Subject-Independent EEG Emotion Recognition Based on Genetically Optimized Projection Dictionary Pair Learning
Abstract
1. Introduction
- In recognition of the variability in EEG-based emotion recognition among individuals, we applied the PDPL algorithm to perform cross-subject analysis, with a specific focus on feature selection.
- The exploration of parameter space in the PDPL algorithm presents a substantial computational burden due to the wide range of parameter adjustments and the resulting extensive combinations. To address this challenge, we propose the utilization of the Genetic Algorithm (GA) for adaptive parameter optimization.
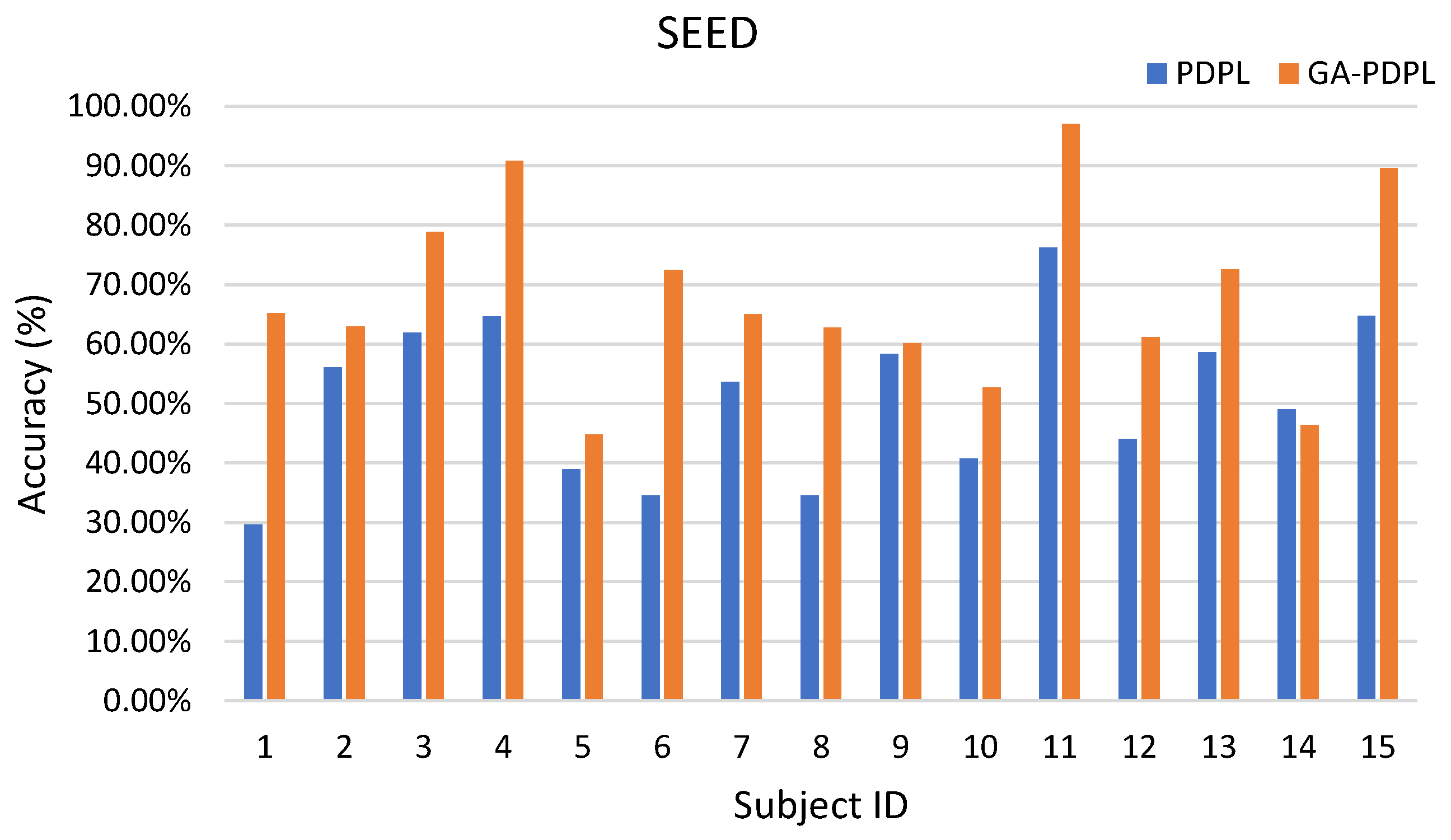
