sackler colloquium: Somatic evolutionary genomics: Mutations during development cause highly variable genetic mosaicism with risk of cancer and neurodegeneration. Proc. Natl. Acad. Sci. USA 2010, 107, 1725–1730. [Google Scholar] [CrossRef] [PubMed]
- Perez-Rodriguez, D.; Kalyva, M.; Leija-Salazar, M.; Lashley, T.; Tarabichi, M.; Chelban, V.; Gentleman, S.; Schottlaender, L.; Franklin, H.; Vasmatzis, G.; et al. Investigation of somatic CNVs in brains of synucleinopathy cases using targeted SNCA analysis and single cell sequencing. Acta Neuropathol. Commun. 2019, 7, 219. [Google Scholar] [CrossRef]
- van Rooij, J.; Arp, P.; Broer, L.; Verlouw, J.; van Rooij, F.; Kraaij, R.; Uitterlinden, A.; Verkerk, A.J.M.H. Reduced penetrance of pathogenic ACMG variants in a deeply phenotyped cohort study and evaluation of ClinVar classification over time. Genet. Med. 2020, 22, 1812–1820. [Google Scholar] [CrossRef]
- Nicolas, G.; Acuña-Hidalgo, R.; Keogh, M.J.; Quenez, O.; Steehouwer, M.; Lelieveld, S.; Rousseau, S.; Richard, A.; Oud, M.S.; Marguet, F.; et al. Somatic variants in autosomal dominant genes are a rare cause of sporadic Alzheimer’s disease. Alzheimers Dement. 2018, 14, 1632–1639. [Google Scholar] [CrossRef]
- Wei, W.; Keogh, M.J.; Aryaman, J.; Golder, Z.; Kullar, P.J.; Wilson, I.; Talbot, K.; Turner, M.R.; McKenzie, C.A.; Troakes, C.; et al. Frequency and signature of somatic variants in 1461 human brain exomes. Genet Med. 2019, 21, 904–912. [Google Scholar] [CrossRef]
