Genetic Markers and Mutations in Primary Spinal Cord Tumors and Their Impact on Clinical Management
Abstract
1. Introduction
2. Tumor-Specific Genetic Profiles Affecting Behavior and Prognosis
2.1. Ependymoma
2.2. Ganglioglioma
2.3. Astrocytoma/Glioma
2.4. Glioblastoma (GBM)
2.5. Hemangioblastoma
3. Molecular Pathways Altered by the Key Mutations and Their Impact on Spinal Cord Tumors
3.1. N-Myc/MYCN
3.2. HOXB13
3.3. BRAF V600E
3.4. H3 K27M (H3F3A/HIST1H3B)
3.5. TERT Promoter Mutations
3.6. TP53
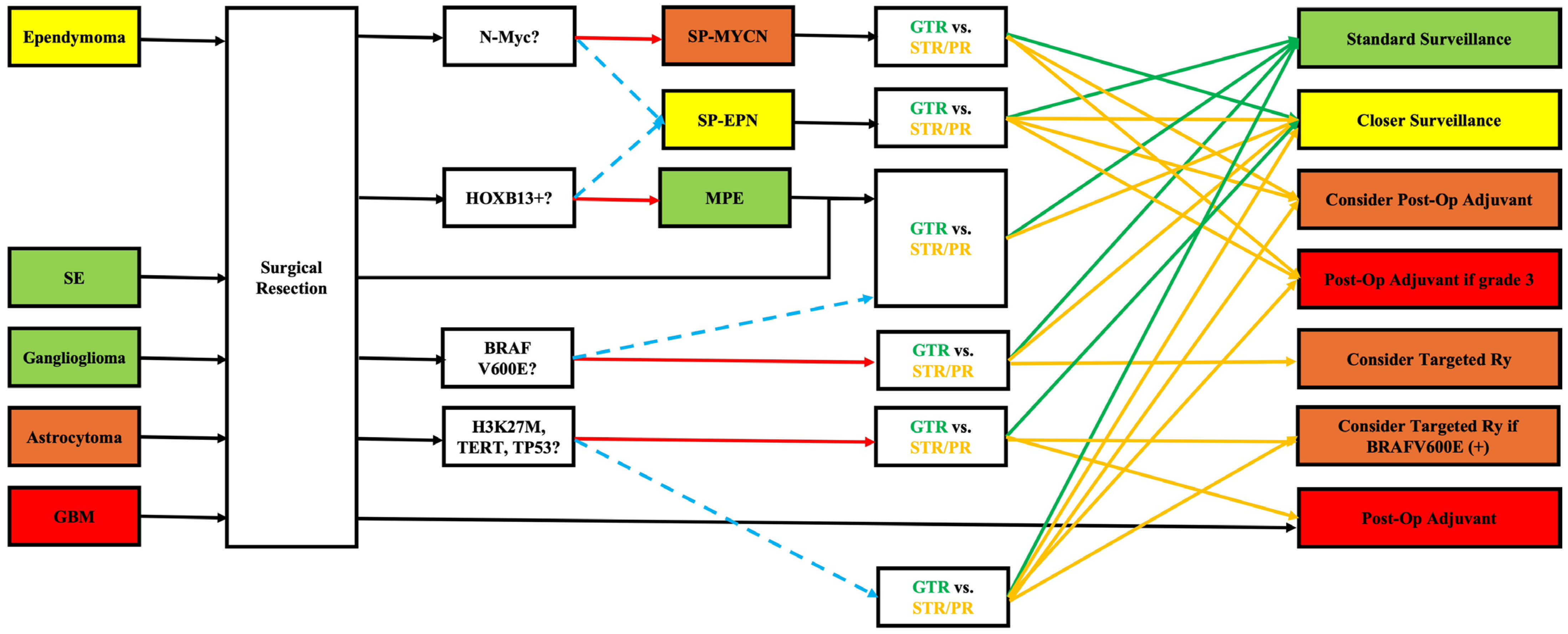
4. Integrating Genetic Data into Clinical Practice
5. Limitations and Future Directions
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dickman, C.A.; Fehlings, M.; Gokaslan, Z.L. Spinal Cord and Spinal Column Tumors: Principles and Practice; Thieme: Stuttgart, Germany, 2006. [Google Scholar]
- Chamberlain, M.C.; Tredway, T.L. Adult primary intradural spinal cord tumors: A review. Curr. Neurol. Neurosci. Rep. 2011, 11, 320–328. [Google Scholar] [CrossRef]
- Nagashima, Y.; Nishimura, Y.; Eguchi, K.; Yamaguchi, J.; Haimoto, S.; Ohka, F.; Takayasu, M.; Saito, R. Recent molecular and genetic findings in intramedullary spinal cord tumors. Neurospine 2022, 19, 262–271. [Google Scholar] [CrossRef]
- Zhao, Z.; Song, Z.; Wang, Z.; Zhang, F.; Ding, Z.; Fan, T. Advances in molecular pathology, diagnosis and treatment of spinal cord astrocytomas. Technol. Cancer Res. Treat. 2024, 23, 15330338241262483. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 who classification of tumors of the central nervous system: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Pratt, D.; Sahm, F.; Aldape, K. DNA methylation profiling as a model for discovery and precision diagnostics in neuro-oncology. Neuro Oncol. 2021, 23, S16–S29. [Google Scholar] [CrossRef]
- Khalid, S.I.; Adogwa, O.; Kelly, R.; Metha, A.; Bagley, C.; Cheng, J.; O’Toole, J. Adult spinal ependymomas: An epidemiologic study. World Neurosurg. 2018, 111, e53–e61. [Google Scholar] [CrossRef]
- Wu, L.; Wang, L.; Zou, W.; Yang, J.; Jia, W.; Xu, Y. Primary spinal anaplastic ependymoma: A single-institute retrospective cohort and systematic review. Front. Oncol. 2023, 13, 1083085. [Google Scholar] [CrossRef] [PubMed]
- Davison, M.A.; Lilly, D.T.; Patel, A.A.; Kashkoush, A.; Chen, X.; Wei, W.; Benzel, E.C.; Prayson, R.A.; Chao, S.; Angelov, L. Clinical presentation and extent of resection impacts progression-free survival in spinal ependymomas. J. Neurooncol. 2024, 167, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, D.R.; Sill, M.; Okonechnikov, K.; Korshunov, A.; Yip, S.; Schutz, P.W.; Scheie, D.; Kruse, A.; Harter, P.N.; Kastelan, M.; et al. MYCN amplification drives an aggressive form of spinal ependymoma. Acta Neuropathol. 2019, 138, 1075–1089. [Google Scholar] [CrossRef]