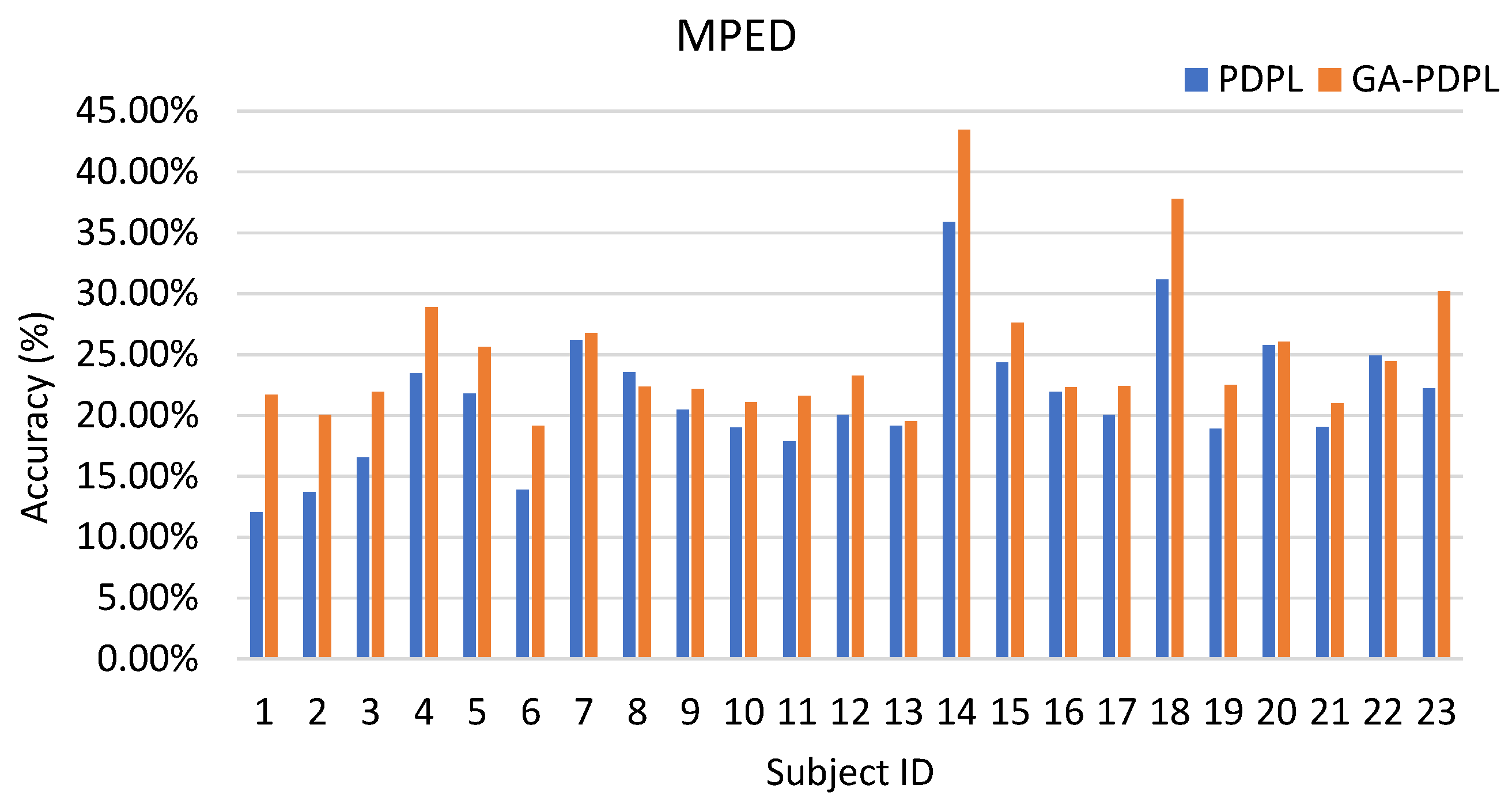
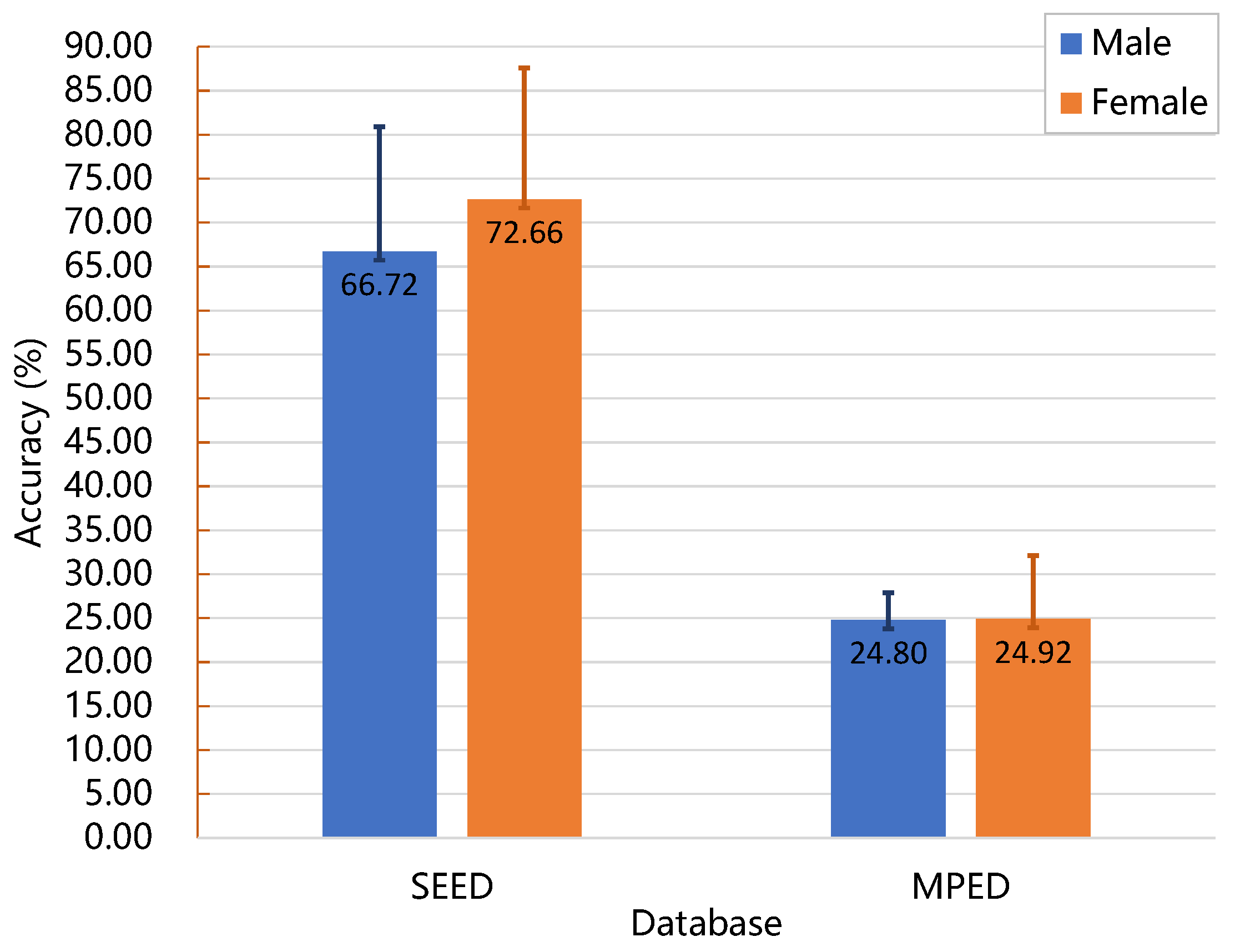
- Our proposed method surpasses conventional machine learning approaches, demonstrating exceptional recognition performance. Specifically, it achieves an average accuracy of 69.89% on the SEED database, 24.11% on the MPED database, 64.34% for the two-class GAMEEMO dataset, and 49.01% for the four-class GAMEEMO dataset. These results shed light on the effectiveness of emotion recognition, particularly for females, providing valuable insights into their emotional susceptibility.
2. Materials and Methods
2.1. EEG Emotion Database
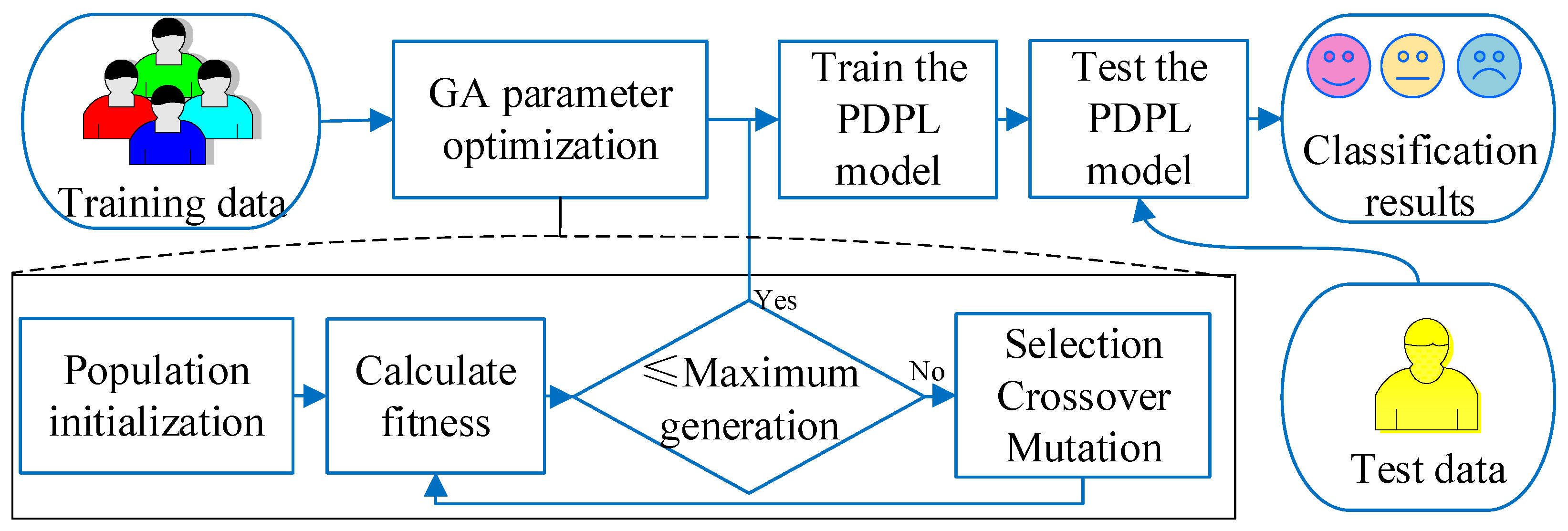
2.2. GA-PDPL for EEG Emotion Recognition
2.2.1. Descriminative Dictionary Learning (DDL)
