Measurement of Functional Brain Network Connectivity in People with Orthostatic Tremor
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
2.2. MRI Data Collection
2.2.1. Sites and Procedures
2.2.2. Scanning Parameters
2.3. MRI Data Processing
2.3.1. Structural Data
2.3.2. Functional Data
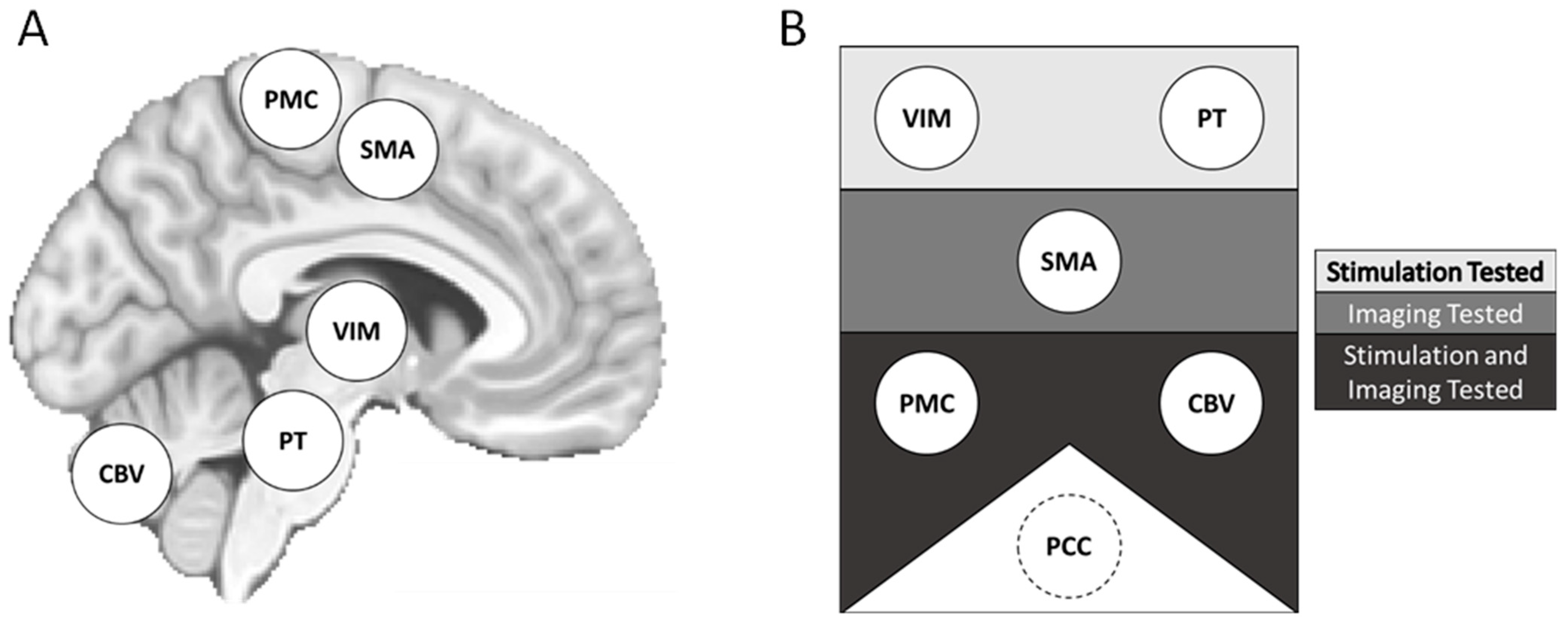
2.4. Transcranial Magnetic Stimulation
2.5. 1000 Functional Connectomes
2.6. Standardizing Datasets
2.7. Resting-State Functional Connectivity: ROI-ROI Analysis
2.8. RSFC: ROI-Whole Brain Analysis
2.9. Statistics
2.10. Multiple Comparisons
3. Results
3.1. Participants
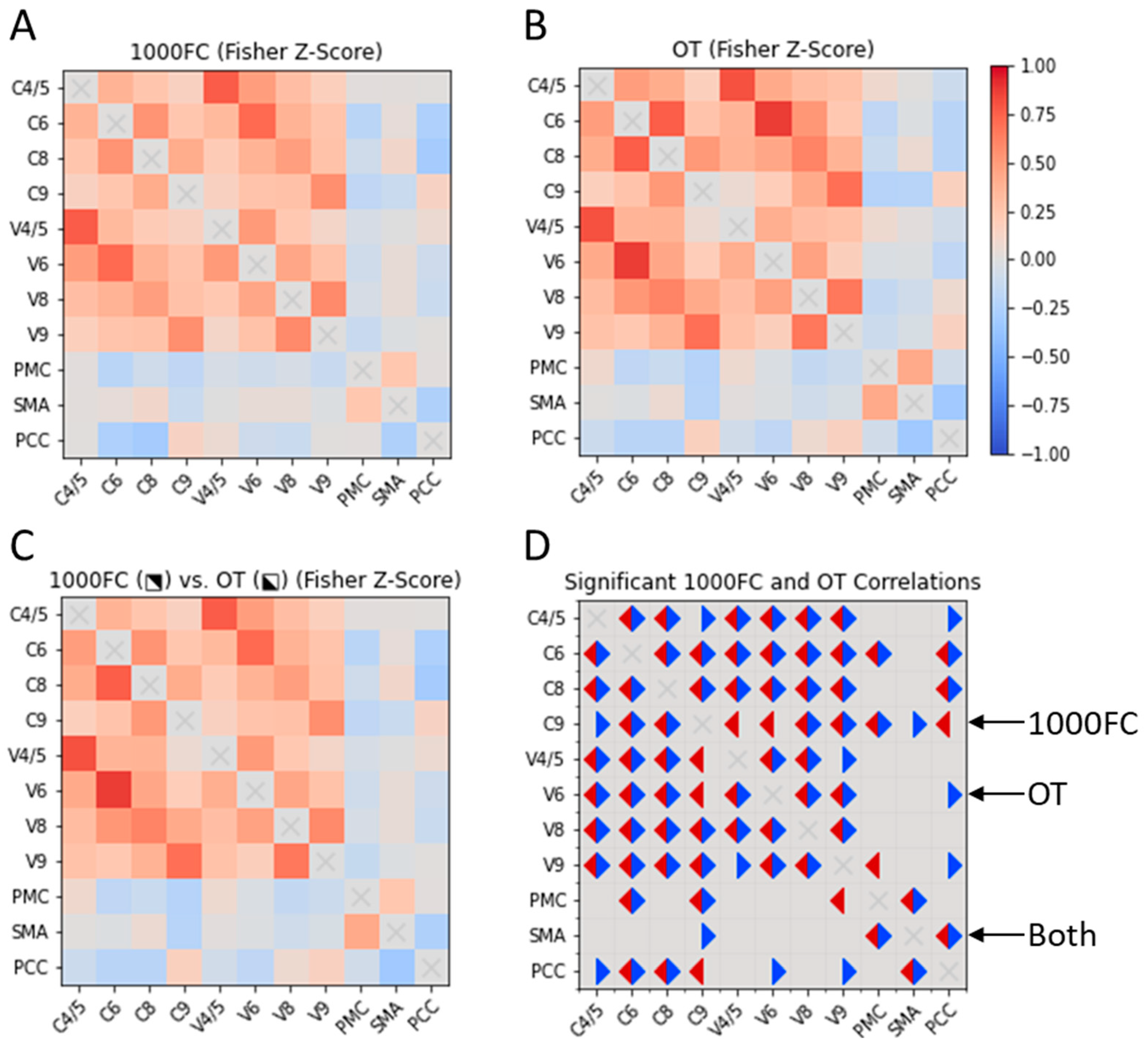
3.2. Correlation of RSFC Patterns between Seed Regions in OT and 1000FC
3.2.1. ROI-ROI Correlations
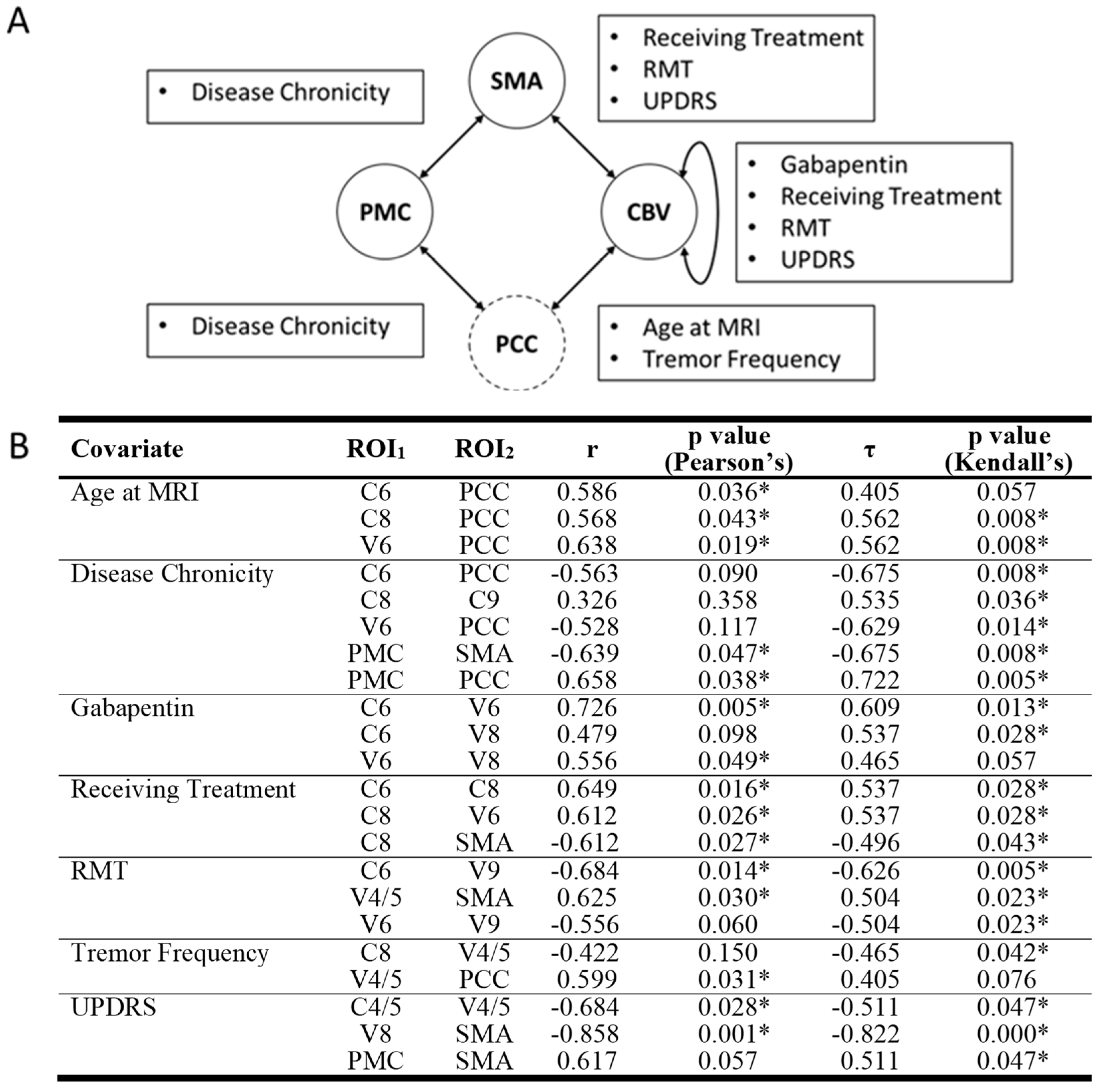
3.2.2. Correlations of ROI Pairs on OT Demographics and Disease Variables
3.3. Whole-Brain Resting-State Functional Connectivity of Putative CON Components
3.3.1. RSFC of the PMC
3.3.2. RSFC of the SMA
3.3.3. RSFC of the Cerebellum
3.3.4. Small-Volume ROIs
RSFC of the PT
RSFC of the VIM
Spatial Similarity with Normative RSFC: PT and VIM
3.3.5. Resting-State Functional Connectivity of a Non-Motor Network (DMN)
4. Discussion
4.1. Central Oscillatory Network
4.2. Functional Connectivity in OT beyond Motor Regions
4.3. Network Integrity
4.4. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhatia, K.P.; Bain, P.; Bajaj, N.; Elble, R.J.; Hallett, M.; Louis, E.D.; Raethjen, J.; Stamelou, M.; Testa, C.M.; Deuschl, G.; et al. Consensus Statement on the classification of tremors. from the task force on tremor of the International Parkinson and Movement Disorder Society. Mov. Disord. 2017, 33, 75–87. [Google Scholar] [CrossRef]