- Swanson, A.A.; Raghunathan, A.; Jenkins, R.B.; Messing-Jünger, M.; Pietsch, T.; Clarke, M.J.; Kaufmann, T.J.; Giannini, C. Spinal cord ependymomas with MYCN amplification show aggressive clinical behavior. J. Neuropathol. Exp. Neurol. 2019, 78, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Gu, W.; Shou, J.; Xiong, J.; Liu, X.; Sun, B.; Yang, D.; Xie, R. The molecular feature of hox gene family in the intramedullary spinal tumors. Spine 2017, 42, 291–297. [Google Scholar] [CrossRef]
- Purkait, S.; Praeger, S.; Felsberg, J.; Pauck, D.; Kaulich, K.; Wolter, M.; Koppstein, D.; Reifenberger, G. Strong nuclear expression of hoxb13 is a reliable surrogate marker for DNA methylome profiling to distinguish myxopapillary ependymoma from spinal ependymoma. Acta Neuropathol. 2025, 149, 29. [Google Scholar] [CrossRef]
- Yagi, T.; Mizuno, M.; Kageyama, H.; Tatebayashi, K.; Endo, T.; Takeshima, Y.; Iwasaki, M.; Kurokawa, R.; Takai, K.; Nishikawa, M.; et al. Spinal cord subependymoma: A subanalysis of the neurospinal society of japan’s multicenter study of intramedullary spinal cord tumors. Neurospine 2023, 20, 735–746. [Google Scholar] [CrossRef]
- Lang, F.F.; Epstein, F.J.; Ransohoff, J.; Allen, J.C.; Wisoff, J.; Abbott, I.R.; Miller, D.C. Central nervous system gangliogliomas. Part 2: Clinical outcome. J. Neurosurg. 1993, 79, 867–873. [Google Scholar] [CrossRef] [PubMed]
- Song, J.Y.; Kim, J.H.; Cho, Y.H.; Kim, C.J.; Lee, E.J. Treatment and outcomes for gangliogliomas: A single-center review of 16 patients. Brain Tumor Res. Treat. 2014, 2, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Jyothirmayi, R.; Madhavan, J.; Nair, M.K.; Rajan, B. Conservative surgery and radiotherapy in the treatment of spinal cord astrocytoma. J. Neurooncol. 1997, 33, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Neyazi, B.; Haghikia, A.; Mawrin, C.; Hattingen, E.; Vordermark, D.; Sandalcioglu, I.E. Spinal intramedullary tumors. Dtsch. Arztebl. Int. 2024, 121, 840–846. [Google Scholar] [CrossRef]
- Tonn, J.-C.; Teske, N.; Karschnia, P. Astrocytomas of the spinal cord. Neuro Oncol. Adv. 2024, 6, iii48–iii56. [Google Scholar] [CrossRef]
- Zhang, F.; Cheng, L.; Ding, Z.; Wang, S.; Zhao, X.; Zhao, Z.; Liang, C.; Wu, K.; Zhang, D.; Wang, Y.; et al. Does h3k27m mutation impact survival outcome of high-grade spinal cord astrocytoma? Neurospine 2023, 20, 1480–1489. [Google Scholar] [CrossRef]
- Akinduro, O.O.; Garcia, D.P.; Higgins, D.M.O.; Vivas-Buitrago, T.; Jentoft, M.; Solomon, D.A.; Daniels, D.J.; Pennington, Z.; Sherman, W.J.; Delgardo, M.; et al. A multicenter analysis of the prognostic value of histone h3 k27m mutation in adult high-grade spinal glioma. J. Neurosurg. Spine 2021, 35, 834–843. [Google Scholar] [CrossRef]