2.2.2. PDPL Model
- (1)
- Fix and , update ,
- (2)
- Fix , update and ,
2.2.3. The GA for the Parameter Optimization of the PDPL
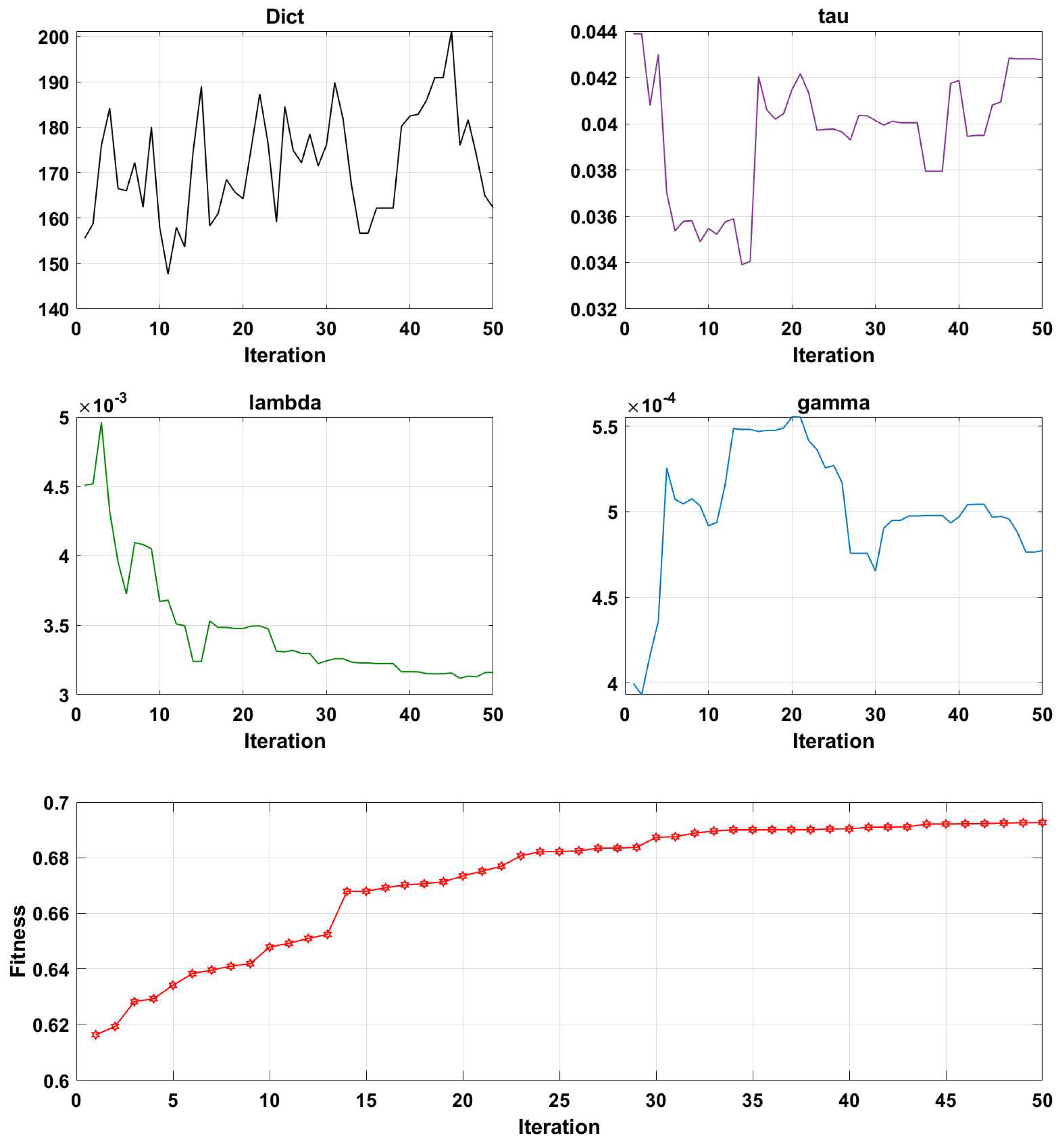
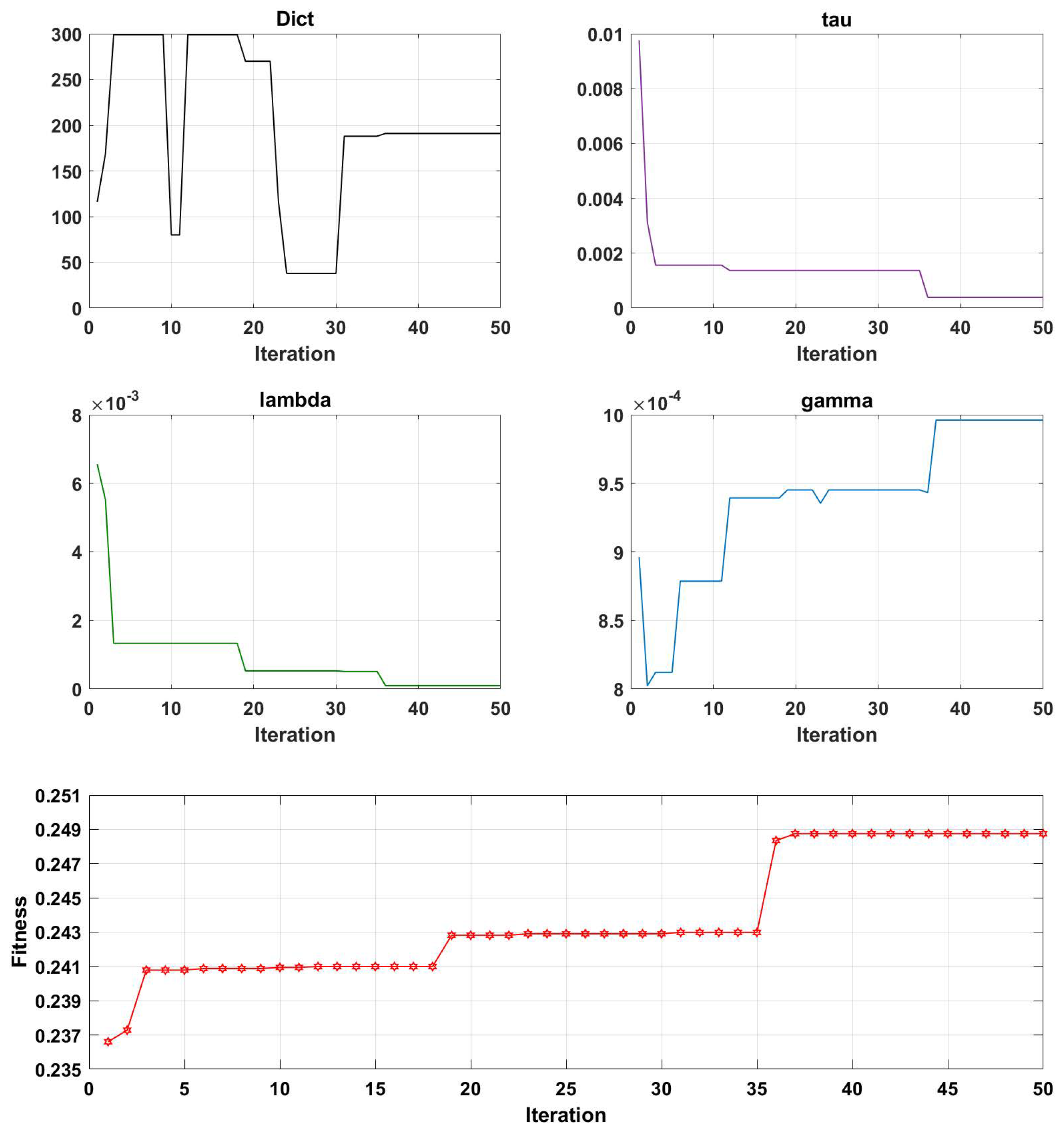
- Initialization: An initial population of solutions is generated by randomly assigning values to the parameters. The initialization parameters, which include the maximum genetic algebra, population size, crossover function, mutation probability, and t parameter of PDPL, are presented in Table 2. Furthermore, the GA optimization process involves tuning four PDPL parameters: m, , , and . The threshold ranges and coding methods for these parameters are provided in Table 3.