- Bhatti, D.; Thompson, R.; Xia, Y.; Hellman, A.; Schmaderer, L.; Suing, K.; McKune, J.; Penke, C.; Iske, R.; Roeder, B.J.; et al. Comprehensive, blinded assessment of balance in orthostatic tremor. Park. Relat. Disord. 2018, 47, 22–25. [Google Scholar] [CrossRef]
- Benito-León, J.; Domingo-Santos, Á. Orthostatic Tremor: An Update on a Rare Entity. Tremor Other Hyperkinetic Mov. 2016, 6, 411. [Google Scholar] [CrossRef]
- Bhatti, D.E.; Srikanth-Mysore, C.; Bertoni, J.; Torres-Russotto, D. Ataxia is common in individuals with orthostatic tremor. Mov. Disord. 2013, 28, S349. [Google Scholar] [CrossRef]
- Bhatti, D.; Thompson, R.; Malgireddy, K.; Syed, N.; Bayer, B.; Bessette, D.; Fleisher, M.; Murman, D.; Torres-Russotto, D. Anxiety spectrum disorders are common in patients with orthostatic tremor. Clin. Park. Relat. Disord. 2019, 1, 10–12. [Google Scholar] [CrossRef]
- Gallea, C.; Popa, T.; García-Lorenzo, D.; Valabregue, R.; Legrand, A.-P.; Apartis, E.; Marais, L.; Degos, B.; Hubsch, C.; Fernández-Vidal, S.; et al. Orthostatic tremor: A cerebellar pathology? Brain 2016, 139, 2182–2197. [Google Scholar] [CrossRef]
- Norton, J.A.; Wood, D.E.; Day, B.L. Is the spinal cord the generator of 16-Hz orthostatic tremor? Neurology 2004, 62, 632–634. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Ashby, P.; Lang, A. Orthostatic tremor arises from an oscillator in the posterior fossa. Mov. Disord. 2001, 16, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Whitney, D.; Bhatti, D.; Torres-Russotto, D. Orthostatic Tremor: Pathophysiology Guiding Treatment. Curr. Treat. Options Neurol. 2018, 20, 35. [Google Scholar] [CrossRef] [PubMed]