- Alvi, M.A.; Ida, C.M.; Paolini, M.A.; Kerezoudis, P.; Meyer, J.; Fritcher, E.G.B.; Goncalves, S.; Meyer, F.B.; Bydon, M.; Raghunathan, A. Spinal cord high-grade infiltrating gliomas in adults: Clinico-pathological and molecular evaluation. Mod. Pathol. 2019, 32, 1236–1243. [Google Scholar] [CrossRef]
- Chai, R.C.; Zhang, Y.W.; Liu, Y.Q.; Chang, Y.Z.; Pang, B.; Jiang, T.; Jia, W.Q.; Wang, Y.Z. The molecular characteristics of spinal cord gliomas with or without h3 k27m mutation. Acta Neuropathol. Commun. 2020, 8, 40. [Google Scholar] [CrossRef]
- Arita, H.; Matsushita, Y.; Machida, R.; Yamasaki, K.; Hata, N.; Ohno, M.; Yamaguchi, S.; Sasayama, T.; Tanaka, S.; Higuchi, F.; et al. TERT promoter mutation confers favorable prognosis regardless of 1p/19q status in adult diffuse gliomas with idh1/2 mutations. Acta Neuropathol. Commun. 2020, 8, 201. [Google Scholar] [CrossRef]
- Gorria, T.; Crous, C.; Pineda, E.; Hernandez, A.; Domenech, M.; Sanz, C.; Jares, P.; Muñoz-Mármol, A.M.; Arpí-Llucía, O.; Melendez, B.; et al. The c250t mutation of TERTp might grant a better prognosis to glioblastoma by exerting less biological effect on telomeres and chromosomes than the c228t mutation. Cancers 2024, 16, 735. [Google Scholar] [CrossRef]
- Heidenreich, B.; Rachakonda, P.S.; Hosen, I.; Volz, F.; Hemminki, K.; Weyerbrock, A.; Kumar, R. TERT promoter mutations and telomere length in adult malignant gliomas and recurrences. Oncotarget 2015, 6, 10617–10633. [Google Scholar] [CrossRef]
- Lee, Y.; Park, C.K.; Park, S.H. Prognostic impact of TERT promoter mutations in adult-type diffuse gliomas based on who2021 criteria. Cancers 2024, 16, 2032. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.K.; Wang, J.; Guo, Y.; Sun, Z.X.; Wang, G.H. Identification of differentially expressed genes and fusion genes associated with malignant progression of spinal cord gliomas by transcriptome analysis. Sci. Rep. 2019, 9, 13583. [Google Scholar] [CrossRef] [PubMed]
- Jokovic, M.; Somma, T.; Ilic, R.; Guizzardi, G.; Stanimirovic, A.; Raicevic, S.; Milicevic, M.; Grujicic, D.; Solari, D. Primary spinal glioblastoma multiforme. Single center experience and literature review. Interdiscip. Neurosurg. 2021, 24, 101109. [Google Scholar] [CrossRef]
- Lucas Negromonte Guerra, P.; da Silva, I.C.S.; Júnior, D.L.B.; Lopes, A.A.P.; de Sá Carneiro Filho, G.; de Carvalho Júnior, E.V. Epidemiological and clinical characteristics of primary spinal cord glioblastomas: A systematic review and meta-analysis. J. Clin. Neurosci. 2024, 130, 110862. [Google Scholar] [CrossRef]