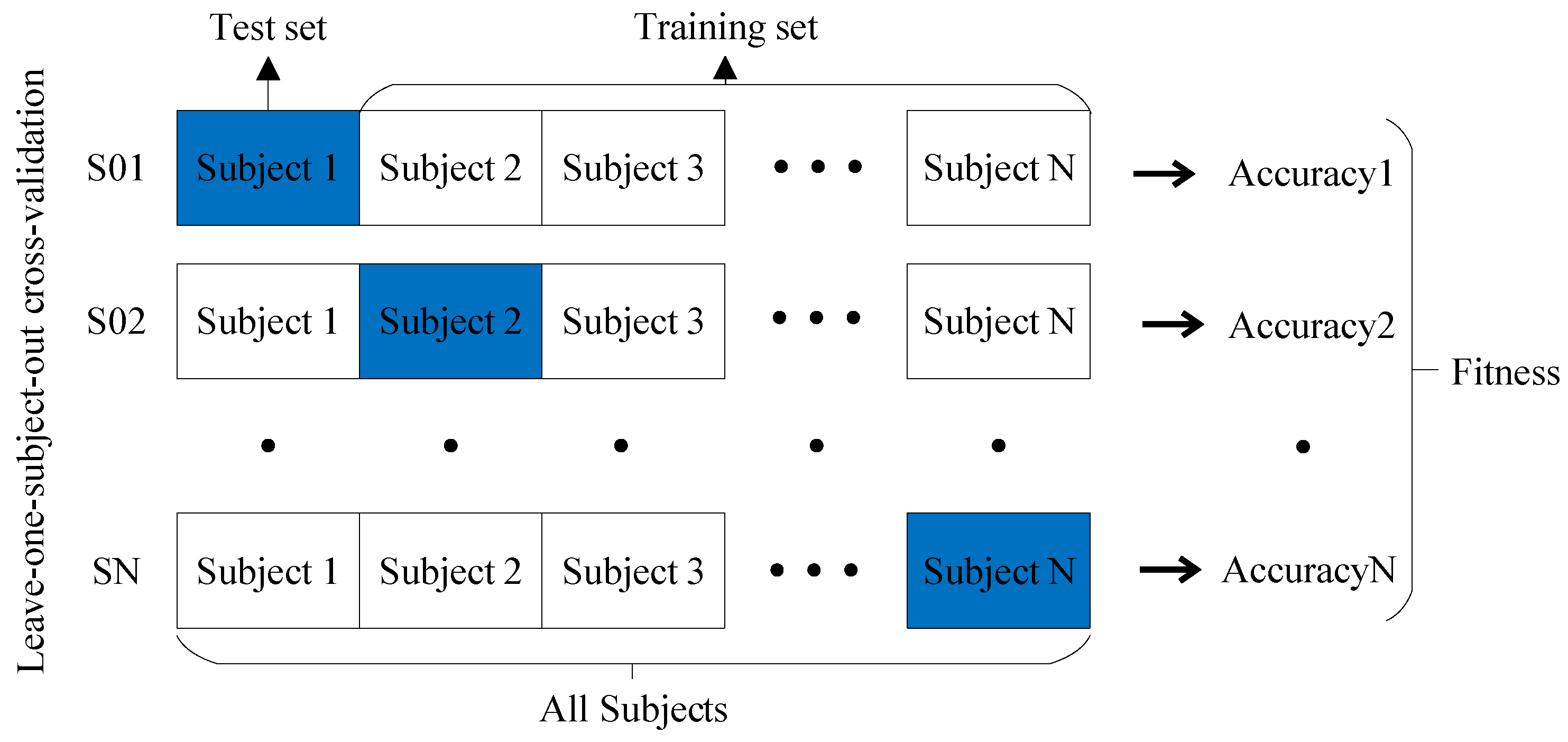
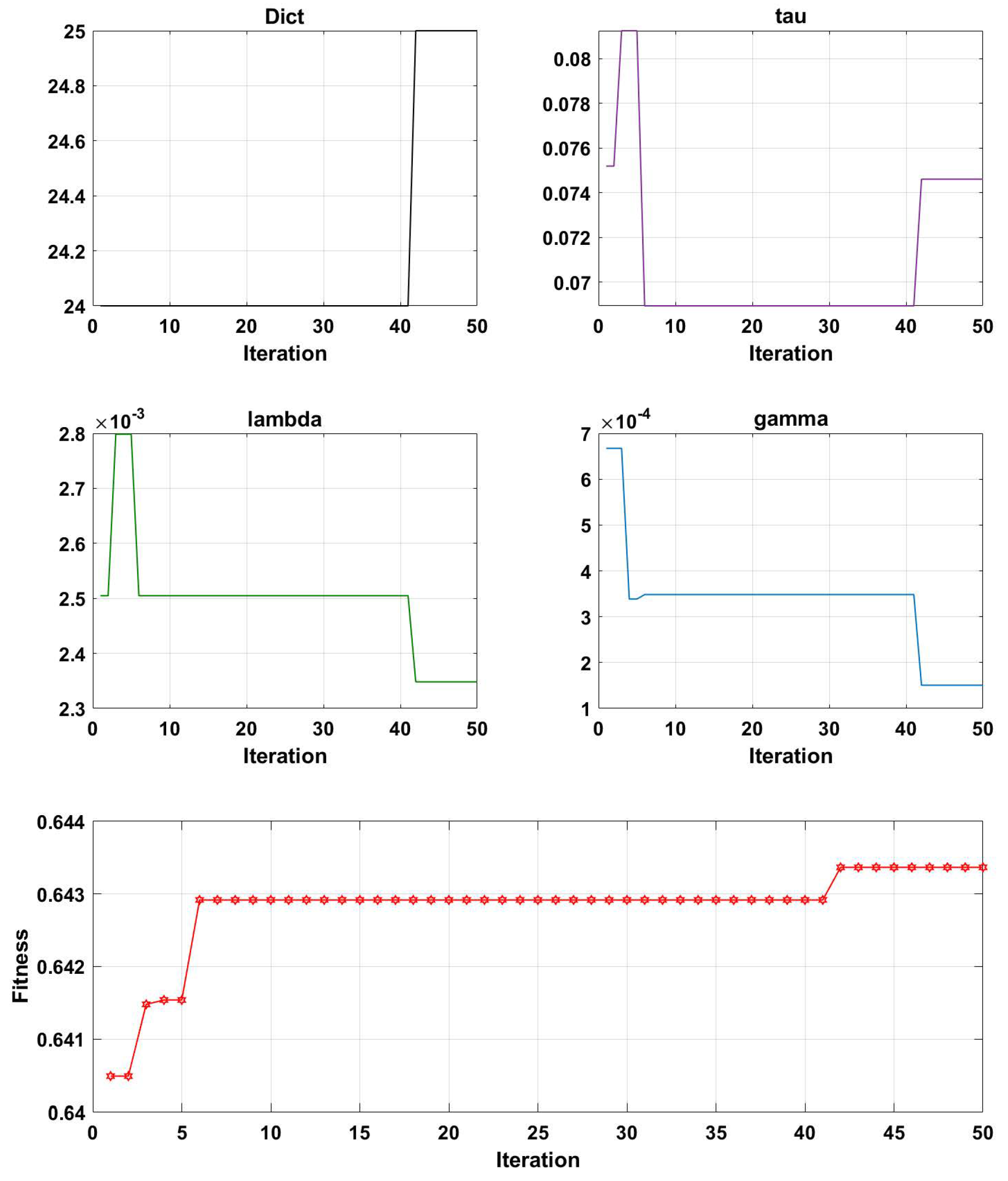
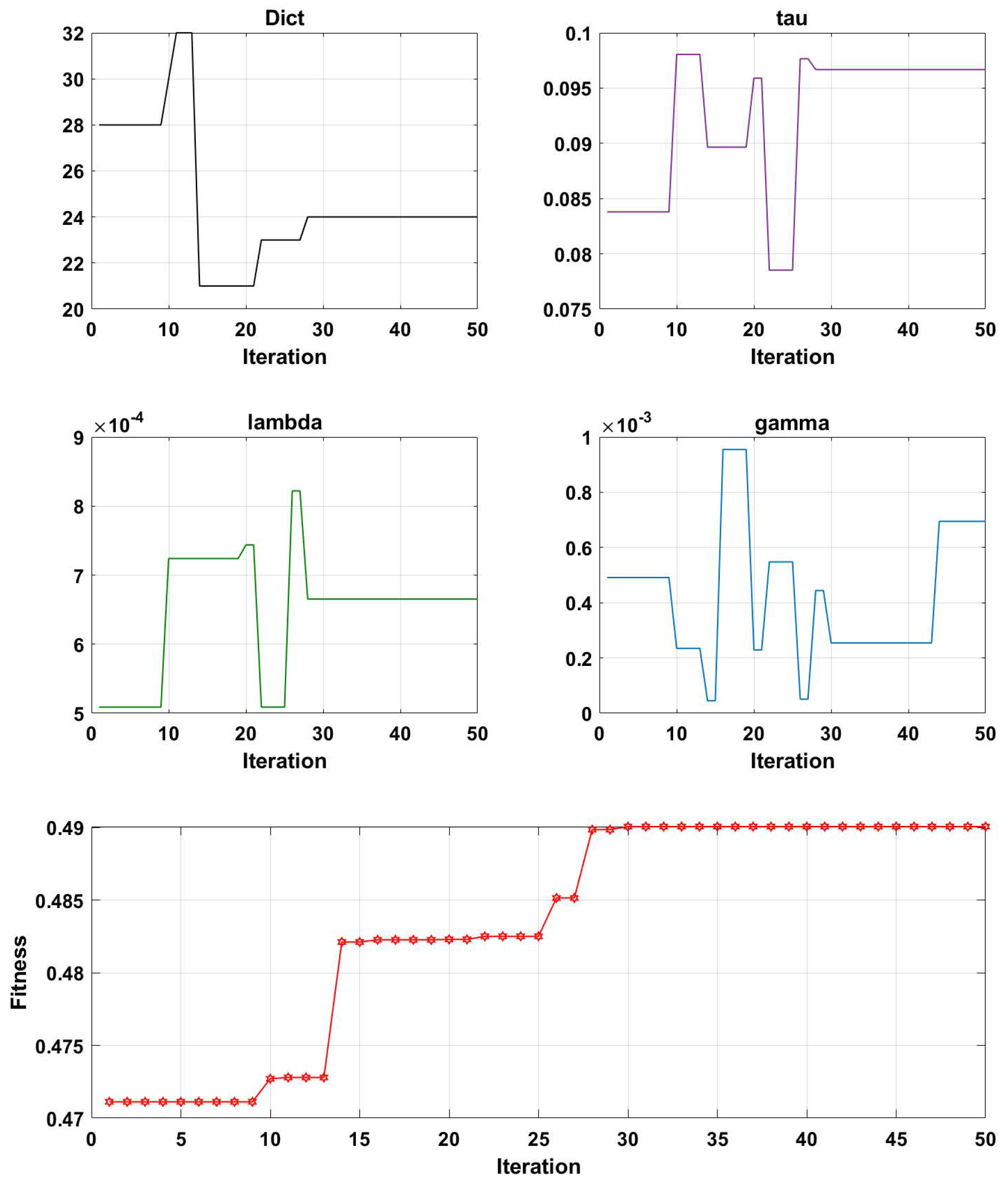
- Evaluation: The fitness of each solution in the population is assessed using the projection dictionary pair learning (PDPL) algorithm and a fitness function. In this study, we defined the fitness function as the accuracy of the PDPL recognition on the test set. The calculation method for the fitness is illustrated in Figure 2 and can be found in Formula (14). To incorporate the research background, which was unrelated to the subjects, we introduced a leave-one-out subject cross test into the fitness calculation. The final fitness value was determined by averaging the accuracy across all subjects.
- Selection: Choose a subset of solutions to serve as parents for the next generation based on their fitness scores.
- Crossover: Generate new solutions by combining the parameters of the selected parents through crossover.
- Mutation: Introduce random changes to the parameters of some solutions to explore different areas of the search space.
- Evaluation: Assess the fitness of the newly created solutions resulting from crossover and mutation.
- Replacement: Select the top-performing solutions from both the previous and new generations to form the subsequent generation.
- Termination: Stop the algorithm when a specified termination criterion is met, such as reaching the maximum number of generations or achieving the desired level of fitness.
- Output: Provide the best solution obtained by the genetic algorithm (GA), which corresponds to the optimal parameter values for the projection dictionary pair learning algorithm.
3. Results and Discussion
3.1. Recognition Results of the GA-PDPL Method on Three Databases
3.2. Parameter Optimization Analysis of the GA-PDPL Method
3.3. Emotion Recognition Performance of the GA-PDPL Method with Regard to Sex
3.4. Training and Testing Time of the GA-PDPL
3.5. Comparison of the GA-PDPL Method and SOTA Method
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xue, Y.; Zheng, W.; Zong, Y.; Chang, H.; Jiang, X. Adaptive Hierarchical Graph Convolutional Network for EEG Emotion Recognition. In Proceedings of the 2022 International Joint Conference on Neural Networks (IJCNN), Padua, Italy, 18–23 July 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 1–8. [Google Scholar]
- Chang, H.; Zong, Y.; Zheng, W.; Tang, C.; Zhu, J.; Li, X. Depression Assessment Method: An EEG Emotion Recognition Framework Based on Spatiotemporal Neural Network. Front. Psychiatry 2022, 12, 2620. [Google Scholar] [CrossRef] [PubMed]
- Al-Ezzi, A.; Kamel, N.; Faye, I.; Gunaseli, E. Review of EEG, ERP, and brain connectivity estimators as predictive biomarkers of social anxiety disorder. Front. Psychol. 2020, 11, 730. [Google Scholar] [CrossRef] [PubMed]
- Meyer, T.; Smeets, T.; Giesbrecht, T.; Quaedflieg, C.W.; Smulders, F.T.; Meijer, E.H.; Merckelbach, H.L. The role of frontal EEG asymmetry in post-traumatic stress disorder. Biol. Psychol. 2015, 108, 62–77. [Google Scholar] [CrossRef]
- Liu, B.; Chang, H.; Peng, K.; Wang, X. An End-to-End Depression Recognition Method Based on EEGNet. Front. Psychiatry 2022, 13, 864393. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Chang, H.; Zhou, X.; Wang, J.; Wang, L.; Shao, Y. E2ENNet: An end-to-end neural network for emotional brain-computer interface. Front. Comput. Neurosci. 2022, 16, 942979. [Google Scholar] [CrossRef]
