Automated Classification of Brain Tumors from Magnetic Resonance Imaging Using Deep Learning
Abstract
1. Introduction
- This study presents a novel CNN approach for classifying three types of brain tumors: glioma, meningioma, and pituitary tumors.
- The objective is to show that the presented approach can outperform more complex methods with limited resources for deployment and training. The study evaluates the network’s ability to generalize for clinical research and further deployment.
- The presented investigation suggests that the proposed methodology outperforms existing approaches, as evidenced by achieving the highest accuracy score on the Kaggle dataset. Furthermore, comparisons were made with pre-trained models and previous methods to reveal the prediction performance of the presented approach.
2. Literature Review
3. Material and Methods
3.1. Dataset
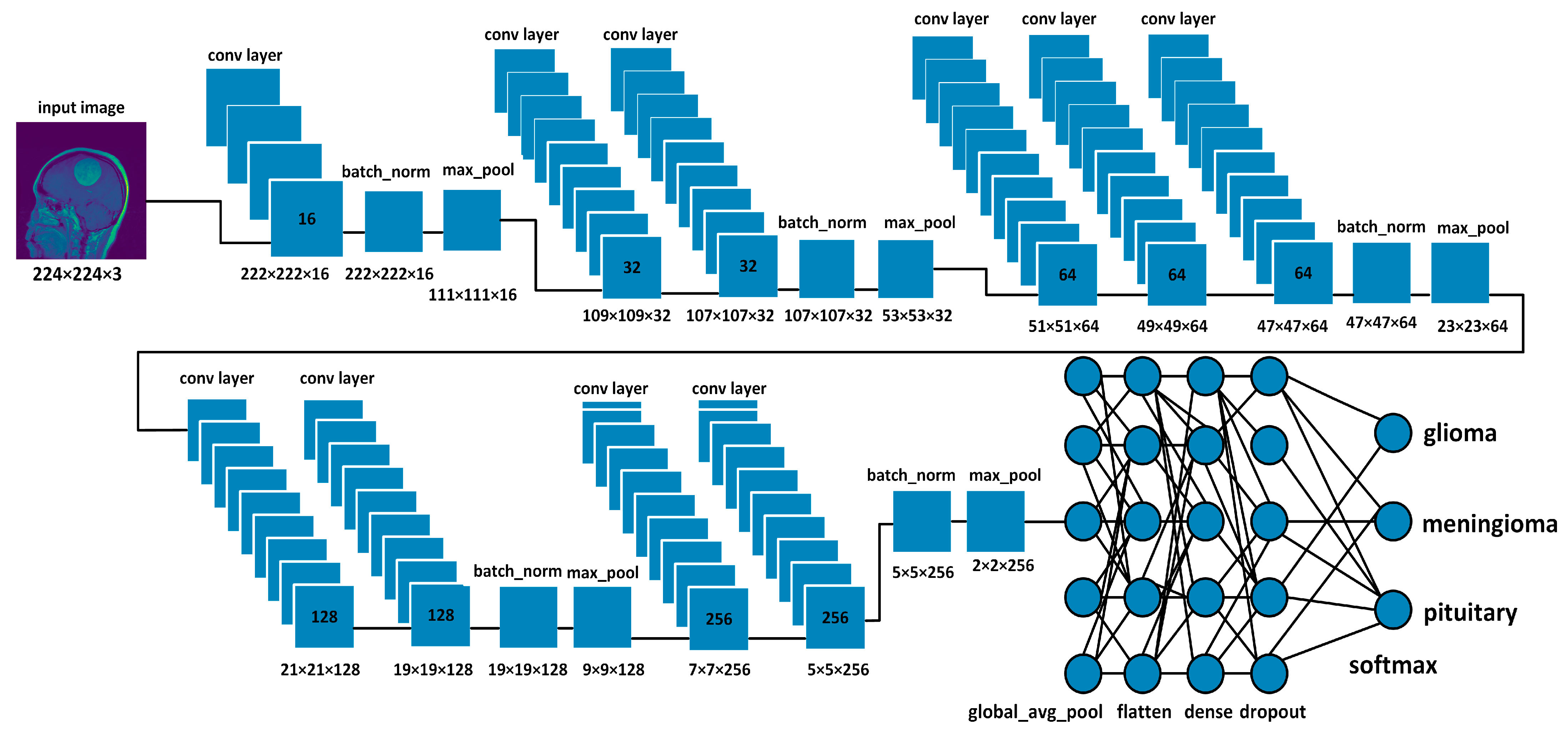
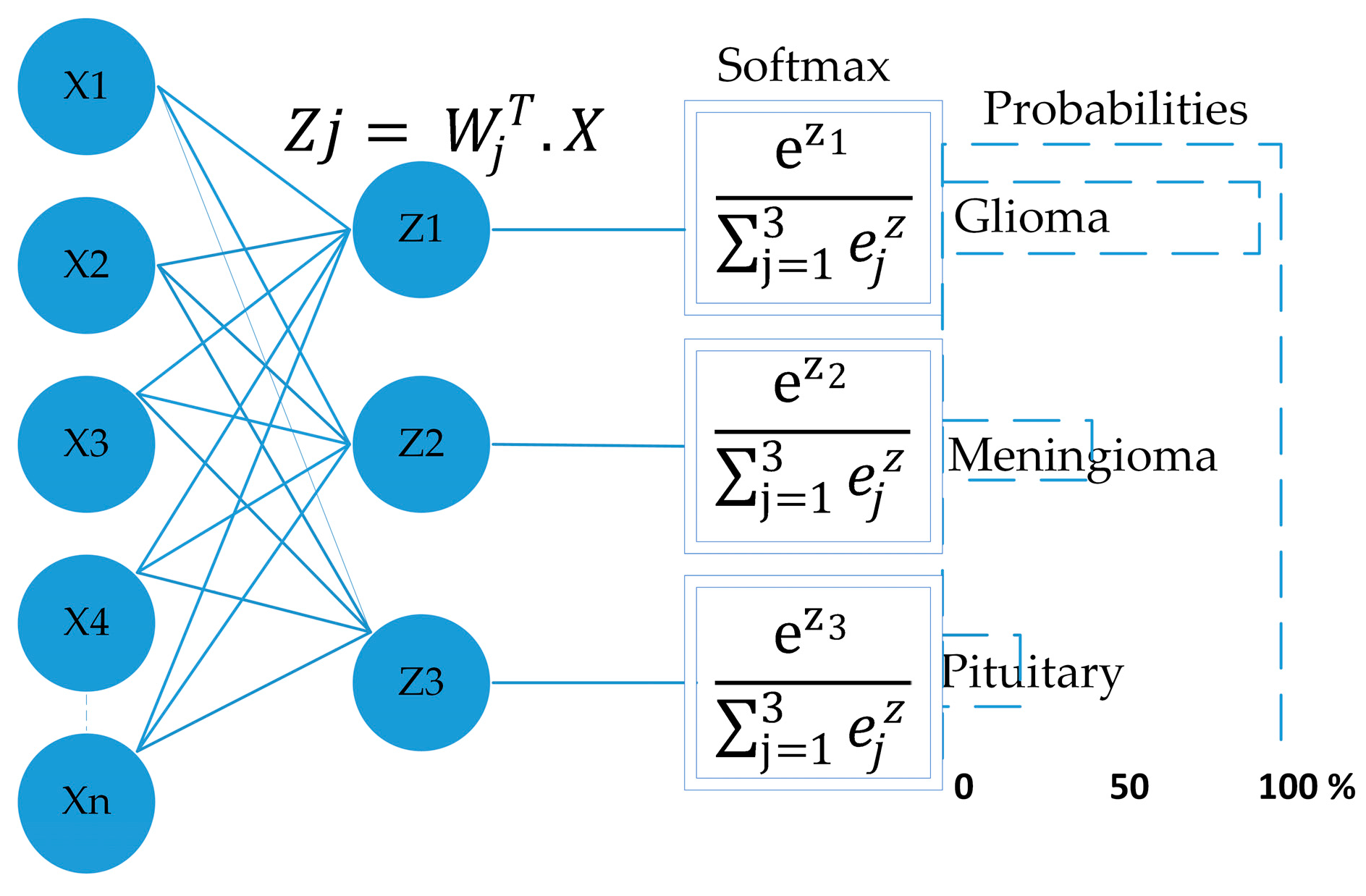
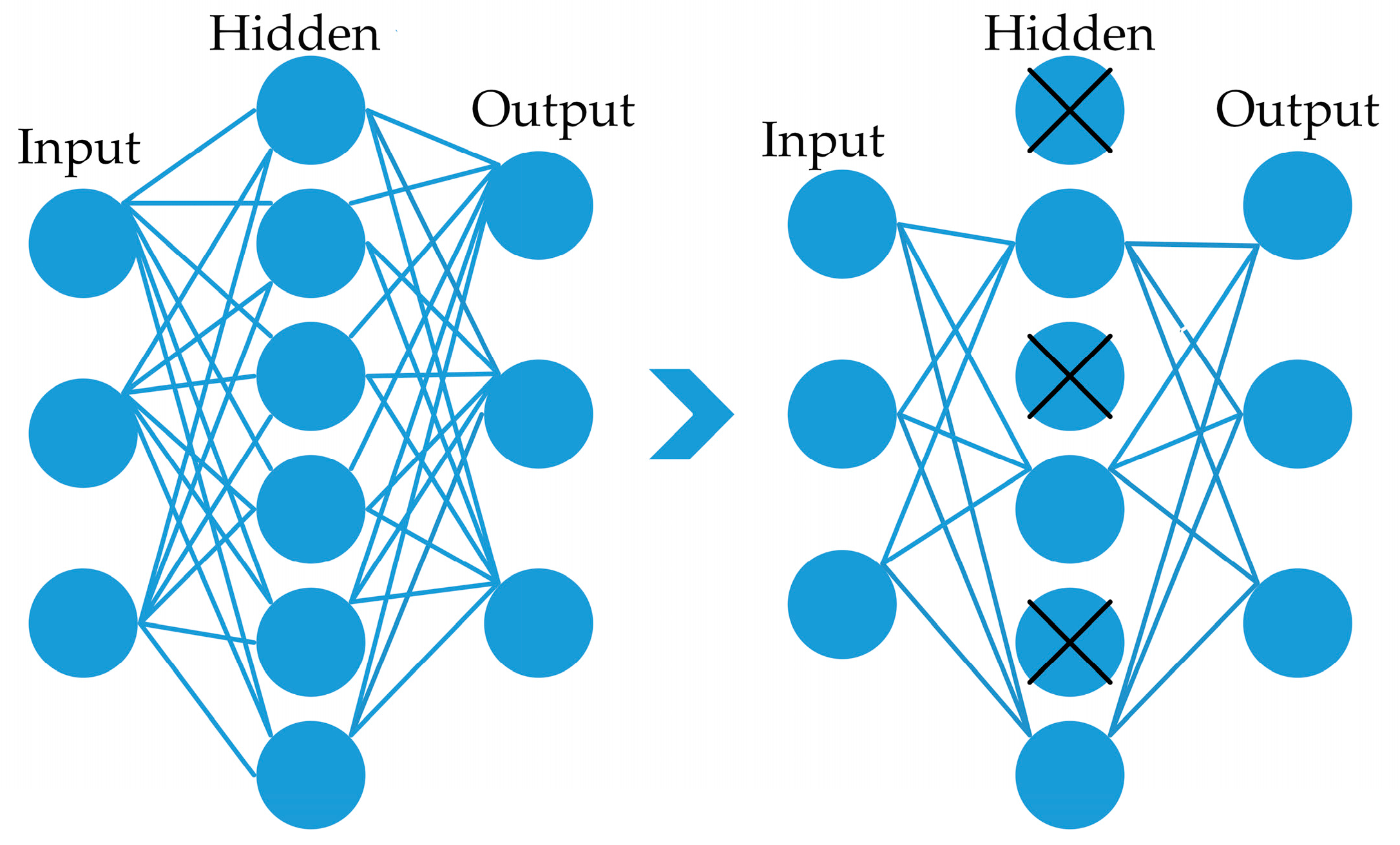
3.2. Network Architectures
3.2.1. Proposed Model
3.2.2. Optimization Approaches
| Algorithm 1: Pseudocode: For the Adam algorithm. |
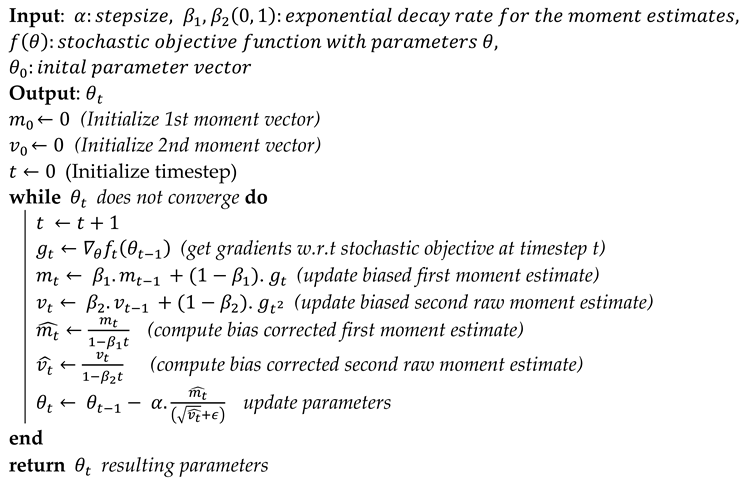 |