- Muthuraman, M.; Hellriegel, H.; Paschen, S.; Hofschulte, F.; Reese, R.; Volkmann, J.; Witt, K.; Deuschl, G.; Raethjen, J. The central oscillatory network of orthostatic tremor. Mov. Disord. 2013, 28, 1424–1430. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.M.; Fox, P.T.; Miller, K.L.; Glahn, D.C.; Mackay, C.E.; Filippini, N.; Watkins, K.; Toro, R.; Laird, A.; Beckmann, C.F. Correspondence of the brain’s functional architecture during activation and rest. Proc. Natl. Acad. Sci. USA 2009, 106, 13040–13045. [Google Scholar] [CrossRef]
- Sporns, O. From simple graphs to the connectome: Networks in neuroimaging. NeuroImage 2012, 62, 881–886. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Raichle, M.E. Disease and the brain’s dark energy. Nat. Rev. Neurol. 2010, 6, 15–28. [Google Scholar] [CrossRef]
- Raichle, M.E. A brief history of human brain mapping. Trends Neurosci. 2009, 32, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Chiang, H.-L.; Tai, Y.-C.; McMaster, J.; Fung, V.S.; Mahant, N. Primary orthostatic tremor: Is deep brain stimulation better than spinal cord stimulation? J. Neurol. Neurosurg. Psychiatry 2017, 88, 804–805. [Google Scholar] [CrossRef] [PubMed]
- Guridi, J.; Rodriguez-Oroz, M.C.; Arbizu, J.; Alegre, M.; Prieto, E.; Landecho, I.; Manrique, M.; Artieda, J.; Obeso, J.A. Successful thalamic deep brain stimulation for orthostatic tremor. Mov. Disord. 2008, 23, 1808–1811. [Google Scholar] [CrossRef]