- Ezzat, B.; Young, T.; Schüpper, A.J.; Kalagara, R.; Zhang, J.Y.; Lemonick, M.; Bhanot, P.; Quinones, A.; Choudhri, T.; Germano, I.M. Molecular profile and clinical outcome of adult primary spinal cord glioblastoma: A systematic review. J. Neurosurg. Spine 2024, 41, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Wach, J.; Basaran, A.E.; Vychopen, M.; Tihan, T.; Wostrack, M.; Butenschoen, V.M.; Meyer, B.; Siller, S.; Schmidt, N.O.; Onken, J.; et al. Local tumor control and neurological outcomes after surgery for spinal hemangioblastomas in sporadic and von hippel-lindau disease: A multicenter study. Neuro Oncol. 2025, 27, 1567–1578. [Google Scholar] [CrossRef]
- Cerretti, G.; Pessina, F.; Franceschi, E.; Barresi, V.; Salvalaggio, A.; Padovan, M.; Manara, R.; Di Nunno, V.; Bono, B.C.; Librizzi, G.; et al. Spinal ependymoma in adults: From molecular advances to new treatment perspectives. Front. Oncol. 2023, 13, 1301179. [Google Scholar] [CrossRef]
- Connolly, I.D.; Ali, R.; Li, Y.; Gephart, M.H. Genetic and molecular distinctions in spinal ependymomas: A review. Clin. Neurol. Neurosurg. 2015, 139, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Kresbach, C.; Neyazi, S.; Schüller, U. Updates in the classification of ependymal neoplasms: The 2021 who classification and beyond. Brain Pathol. 2022, 32, e13068. [Google Scholar] [CrossRef] [PubMed]
- Raffeld, M.; Abdullaev, Z.; Pack, S.D.; Xi, L.; Nagaraj, S.; Briceno, N.; Vera, E.; Pittaluga, S.; Neto, O.L.A.; Quezado, M.; et al. High level MYCN amplification and distinct methylation signature define an aggressive subtype of spinal cord ependymoma. Acta Neuropathol. Commun. 2020, 8, 101. [Google Scholar] [CrossRef] [PubMed]
- Gomez, D.R.; Missett, B.T.; Wara, W.M.; Lamborn, K.R.; Prados, M.D.; Chang, S.; Berger, M.S.; Haas-Kogan, D.A. High failure rate in spinal ependymomas with long-term follow-up. Neuro Oncol. 2005, 7, 254–259. [Google Scholar] [CrossRef]
- Yao, K.; Duan, Z.; Wang, Y.; Zhang, M.; Fan, T.; Wu, B.; Qi, X. Detection of h3k27m mutation in cases of brain stem subependymoma. Hum. Pathol. 2019, 84, 262–269. [Google Scholar] [CrossRef]
- Thomas, C.; Thierfelder, F.; Träger, M.; Soschinski, P.; Müther, M.; Edelmann, D.; Förster, A.; Geiler, C.; Kim, H.Y.; Filipski, K.; et al. TERT promoter mutation and chromosome 6 loss define a high-risk subtype of ependymoma evolving from posterior fossa subependymoma. Acta Neuropathol. 2021, 141, 959–970. [Google Scholar] [CrossRef]
- Gessi, M.; Dörner, E.; Dreschmann, V.; Antonelli, M.; Waha, A.; Giangaspero, F.; Gnekow, A.; Pietsch, T. Intramedullary gangliogliomas: Histopathologic and molecular features of 25 cases. Hum. Pathol. 2016, 49, 107–113. [Google Scholar] [CrossRef][Green Version]