3.3. Pre-Trained Models
3.3.1. VGG16
3.3.2. VGG19
3.3.3. ResNet50
3.3.4. InceptionV3
3.3.5. MobileNetV2
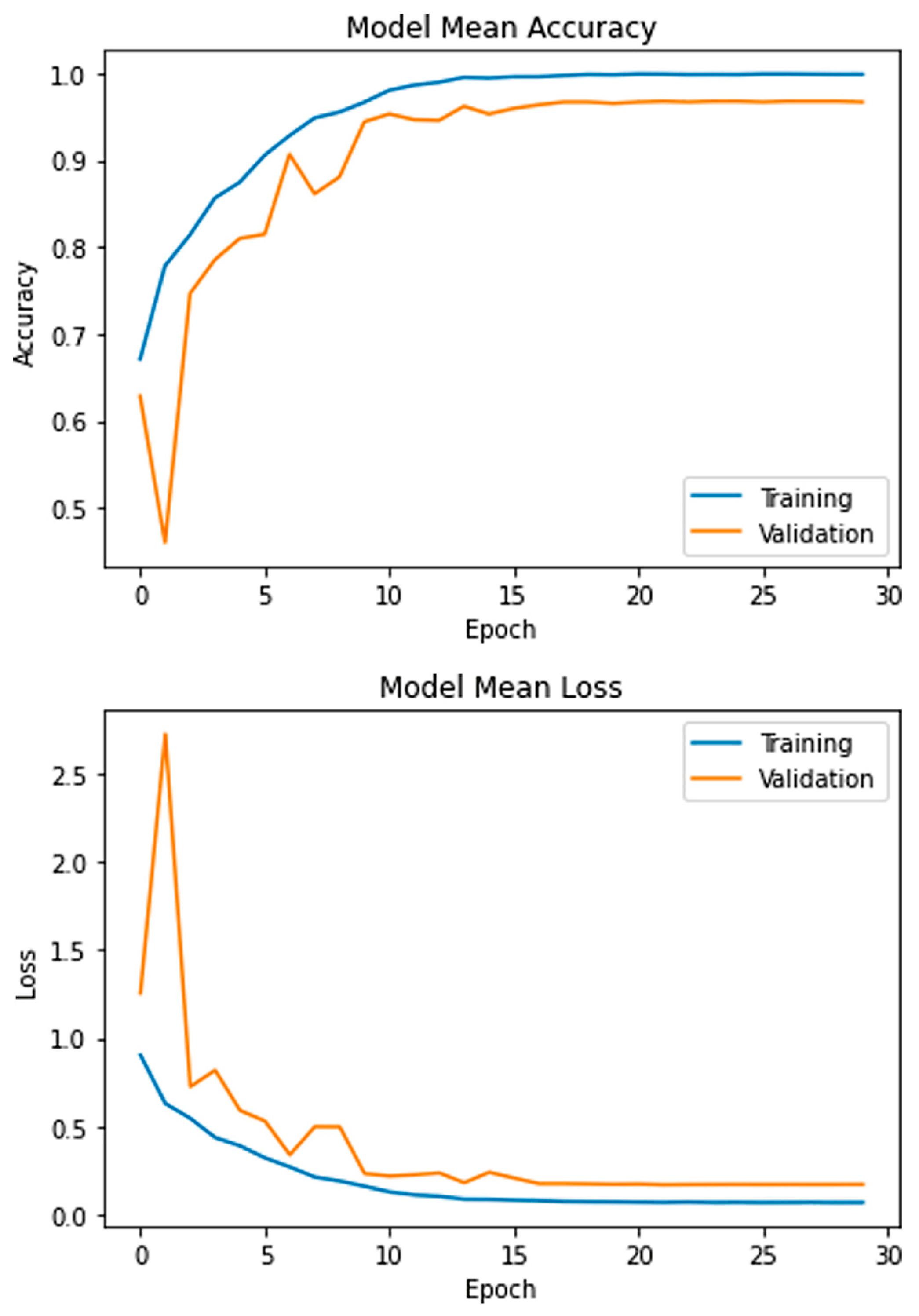
4. Experimental Results
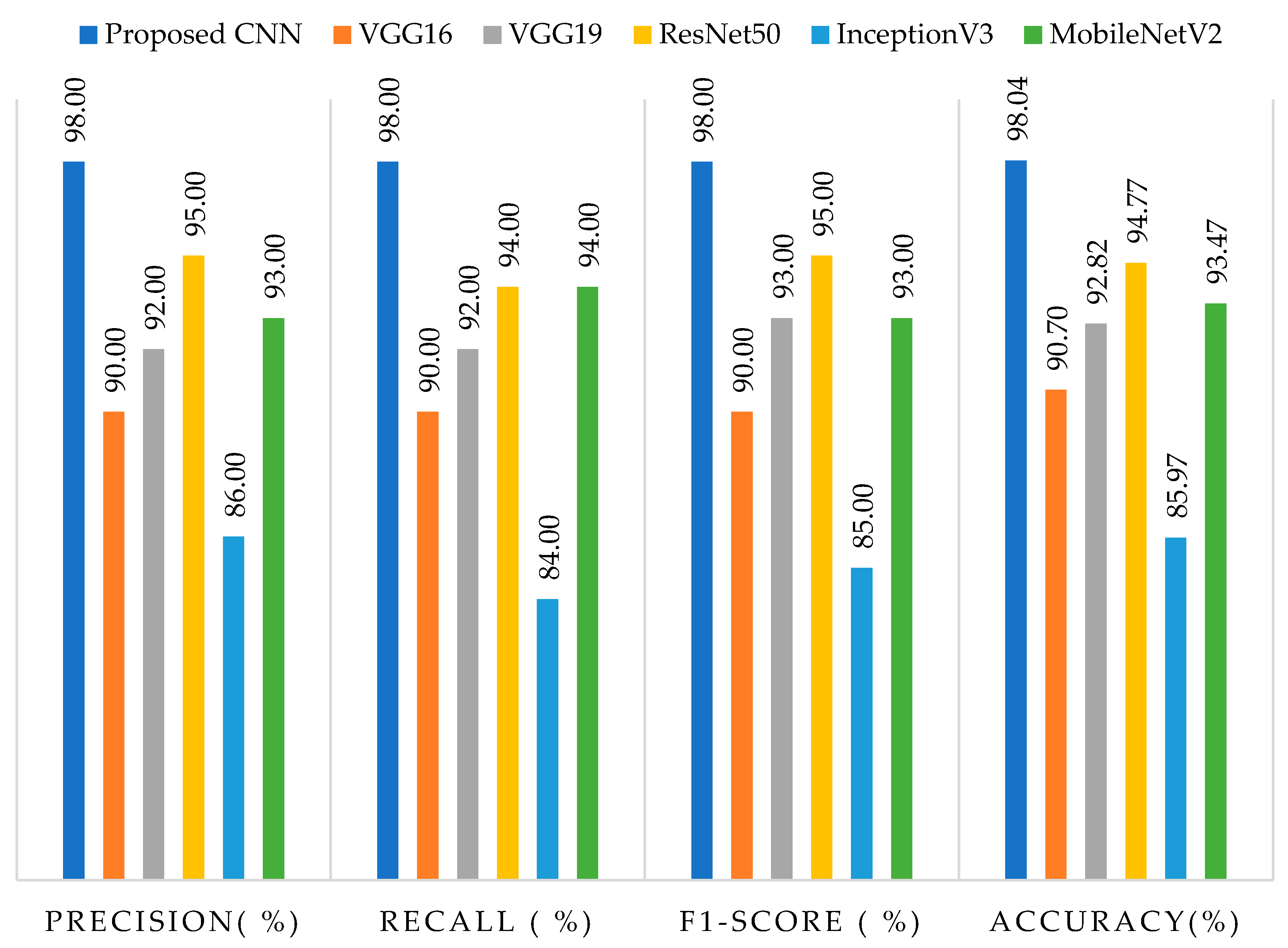
4.1. Evaluation Matrix
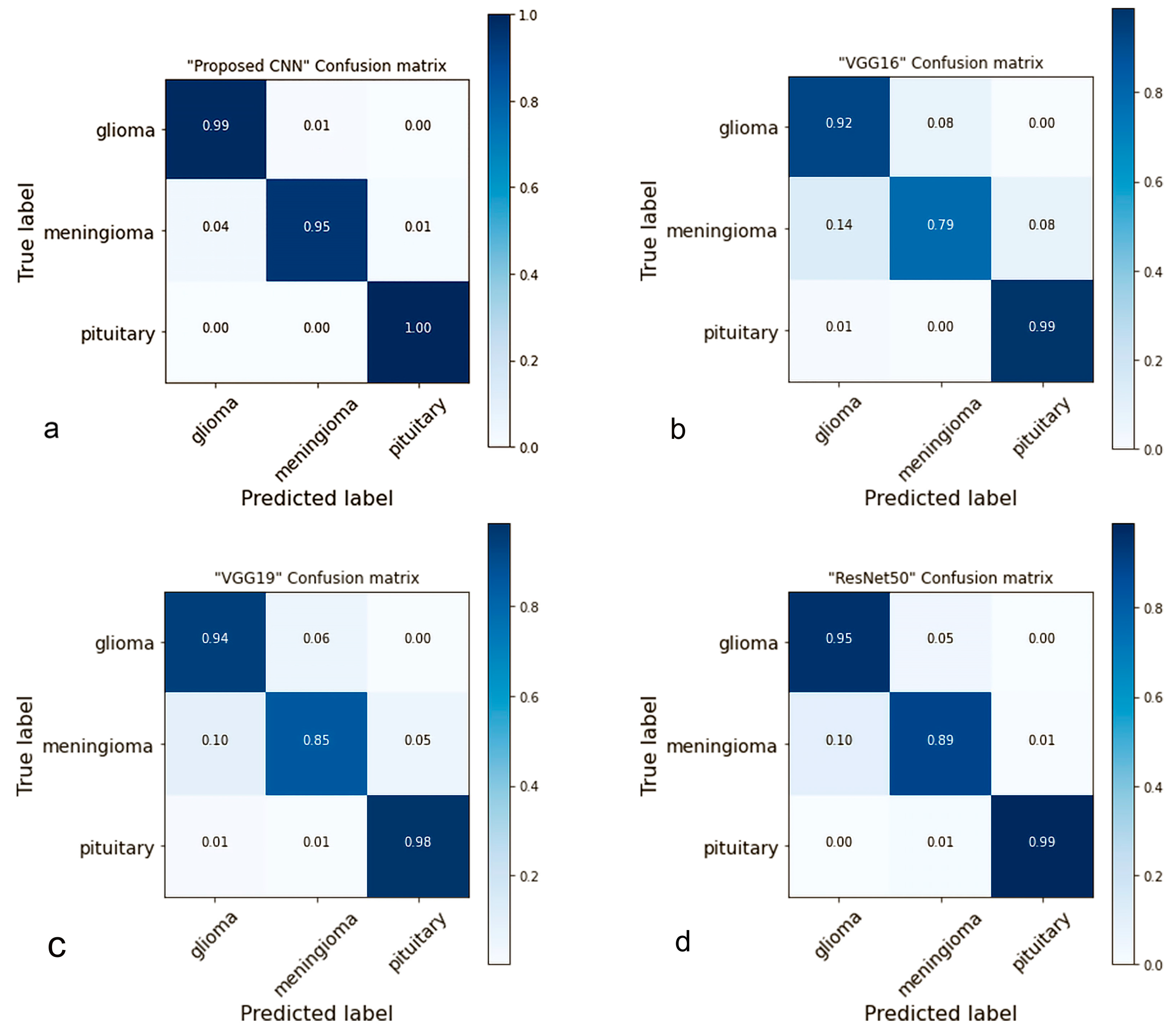
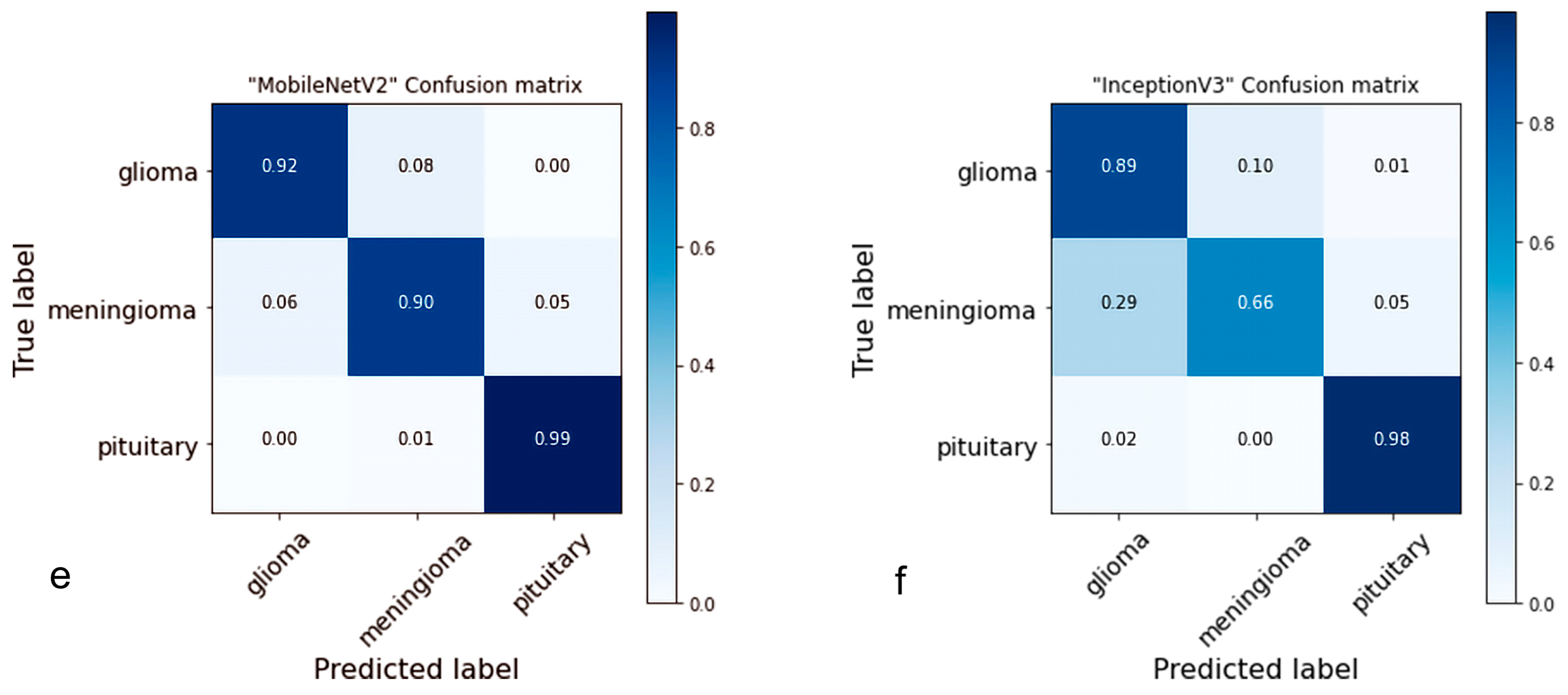
4.2. Confusion Matrix
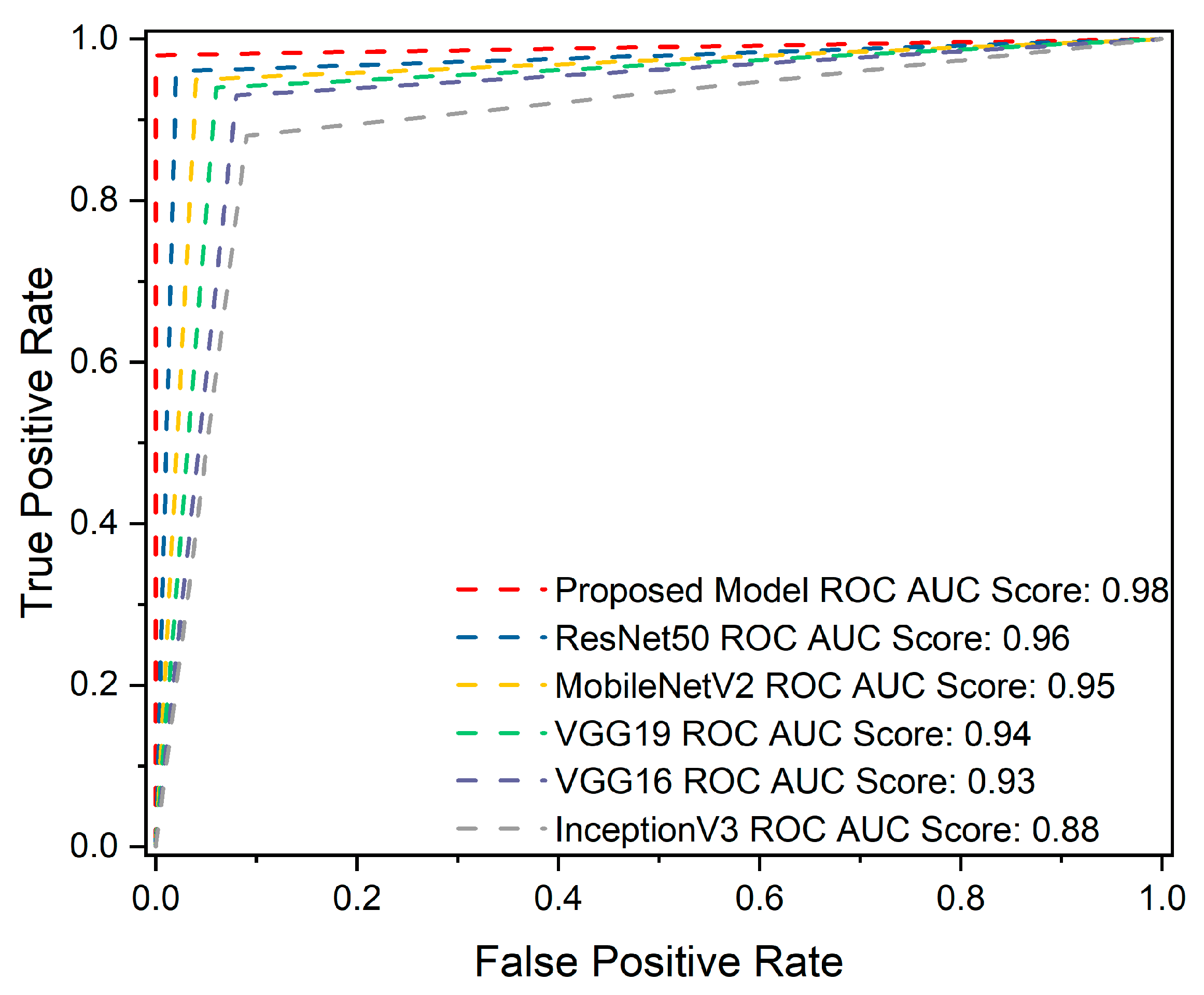
4.3. ROC Curve Analysis
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Khazaei, Z.; Goodarzi, E.; Borhaninejad, V.; Iranmanesh, F.; Mirshekarpour, H.; Mirzaei, B.; Naemi, H.; Bechashk, S.M.; Darvishi, I.; Sarabi, R.E.; et