- Merola, A.; Fasano, A.; Hassan, A.; Ostrem, J.L.; Contarino, M.F.; Lyons, M.; Krauss, J.K.; Wolf, M.E.; Klassen, B.T.; van Rootselaar, A.; et al. Thalamic deep brain stimulation for orthostatic tremor: A multicenter international registry. Mov. Disord. 2017, 32, 1240–1244. [Google Scholar] [CrossRef]
- Muthuraman, M.; Groppa, S.; Deuschl, G. Cerebello-cortical networks in orthostatic tremor. Brain 2016, 139, 2104–2106. [Google Scholar] [CrossRef]
- Schöberl, F.; Feil, K.; Xiong, G.; Bartenstein, P.; la Fougére, C.; Jahn, K.; Brandt, T.; Strupp, M.; Dieterich, M.; Zwergal, A. Pathological ponto-cerebello-thalamo-cortical activations in primary orthostatic tremor during lying and stance. Brain 2016, 140, 83–97. [Google Scholar] [CrossRef]
- Tsai, C.H.; Semmler, J.G.; Kimber, T.E.; Thickbroom, G.; Stell, R.; Mastaglia, F.L.; Thompson, P.D. Modulation of primary orthostatic tremor by magnetic stimulation over the motor cortex. J. Neurol. Neurosurg. Psychiatry 1998, 64, 33–36. [Google Scholar] [CrossRef]
- Krieg, S.M.; Lioumis, P.; Mäkelä, J.P.; Wilenius, J.; Karhu, J.; Hannula, H.; Savolainen, P.; Lucas, C.W.; Seidel, K.; Laakso, A.; et al. Protocol for motor and language mapping by navigated TMS in patients and healthy volunteers; workshop report. Acta Neurochir. 2017, 159, 1187–1195. [Google Scholar] [CrossRef] [PubMed]
- Casey, B.J.; Cannonier, T.; Conley, M.I.; Cohen, A.O.; Barch, D.M.; Heitzeg, M.M.; Soules, M.E.; Teslovich, T.; Dellarco, D.V.; Garavan, H.; et al. The Adolescent Brain Cognitive Development (ABCD) study: Imaging acquisition across 21 sites. Dev. Cogn. Neurosci. 