- Di Nunno, V.; Gatto, L.; Tosoni, A.; Bartolini, S.; Franceschi, E. Implications of BRAF V600E mutation in gliomas: Molecular considerations, prognostic value and treatment evolution. Front. Oncol. 2022, 12, 1067252. [Google Scholar] [CrossRef]
- Garnier, L.; Ducray, F.; Verlut, C.; Mihai, M.I.; Cattin, F.; Petit, A.; Curtit, E. Prolonged response induced by single agent vemurafenib in a BRAF V600E spinal ganglioglioma: A case report and review of the literature. Front. Oncol. 2019, 9, 177. [Google Scholar] [CrossRef] [PubMed]
- Amelot, A.; Terrier, L.M.; Mathon, B.; Joubert, C.; Picart, T.; Jecko, V.; Bauchet, L.; Bernard, F.; Castel, X.; Chenin, L.; et al. Natural course and prognosis of primary spinal glioblastoma: A nationwide study. Neurology 2023, 100, e1497–e1509. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Hou, C.; Chen, H.; Zong, X.; Zong, P. Genetics and epigenetics of glioblastoma: Applications and overall incidence of idh1 mutation. Front. Oncol. 2016, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Aishwarya, S.; Li, J.P.; Pan, D.X.; Shi, J.P. Gene expression profiling of glioblastoma to recognize potential biomarker candidates. Front. Genet. 2022, 13, 832742. [Google Scholar] [CrossRef]
- Klingler, J.H.; Gläsker, S.; Bausch, B.; Urbach, H.; Krauss, T.; Jilg, C.A.; Steiert, C.; Puzik, A.; Neumann-Haefelin, E.; Kotsis, F.; et al. Hemangioblastoma and von hippel-lindau disease: Genetic background, spectrum of disease, and neurosurgical treatment. Childs Nerv. Syst. 2020, 36, 2537–2552. [Google Scholar] [CrossRef]
- Degirmenci, U.; Wang, M.; Hu, J. Targeting aberrant RAS/RAF/MEK/ERK signaling for cancer therapy. Cells 2020, 9, 198. [Google Scholar] [CrossRef]
- Stafford, J.M.; Lee, C.H.; Voigt, P.; Descostes, N.; Saldaña-Meyer, R.; Yu, J.R.; Leroy, G.; Oksuz, O.; Chapman, J.R.; Suarez, F.; et al. Multiple modes of prc2 inhibition elicit global chromatin alterations in h3k27m pediatric glioma. Sci. Adv. 2018, 4, eaau5935. [Google Scholar] [CrossRef]
- Kouznetsova, V.L.; Tchekanov, A.; Li, X.; Yan, X.; Tsigelny, I.F. Polycomb repressive 2 complex-molecular mechanisms of function. Protein Sci. 2019, 28, 1387–1399. [Google Scholar] [CrossRef]
- Alohali, S.; Payne, A.E.; Pusztaszeri, M.; Rajab, M.; Forest, V.I.; Hier, M.P.; Tamilia, M.; Payne, R.J. Effect of having concurrent mutations on the degree of aggressiveness in patients with thyroid cancer positive for TERT promoter mutations. Cancers 2023, 15, 413. [Google Scholar] [CrossRef]
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. DNA methylation-based classification of central nervous system tumours. Nature 2018, 555, 469–474. [Google Scholar] [CrossRef]