- Li, X.; Zheng, W.; Zong, Y.; Chang, H.; Lu, C. Attention-based Spatio-Temporal graphic LSTM for EEG emotion recognition. In Proceedings of the 2021 International Joint Conference on Neural Networks (IJCNN), Shenzhen, China, 18–22 July 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 1–8. [Google Scholar]
- Abdulrahman, A.; Baykara, M. A Comprehensive Review for Emotion Detection Based on EEG Signals: Challenges, Applications, and Open Issues. Trait. Signal 2021, 38, 1189–1200. [Google Scholar] [CrossRef]
- Murugappan, M.; Rizon, M.; Nagarajan, R.; Yaacob, S.; Hazry, D.; Zunaidi, I. Time-frequency analysis of EEG signals for human emotion detection. In Proceedings of the 4th Kuala Lumpur International Conference on Biomedical Engineering 2008: BIOMED 2008, Kuala Lumpur, Malaysia, 25–28 June 2008; Springer: Berlin/Heidelberg, Germany, 2008; pp. 262–265. [Google Scholar]
- Murugappan, M.; Murugappan, S. Human emotion recognition through short time Electroencephalogram (EEG) signals using Fast Fourier Transform (FFT). In Proceedings of the 2013 IEEE 9th International Colloquium on Signal Processing and Its Applications, Kuala Lumpur, Malaysia, 8–10 March 2013; IEEE: Piscataway, NJ, USA, 2013; pp. 289–294. [Google Scholar]
- Dongwei, C.; Fang, W.; Zhen, W.; Haifang, L.; Junjie, C. EEG-based emotion recognition with brain network using independent components analysis and granger causality. In Proceedings of the 2013 International Conference on Computer Medical Applications (ICCMA), Sousse, Tunisia, 20–22 January 2013; IEEE: Piscataway, NJ, USA, 2013; pp. 1–6. [Google Scholar]
- Wang, X.W.; Nie, D.; Lu, B.L. EEG-based emotion recognition using frequency domain features and support vector machines. In Proceedings of the Neural Information Processing: 18th International Conference, ICONIP 2011, Shanghai, China, 13–17 November 2011; Springer: Berlin/Heidelberg, Germany, 2011. Part I 18. pp. 734–743. [Google Scholar]
- Matiko, J.W.; Beeby, S.P.; Tudor, J. Fuzzy logic based emotion classification. In Proceedings of the 2014 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Florence, Italy, 4–9 May 2014; IEEE: Piscataway, NJ, USA, 2014; pp. 4389–4393. [Google Scholar]
- Fu, R.; Wang, H.; Zhao, W. Dynamic driver fatigue detection using hidden Markov model in real driving condition. Expert Syst. Appl. 2016, 63, 397–411. [Google Scholar] [CrossRef]
- Zhang, J.; Yin, Z.; Chen, P.; Nichele, S. Emotion recognition using multi-modal data and machine learning techniques: A tutorial and review. Inf. Fusion 2020, 59, 103–126. [Google Scholar] [CrossRef]
- Song, T.; Zheng, W.; Lu, C.; Zong, Y.; Zhang, X.; Cui, Z. MPED: A multi-modal physiological emotion database for discrete emotion recognition. IEEE Access 2019, 7, 12177–12191. [Google Scholar] [CrossRef]
- Bazgir, O.; Mohammadi, Z.; Habibi, S.A.H. Emotion recognition with machine learning using EEG signals. In Proceedings of the 2018 25th National and 3rd International Iranian Conference on Biomedical Engineering (ICBME), Qom, Iran, 29–30 November 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 1–5. [Google Scholar]
- Islam, M.R.; Moni, M.A.; Islam, M.M.; Rashed-Al-Mahfuz, M.; Islam, M.S.; Hasan, M.K.; Hossain, M.S.; Ahmad, M.; Uddin, S.; Azad, A.; et al. Emotion recognition from EEG signal focusing on deep learning and shallow learning techniques. IEEE Access 2021, 9, 94601–94624. [Google Scholar] [CrossRef]