2018, 32, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Morgan, P.S.; Ashburner, J.; Smith, J.; Rorden, C. The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J. Neurosci. Methods 2016, 264, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Avants, B.B.; Tustison, N.J.; Song, G.; Cook, P.A.; Klein, A.; Gee, J.C. A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage 2011, 54, 2033–2044. [Google Scholar] [CrossRef]
- Reuter, M.; Schmansky, N.J.; Rosas, H.D.; Fischl, B. Within-Subject Template Estimation for Unbiased Longitudinal Image Analysis. NeuroImage 2012, 61, 1402–1418. [Google Scholar] [CrossRef] [PubMed]
- Reuter, M.; Fischl, B. Avoiding Asymmetry-Induced Bias in Longitudinal Image Processing. NeuroImage 2011, 57, 19–21. [Google Scholar] [CrossRef] [PubMed]
- Cox, R.W. AFNI: Software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 1996, 29, 162–173. [Google Scholar] [CrossRef]
- Taylor, P.A.; Chen, G.; Glen, D.R.; Rajendra, J.K.; Reynolds, R.C.; Cox, R.W. FMRI processing with AFNI: Some comments and corrections on “Exploring the Impact of Analysis Software on Task fMRI Results”. bioRxiv 2018, 308643. [Google Scholar] [CrossRef]
- Dale, A.M.; Fischl, B.; Sereno, M.I. Cortical surface-based analysis. I. Segmentation and Surface Reconstruction. NeuroImage 1999, 9, 179–194. [Google Scholar] [CrossRef]
- Dale, A.M.; Sereno, M.I. Improved Localizadon of Cortical Activity by Combining EEG and MEG with MRI Cortical Surface Reconstruction: A Linear Approach. J. Cogn. Neurosci. 1993, 5, 162–176. [Google Scholar] [CrossRef]