- Lobon, I.; Solís-Moruno, M.; Juan, D.; Muhaisen, A.; Abascal, F.; Esteller-Cucala, P.; García-Pérez, R.; Martí, M.J.; Tolosa, E.; Ávila, J.; et al. Somatic Mutations detected in Parkinson disease could affect genes with a role in synaptic and neuronal processes. Front. Aging 2022, 3, 851039. [Google Scholar] [CrossRef] [PubMed]
- Verheijen, B.M.; Vermulst, M.; van Leeuwen, F.W. Somatic mutations in neurons during aging and neurodegeneration. Acta Neuropathol. 2018, 135, 811–826. [Google Scholar] [CrossRef] [PubMed]
- Turan, Z.G.; Richter, V.; Bochmann, J.; Parvizi, P.; Yapar, E.; Işıldak, U.; Waterholter, S.K.; Leclere-Turbant, S.; Son, Ç.D.; Duyckaerts, C.; et al. Somatic copy number variant load in neurons of healthy controls and Alzheimer’s disease patients. Acta Neuropathol. Commun. 2022, 10, 175. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, J.H. Brain somatic mutations as RNA therapeutic targets in neurological disorders. Ann. N. Y. Acad. Sci. 2022, 1514, 11–20. [Google Scholar] [CrossRef]
- Jia, J.; Zhang, Y.; Shi, Y.; Yin, X.; Wang, S.; Li, Y.; Zhao, T.; Liu, W.; Zhou, A.; Jia, L. A 19-year-old adolescent with probable Alzheimer’s disease. J. Alzheimer’s Dis. 2023, 91, 915–922. [Google Scholar] [CrossRef]



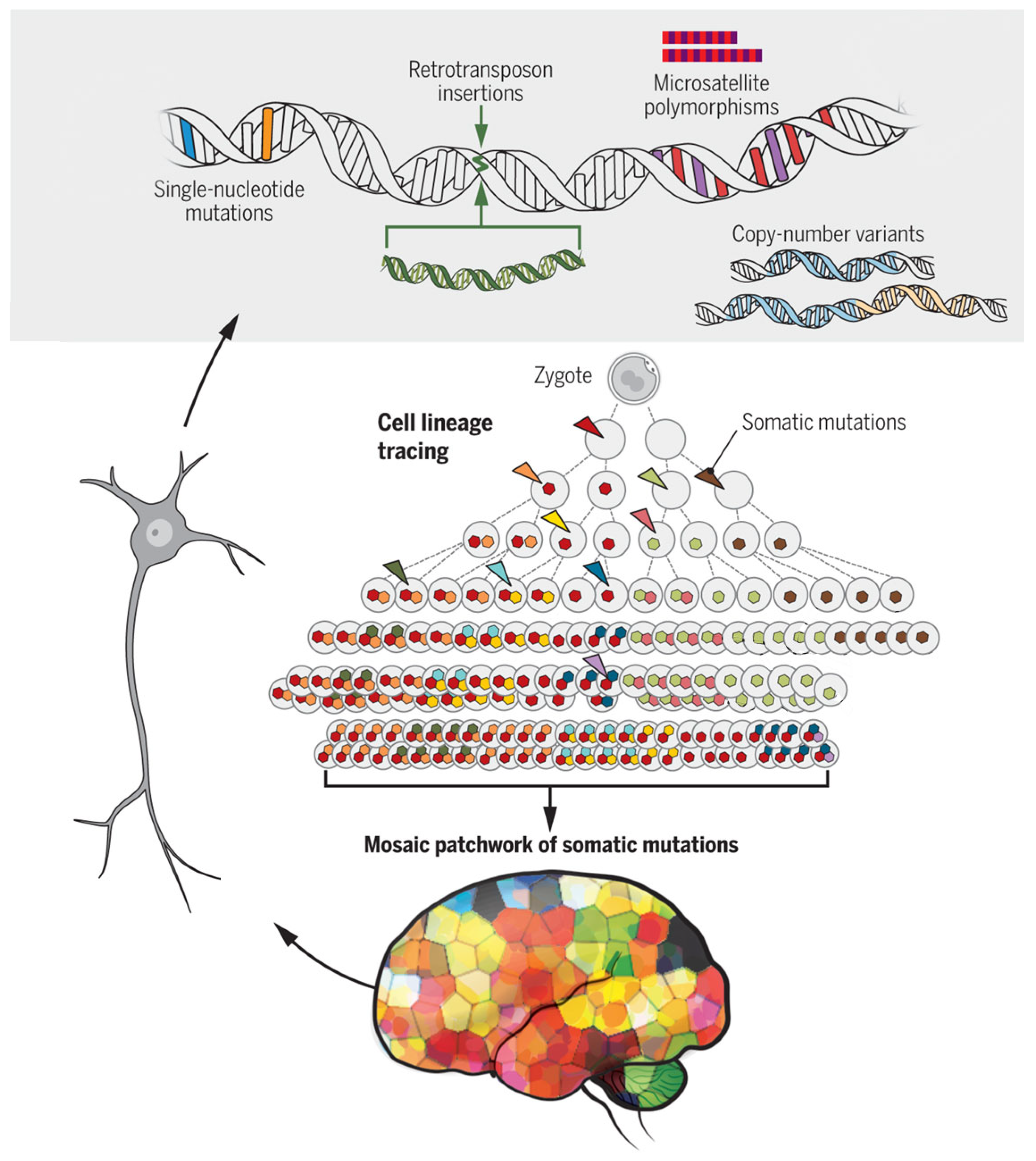