- Zhang, J.; Li, C.; Kosov, S.; Grzegorzek, M.; Shirahama, K.; Jiang, T.; Sun, C.; Li, Z.; Li, H. LCU-Net: A novel low-cost U-Net for environmental microorganism image segmentation. Pattern Recognit. 2021, 115, 107885. [Google Scholar] [CrossRef]
- Abdulrahman, A.; Baykara, M.; Alakus, T.B. A Novel Approach for Emotion Recognition Based on EEG Signal Using Deep Learning. Appl. Sci. 2022, 12, 10028. [Google Scholar] [CrossRef]
- Abdulrahman, A.; Baykara, M. Feature extraction approach based on statistical methods and wavelet packet decomposition for emotion recognition using EEG signals. In Proceedings of the 2021 International Conference on INnovations in Intelligent SysTems and Applications (INISTA), Kocaeli, Turkey, 25–27 August 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 1–7. [Google Scholar]
- Li, J.; Qiu, S.; Shen, Y.Y.; Liu, C.L.; He, H. Multisource transfer learning for cross-subject EEG emotion recognition. IEEE Trans. Cybern. 2019, 50, 3281–3293. [Google Scholar] [CrossRef]
- Li, W.; Huan, W.; Hou, B.; Tian, Y.; Zhang, Z.; Song, A. Can emotion be transferred?—A review on transfer learning for EEG-Based Emotion Recognition. IEEE Trans. Cogn. Dev. Syst. 2021, 14, 833–846. [Google Scholar] [CrossRef]
- Das, A.; Mondal, P.; Pal, U.; Ferrer, M.A.; Blumenstein, M. Fast and efficent multimodal eye biometrics using projective dictionary pair learning. In Proceedings of the 2016 IEEE Congress on Evolutionary Computation (CEC), Vancouver, BC, Canada, 24–29 July 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 1402–1408. [Google Scholar]
- Ameri, R.; Alameer, A.; Ferdowsi, S.; Nazarpour, K.; Abolghasemi, V. Labeled projective dictionary pair learning: Application to handwritten numbers recognition. Inf. Sci. 2022, 609, 489–506. [Google Scholar] [CrossRef]
- Cai, W.; Gao, M.; Jiang, Y.; Gu, X.; Ning, X.; Qian, P.; Ni, T. Hierarchical domain adaptation projective dictionary pair learning model for EEG classification in IoMT systems. IEEE Trans. Comput. Soc. Syst. 2022. [Google Scholar] [CrossRef]
- Gu, S.; Zhang, L.; Zuo, W.; Feng, X. Projective dictionary pair learning for pattern classification. Adv. Neural Inf. Process. Syst. 2014, 27, 1–9. [Google Scholar]
- Sehgal, A.; La, H.; Louis, S.; Nguyen, H. Deep reinforcement learning using genetic algorithm for parameter optimization. In Proceedings of the 2019 Third IEEE International Conference on Robotic Computing (IRC), Naples, Italy, 25–27 February 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 596–601. [Google Scholar]
- Dilip, D.G.; Panda, S.; Mathew, J. Characterization and parametric optimization of micro-hole surfaces in micro-EDM drilling on Inconel 718 superalloy using genetic algorithm. Arab. J. Sci. Eng. 2020, 45, 5057–5074. [Google Scholar] [CrossRef]
- Syarif, I.; Prugel-Bennett, A.; Wills, G. SVM parameter optimization using grid search and genetic algorithm to improve classification performance. TELKOMNIKA (Telecommun. Comput. Electron. Control) 2016, 14, 1502–1509. [Google Scholar] [CrossRef]
- Holland, J.H. Genetic algorithms. Sci. Am. 1992, 267, 66–73. [Google Scholar] [CrossRef]
- Katoch, S.; Chauhan, S.S.; Kumar, V. A review on genetic algorithm: Past, present, and future. Multimed. Tools Appl. 2021, 80, 8091–8126. [Google Scholar] [CrossRef] [PubMed]
- Duan, R.N.; Zhu, J.Y.; Lu, B.L. Differential entropy feature for EEG-based emotion classification. In Proceedings of the 2013 6th International IEEE/EMBS Conference on Neural Engineering (NER), San Diego, CA, USA, 6–8 November 2013; IEEE: Piscataway, NJ, USA, 2013; pp. 81–84. [Google Scholar]