- Reuter, M.; Rosas, H.D.; Fischl, B. Highly accurate inverse consistent registration: A Robust Approach. NeuroImage 2010, 53, 1181–1196. [Google Scholar] [CrossRef] [PubMed]
- Power, J.D.; Mitra, A.; Laumann, T.O.; Snyder, A.Z.; Schlaggar, B.L.; Petersen, S.E. Methods to detect, characterize, and remove motion artifact in resting state fMRI. NeuroImage 2014, 84, 320–341. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, J.L.; Woldorff, M.G.; Parsons, L.M.; Liotti, M.; Freitas, C.S.; Rainey, L.; Kochunov, P.V.; Nickerson, D.; Mikiten, S.A.; Fox, P.T. Automated Talairach Atlas labels for functional brain mapping. Hum. Brain Mapp. 2000, 10, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Maldjian, J.A.; Laurienti, P.J.; Kraft, R.A.; Burdette, J.H. An automated method for neuroanatomic and cytoarchitectonic atlas-based interrogation of fMRI data sets. NeuroImage 2003, 19, 1233–1239. [Google Scholar] [CrossRef] [PubMed]
- Nexstim NBS System User Manual 2016. Available online: http://www.neurosurgeryresident.net/Op.%20Operative%20Techniques/00.%20Catalogs,%20Brochures,%20Manuals/Nexstim/NX81491%20NBS%20System%20User%20Manual%204.3_USA.pdf (accessed on 10 June 2020).
- Biswal, B.B.; Mennes, M.; Zuo, X.-N.; Gohel, S.; Kelly, C.; Smith, S.M.; Beckmann, C.F.; Adelstein, J.S.; Buckner, R.L.; Colcombe, S.; et al. Toward discovery science of human brain function. Proc. Natl. Acad. Sci. USA 2010, 107, 4734–4739. [Google Scholar] [CrossRef] [PubMed]
- Buckner, R. International Neuroimaging Data-Sharing Initiative n.d. Available online: http://fcon_1000.projects.nitrc.org/ (accessed on 26 June 2023).