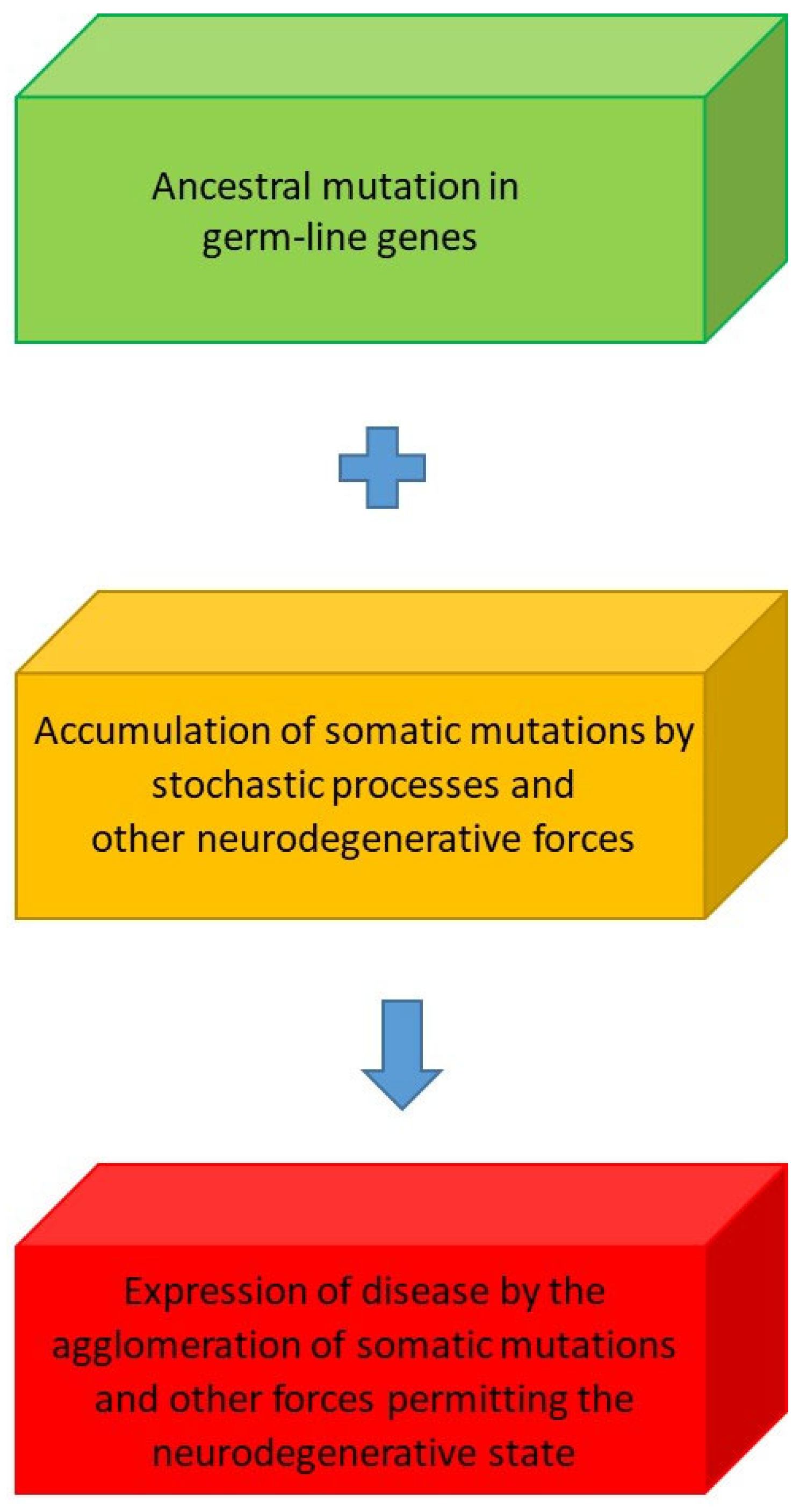

| Probabilistic State Designation | Processes |
|---|---|
| A | Gene mutations (APP, PS1 & 2) |
| B | Cerebrovascular risk factors (hypertension, stroke, diabetes mellitus, or dyslipidemia) |
| Γ | APOE ε4/ε4 |
| Δ | Head injury |
| E | Age |
| Z | Brain health and lifestyle (education, exercise, smoking, diet, alcohol, body weight, sleep, social network and engagement and cognitive activity) |
| H | Deafness |
| Θ | Socioeconomic status |
| I | Ethnicity |
| K | Biophysical (microenvironment, pH, immune reactions, glial responses and phagosome function) |
| Λ | Unknown (yet to be determined) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panegyres, P.K. Probability and Neurodegeneration: Alzheimer’s Disease and Huntington’s Disease. Brain Sci. 2025, 15, 814. https://doi.org/10.3390/brainsci15080814
Panegyres PK. Probability and Neurodegeneration: Alzheimer’s Disease and Huntington’s Disease. Brain Sciences. 2025; 15(8):814. https://doi.org/10.3390/brainsci15080814
Chicago/Turabian StylePanegyres, Peter K. 2025. "Probability and Neurodegeneration: Alzheimer’s Disease and Huntington’s Disease" Brain Sciences 15, no. 8: 814. https://doi.org/10.3390/brainsci15080814
APA StylePanegyres, P. K. (2025). Probability and Neurodegeneration: Alzheimer’s Disease and Huntington’s Disease. Brain Sciences, 15(8), 814. https://doi.org/10.3390/brainsci15080814