- Alakus, T.B.; Gonen, M.; Turkoglu, I. Database for an emotion recognition system based on EEG signals and various computer games–GAMEEMO. Biomed. Signal Process. Control 2020, 60, 101951. [Google Scholar] [CrossRef]
- Sani, O.G.; Yang, Y.; Lee, M.B.; Dawes, H.E.; Chang, E.F.; Shanechi, M.M. Mood variations decoded from multi-site intracranial human brain activity. Nat. Biotechnol. 2018, 36, 954–961. [Google Scholar] [CrossRef]
- Deng, Y.; Chang, L.; Yang, M.; Huo, M.; Zhou, R. Gender differences in emotional response: Inconsistency between experience and expressivity. PLoS ONE 2016, 11, e0158666. [Google Scholar] [CrossRef]
- Goshvarpour, A.; Goshvarpour, A. EEG spectral powers and source localization in depressing, sad, and fun music videos focusing on gender differences. Cogn. Neurodyn. 2019, 13, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Kensinger, E.A. Remembering emotional experiences: The contribution of valence and arousal. Rev. Neurosci. 2004, 15, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.Q.; Qiu, J.L.; Tang, H.; Zheng, W.L.; Lu, B.L. Investigating sex differences in classification of five emotions from EEG and eye movement signals. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 6746–6749. [Google Scholar]
- Sugiyama, M.; Nakajima, S.; Kashima, H.; Von Buenau, P.; Kawanabe, M. Direct importance estimation with model selection and its application to covariate shift adaptation. In Advances in Neural Information Processing Systems 20 (NIPS 2007); Citeseer: Princeton, NJ, USA, 2007; Volume 7, pp. 1433–1440. [Google Scholar]
- Kanamori, T.; Hido, S.; Sugiyama, M. A least-squares approach to direct importance estimation. J. Mach. Learn. Res. 2009, 10, 1391–1445. [Google Scholar]
- Chu, W.S.; De la Torre, F.; Cohn, J.F. Selective transfer machine for personalized facial expression analysis. IEEE Trans. Pattern Anal. Mach. Intell. 2016, 39, 529–545. [Google Scholar] [CrossRef]
- Suykens, J.A.; Vandewalle, J. Least squares support vector machine classifiers. Neural Process. Lett. 1999, 9, 293–300. [Google Scholar] [CrossRef]
- Schölkopf, B.; Smola, A.; Müller, K.R. Nonlinear component analysis as a kernel eigenvalue problem. Neural Comput. 1998, 10, 1299–1319. [Google Scholar] [CrossRef]
- Pan, S.J.; Tsang, I.W.; Kwok, J.T.; Yang, Q. Domain adaptation via transfer component analysis. IEEE Trans. Neural Netw. 2010, 22, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Han, J.; Chen, Y.; Sha, X.; Wang, Z.; Hu, B.; Yang, J.; Feng, L.; Ding, Z.; Chen, Y.; et al. A pervasive approach to EEG-based depression detection. Complexity 2018, 2018, 5238028. [Google Scholar] [CrossRef]
- Rigatti, S.J. Random forest. J. Insur. Med. 2017, 47, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Ganin, Y.; Ustinova, E.; Ajakan, H.; Germain, P.; Larochelle, H.; Laviolette, F.; Marchand, M.; Lempitsky, V. Domain-adversarial training of neural networks. J. Mach. Learn. Res. 2016, 17, 2096-2030. [Google Scholar]
- Gong, B.; Shi, Y.; Sha, F.; Grauman, K. Geodesic flow kernel for unsupervised domain adaptation. In Proceedings of the 2012 IEEE Conference on Computer Vision and Pattern Recognition, Providence, RI, USA, 16–21 June 2012; IEEE: Piscataway, NJ, USA, 2012; pp. 2066–2073. [Google Scholar]
| Game Name | Stimuli Type | Positive–Negative | Arousal–Valence |
|---|---|---|---|
| G1 | Boring | Negative | LANV |