- Taylor, P.A.; Saad, Z.S. FATCAT: (An Efficient) Functional and Tractographic Connectivity Analysis Toolbox. Brain Connect. 2013, 3, 523–535. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Benito-León, J.; Louis, E.D.; Manzanedo, E.; Hernández-Tamames, J.A.; Álvarez-Linera, J.; Molina-Arjona, J.A.; Matarazzo, M.; Romero, J.P.; Domínguez-González, C.; Domingo-Santos, Á.; et al. Resting state functional MRI reveals abnormal network connectivity in orthostatic tremor. Medicine 2016, 95, e4310. [Google Scholar] [CrossRef]
- Yamada, K.; Akazawa, K.; Yuen, S.; Goto, M.; Matsushima, S.; Takahata, A.; Nakagawa, M.; Mineura, K.; Nishimura, T. MR Imaging of Ventral Thalamic Nuclei. Am. J. Neuroradiol. 2009, 31, 732–735. [Google Scholar] [CrossRef]
- Roux, F.; Djidjeli, I.; Durand, J. Functional architecture of the somatosensory homunculus detected by electrostimulation. J. Physiol. 2018, 596, 941–956. [Google Scholar] [CrossRef]
- Andrews-Hanna, J.R.; Reidler, J.S.; Sepulcre, J.; Poulin, R.; Buckner, R.L. Functional-Anatomic Fractionation of the Brain’s Default Network. Neuron 2010, 65, 550–562. [Google Scholar] [CrossRef]
- Cox, R.W.; Chen, G.; Glen, D.R.; Reynolds, R.C.; Taylor, P.A. FMRI Clustering in AFNI: False-Positive Rates Redux. Brain Connect. 2017, 7, 152–171. [Google Scholar] [CrossRef] [PubMed]
- Cox, R.W.; Chen, G.; Glen, D.R.; Reynolds, R.C.; Taylor, P.A. fMRI clustering and false-positive rates. Proc. Natl. Acad. Sci. USA 2017, 114, E3370–E3371. [Google Scholar] [CrossRef]
- Eklund, A.; Nichols, T.E.; Knutsson, H. Cluster failure: Why fMRI inferences for spatial extent have inflated false-positive rates. Proc. Natl. Acad. Sci. USA 2016, 113, 7900–7905. [Google Scholar] [CrossRef] [PubMed]
- Altman, D.G.; Bland, J.M. Statistics notes: The normal distribution. BMJ 1995, 310, 298. [Google Scholar] [CrossRef] [PubMed]
- Krithikadatta, J. Normal distribution. J. Conserv. Dent. JCD 2014, 17, 96–97. [Google Scholar] [CrossRef] [PubMed]
- Ganos, C.; Maugest, L.; Apartis, E.; Gasca-Salas, C.; Cáceres-Redondo, M.T.; Erro, R.; Navalpotro-Gómez, I.; Batla, A.; Antelmi, E.; Degos, B.; et al. The long-term outcome of orthostatic tremor. J. Neurol. Neurosurg. Psychiatry 2015, 87, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.; Ahlskog, J.E.; Matsumoto, J.Y.; Milber, J.M.; Bower, J.H.; Wilkinson, J.R. Orthostatic tremor: Clinical, electrophysiologic, and treatment findings in 184 patients. Neurology 2016, 86, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Benito-León, J.; Louis, E.D.; Puertas-Martín, V.; Romero, J.P.; Matarazzo, M.; Molina-Arjona, J.A.; Domínguez-González, C.; Sánchez-Ferro, Á. Cognitive and neuropsychiatric features of orthostatic tremor: A case–control comparison. J. Neurol. Sci. 2015, 361, 137–143. [Google Scholar] [CrossRef]
- Bressler, S.L.; Menon, V. Large-scale brain networks in cognition: Emerging methods and principles. Trends Cogn. Sci. 2010, 14, 277–290. [Google Scholar] [CrossRef]