- Sturm, D.; Orr, B.A.; Toprak, U.H.; Hovestadt, V.; Jones, D.T.W.; Capper, D.; Sill, M.; Buchhalter, I.; Northcott, P.A.; Leis, I.; et al. New brain tumor entities emerge from molecular classification of CNS-PNETS. Cell 2016, 164, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Sahm, F.; Schrimpf, D.; Jones, D.T.; Meyer, J.; Kratz, A.; Reuss, D.; Capper, D.; Koelsche, C.; Korshunov, A.; Wiestler, B.; et al. Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol. 2016, 131, 903–910. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.M.; Shah, R.H.; Pentsova, E.I.; Pourmaleki, M.; Briggs, S.; Distefano, N.; Zheng, Y.; Skakodub, A.; Mehta, S.A.; Campos, C.; et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature 2019, 565, 654–658. [Google Scholar] [CrossRef] [PubMed]
| Tumor Pathology | Tumor Subtype | Grade | Marker | Prognosis (Compared to WT) | Median Progression-Free Survival (Months) | Median Overall Survival (Months) | References |
|---|---|---|---|---|---|---|---|
| Ependymoma | Spinal | 2–3 | WT | Good | 82 | 180 | [7,8,9] |
| Spinal with N-Myc | N-Myc | Worse | 17 | 87 | [10,11] | ||
| Myxopapillary | 2 | HOXB13 | Good—has diagnostic value | 82 | 180 | [12,13] | |
| Subependymoma | 1 | Good | NR | NR | [14] | ||
| Ganglioglioma | 1 | BRAF V600E (rare) | Good, mutation may have therapeutic value | 67 | NR | [15,16] | |
| Astrocytoma | 2–3 | WT | Fair | 96–48 | [17,18,19] | ||
| H3 K27M | Worse | 3–14 | 15 (5–21) | [20,21,22,23] | |||
| TERT | Worse | 7–9 | 14.6–22 | [23,24,25,26,27] | |||
| TP53 | Worse | NR | 11.5–30 | [22,28] | |||
| GALR1 | Unclear, most likely worse | NR | NR | [22,28] | |||
| GRM5 | Unclear, most likely worse | NR | NR | [22,28] | |||
| BRAF V600E | Unknown, may have therapeutic value | NR | NR | [22,28] | |||
| PPM1D | Unclear, most likely worse | NR | NR | [22,28] | |||
| Glioblastoma | 4 | WT | Poor | 6–11 | 13 (10–21) | [29,30,31] | |
| Unknown | NR | NR | |||||
| Hemangioblastoma | 1 | VHL | Slightly worse due to higher recurrence | NR | NR | [32] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Motiei-Langroudi, R. Genetic Markers and Mutations in Primary Spinal Cord Tumors and Their Impact on Clinical Management. Brain Sci. 2025, 15, 1028. https://doi.org/10.3390/brainsci15101028
Motiei-Langroudi R. Genetic Markers and Mutations in Primary Spinal Cord Tumors and Their Impact on Clinical Management. Brain Sciences. 2025; 15(10):1028. https://doi.org/10.3390/brainsci15101028
Chicago/Turabian StyleMotiei-Langroudi, Rouzbeh. 2025. "Genetic Markers and Mutations in Primary Spinal Cord Tumors and Their Impact on Clinical Management" Brain Sciences 15, no. 10: 1028. https://doi.org/10.3390/brainsci15101028
APA StyleMotiei-Langroudi, R. (2025). Genetic Markers and Mutations in Primary Spinal Cord Tumors and Their Impact on Clinical Management. Brain Sciences, 15(10), 1028. https://doi.org/10.3390/brainsci15101028