| G2 | Calm | Positive | LAPV |
| G3 | Horror | Negative | HANV |
| G4 | Funny | Positive | HAPV |
| Parameter | Value |
|---|---|
| Maximum generation | 50 |
| Size of population | 20 |
| Selection function | Stochastic Universal Sampling |
| Rate of individuals to be selected | 0.9 |
| Mutation probability | 0.7 |
| FieldD | m | |||
|---|---|---|---|---|
| len | 9 | 9 | 9 | 9 |
| lb | 1 | 0 | 0 | 0 |
| ub | 310/70 | 0.1 | 0.01 | 0.001 |
| code | gray | gray | gray | gray |
| scale | arithmetic | arithmetic | arithmetic | arithmetic |
| lbin | 0 | 0 | 1 | 1 |
| ubin | 1 | 1 | 1 | 1 |
| Method | PDPL ACC/STD (%) | GA-PDPL ACC/STD (%) |
|---|---|---|
| SEED | 51.02/13.57 | 69.89/14.39 |
| MPED | 21.39/5.41 | 24.87/5.83 |
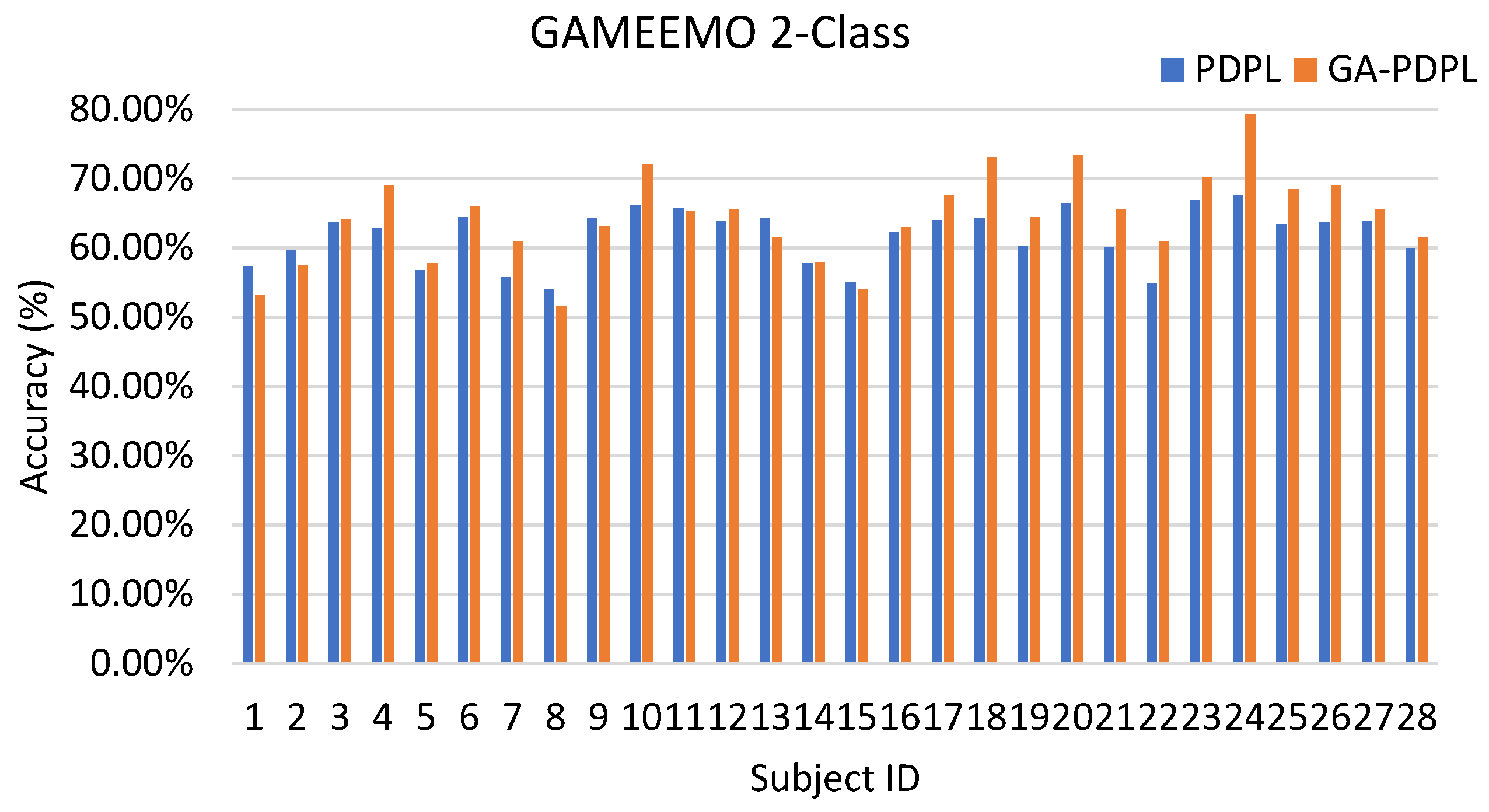
| GAMEEMO (two-class) | 61.76/3.99 | 64.34/6.44 |
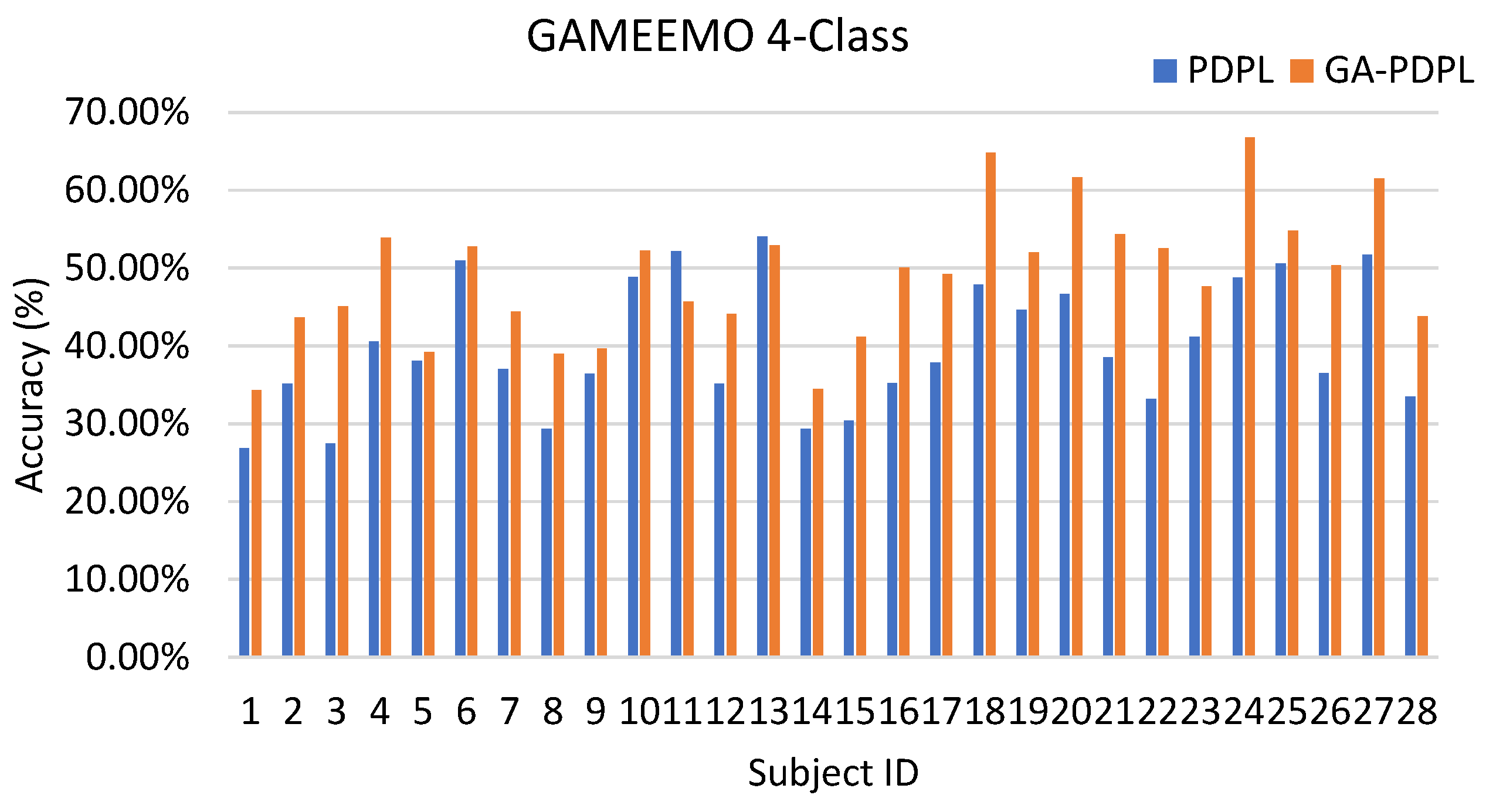
| GAMEEMO (four-class) | 39.92/8.28 | 49.01/8.46 |
| Method | PDPL | GA-PDPL |
|---|---|---|
| SEED | [43.51, 58.54] | [59.52, 76.78] |
| MPED | [19.05, 23.73] | [22.35, 27.39] |
| GAMEEMO (2-class) | [60.21, 63.31] | [61.84, 66.84] |
| GAMEEMO (4-class) | [36.71, 43.13] | [45.72, 52.29] |
| Method | PDPL | GA-PDPL | ||
|---|---|---|---|---|
| Time (s) | Training Time | Testing Time | Training Time | Testing Time |
| SEED | 5.6330 | 0.0208 | 76,902 | 0.001 |
| MPED | 12.6985 | 0.0625 | 335,386 | 0.005 |
| GAMEEMO (2-class) | 1.5734 | 0.0012 | 15,575 | 0.0005 |
| GAMEEMO (4-class) | 1.7660 | 0.0034 | 17,892 | 0.0004 |
| Method | M ± SEM (%) |
|---|---|
| KLIEP [40] * | 45.17 ± 4.59 |
| PDPL [27] * | 51.02 ± 3.50 |
| ULSIF [41] * | 51.18 ± 3.50 |
| STM [42] * | 51.23 ± 3.83 |
| SVM [43] * | 56.73 ± 4.21 |
| KPCA [44] * | 61.28 ± 3.77 |
| TCA [45] * | 63.64 ± 3.84 |
| SA [43] * | 69.00 ± 2.81 |
| GA-PDPL (ours ) | 69.89 ± 3.72 |
| Method | M ± SEM (%) |
|---|---|
| KLIEP [40] * | 18.92 ± 0.95 |
| ULSIF [41] * | 19.63 ± 0.79 |
| TCA [45] * | 19.50 ± 0.75 |
| SVM [43] * | 19.66 ± 0.83 |
| GFK [49] * | 20.27 ± 0.91 |
| SA [43] * | 20.74 ± 0.87 |
| STM [42] * | 20.89 ± 0.75 |
| PDPL [27] * | 21.39 ± 1.13 |
| DANN [48] | 22.36 ± 0.91 |
| A-LSTM [16] | 24.06 ± 0.96 |
| GA-PDPL (ours ) | 24.87 ± 1.22 |
| Method | Two-Class M ± SEM (%) | Four-Class M ± SEM (%) |
|---|---|---|
| KNN [46] * | 58.16 ± 1.45 | 35.46 ± 2.06 |
| Random Forest [47] * | 59.29 ± 2.12 | 38.27 ± 2.95 |
| PDPL [27] * | 61.76 ± 0.75 | 39.92 ± 1.56 |
| SVM [43] * | 63.17 ± 1.27 | 46.62 ± 1.89 |
| GA-PDPL(ours ) | 64.34 ± 1.56 | 49.01 ± 1.60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, J.; Zhu, J.; Song, T.; Chang, H. Subject-Independent EEG Emotion Recognition Based on Genetically Optimized Projection Dictionary Pair Learning. Brain Sci. 2023, 13, 977. https://doi.org/10.3390/brainsci13070977
Su J, Zhu J, Song T, Chang H. Subject-Independent EEG Emotion Recognition Based on Genetically Optimized Projection Dictionary Pair Learning. Brain Sciences. 2023; 13(7):977. https://doi.org/10.3390/brainsci13070977
Chicago/Turabian StyleSu, Jipu, Jie Zhu, Tiecheng Song, and Hongli Chang. 2023. "Subject-Independent EEG Emotion Recognition Based on Genetically Optimized Projection Dictionary Pair Learning" Brain Sciences 13, no. 7: 977. https://doi.org/10.3390/brainsci13070977
APA StyleSu, J., Zhu, J., Song, T., & Chang, H. (2023). Subject-Independent EEG Emotion Recognition Based on Genetically Optimized Projection Dictionary Pair Learning. Brain Sciences, 13(7), 977. https://doi.org/10.3390/brainsci13070977