- Thomas Yeo, B.T.; Krienen, F.M.; Sepulcre, J.; Sabuncu, M.R.; Lashkari, D.; Hollinshead, M.; Roffman, J.L.; Smoller, J.W.; Zöllei, L.; Polimeni, J.R.; et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol. 2011, 106, 1125–1165. [Google Scholar] [CrossRef]
- Katzenschlager, R.; Costa, D.; Gerschlager, W.; O’Sullivan, J.; Zijlmans, J.; Gacinovic, S.; Pirker, W.; Wills, A.; Bhatia, K.; Lees, A.J.; et al. [123I]-FP-CIT-SPECT demonstrates dopaminergic deficit in orthostatic tremor. Ann. Neurol. 2003, 53, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Merola, A.; Torres-Russotto, D.R.; Stebbins, G.T.; Vizcarra, J.A.; Shukla, A.W.; Hassan, A.; Marsili, L.; Krauss, J.K.; Elble, R.J.; Deuschl, G.; et al. Development and Validation of the Orthostatic Tremor Severity and Disability Scale (OT-10). Mov. Disord. 2020, 35, 1796–1801. [Google Scholar] [CrossRef] [PubMed]
| Age at MRI | Sex | Age of Onset | Age at Dx | Tr. Freq. (Hz) | RMT | Tx | Family History | UPDRS |
|---|---|---|---|---|---|---|---|---|
| 70 | F | 67 | 70 | 12 | 32 | G | No | 23 |
| 78 | F | 75 | 75 | 13 | * | — | No | 27 |
| 68 | F | 60 | 64 | 14 | 25 | G | No | * |
| 76 | F | 55 | 59 | 14 | 52 | — | Yes | 8 |
| 61 | F | 46 | 61 | 15 | 33 | G | No | 7 |
| 60 | F | 45 | 52 | 15 | 43 | C | No | 19 |
| 78 | F | 67 | 70 | 15 | 33 | — | No | 4 |
| 70 | F | 64 | 66 | 14 | 28 | C | No | 16 |
| 41 | M | 40 | 40 | 14 | 51 | G + C | No | 14 |
| 79 | F | 49 | 52 | 16 | 54 | G + C | No | 10 |
| 62 | F | 47 | 47 | 15 | 60 | C | No | * |
| 76 | F | 57 | 58 | 14 | 48 | C | No | 28 |
| 67 | F | 30 | 30 | 14 | 10 | C | Yes | * |
| Source Coordinates (MNI) | ||||
|---|---|---|---|---|
| ROI | Source | X | Y | Z |
| Pontine Tegmentum | Schöberl et al., 2017 [19] | +5.0 | −38.0 | −42.0 |
| Ventral intermediate Nucleus (Thalamus) | Yamada et al., 2010 [41] | +/−11.5 | −15.5 | −1.0 |
| Thumb | Roux et al., 2018 [42] | +46.6 | −22.8 | +56.2 |
| Supplementary Motor Area | Gallea et al., 2016 [6] | −9.0 | +8.0 | +56.0 |
| Posterior Cingulate Cortex | Andrews-Hanna et al., 2010 [43] | −8.0 | −56.0 | +26.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phipps, C.J.; Whitney, D.; Shou, J.; Torres-Russotto, D.; Warren, D.E. Measurement of Functional Brain Network Connectivity in People with Orthostatic Tremor. Brain Sci. 2024, 14, 219. https://doi.org/10.3390/brainsci14030219
Phipps CJ, Whitney D, Shou J, Torres-Russotto D, Warren DE. Measurement of Functional Brain Network Connectivity in People with Orthostatic Tremor. Brain Sciences. 2024; 14(3):219. https://doi.org/10.3390/brainsci14030219
Chicago/Turabian StylePhipps, Connor J., David Whitney, James Shou, Diego Torres-Russotto, and David E. Warren. 2024. "Measurement of Functional Brain Network Connectivity in People with Orthostatic Tremor" Brain Sciences 14, no. 3: 219. https://doi.org/10.3390/brainsci14030219
APA StylePhipps, C. J., Whitney, D., Shou, J., Torres-Russotto, D., & Warren, D. E. (2024). Measurement of Functional Brain Network Connectivity in People with Orthostatic Tremor. Brain Sciences, 14(3), 219. https://doi.org/10.3390/brainsci14030219