al. The association between incidence and mortality of brain cancer and human development index (HDI): An ecological study. BMC Public Health 2020, 20, 1696. [Google Scholar] [CrossRef]
- GLOBOCAN. The Global Cancer Observatory—All Cancers; International Agency for Research on Cancer—WHO: Lyon, France, 2020; Volume 419, pp. 199–200. Available online: http://gco.iarc.fr/today/home (accessed on 10 February 2022).
- Kalpana, R.; Chandrasekar, P. An optimized technique for brain tumor classification and detection with radiation dosage calculation in MR image. Microprocess. Microsyst. 2019, 72, 102903. [Google Scholar] [CrossRef]
- Malignant Brain Tumour (Cancerous). NHS Inform. Available online: https://www.nhsinform.scot/illnesses-and-conditions/cancer/cancer-types-in-adults/malignant-brain-tumour-cancerous (accessed on 15 February 2023).
- Gliomas. Johns Hopkins Medicine. Available online: https://www.hopkinsmedicine.org/health/conditions-and-diseases/gliomas (accessed on 12 February 2023).
- Pituitary Tumors—Symptoms and Causes—Mayo Clinic. Available online: https://www.mayoclinic.org/diseases-conditions/pituitary-tumors/symptoms-causes/syc-20350548 (accessed on 12 February 2023).
- Meningioma. Johns Hopkins Medicine. Available online: https://www.hopkinsmedicine.org/health/conditions-and-diseases/meningioma (accessed on 12 February 2023).
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Markman, S.K.; Narasimhan, J. Chronic Pain–Brain, Spinal Cord, and Nerve Disorders–Merck Manuals Consumer Version. 2014. Available online: https://web.archive.org/web/20160812032003/http://www.merckmanuals.com/home/brain,-spinal-cord,-and-nerve-disorders/tumors-of-the-nervous-system/brain-tumors (accessed on 11 December 2022).
- American Brain Tumor Association. Mood Swings and Cognitive Changes. 2014. Available online: https://web.archive.org/web/20160802203516/http://www.abta.org/brain-tumor-information/symptoms/mood-swings.html (accessed on 11 December 2022).
- Glioma: What Is It, Causes, Symptoms, Treatment & Outlook. Available online: https://my.clevelandclinic.org/health/diseases/21969-glioma (accessed on 18 March 2023).
- Glioma–Symptoms and Causes–Mayo Clinic. Available online: https://www.mayoclinic.org/diseases-conditions/glioma/symptoms-causes/syc-20350251 (accessed on 18 March 2023).
- Meningioma–Symptoms and Causes–Mayo Clinic. Available online: https://www.mayoclinic.org/diseases-conditions/meningioma/symptoms-causes/syc-20355643 (accessed on 18 March 2023).
- Afshar, P.; Plataniotis, K.N.; Mohammadi, A. Capsule Networks for Brain Tumor Classification Based on MRI Images and Coarse Tumor Boundaries. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 1368–1372. [Google Scholar]
- Rogers, L.; Barani, I.; Chamberlain, M.; Kaley, T.J.; McDermott, M.; Raizer, J.; Schiff, D.; Weber, D.C.; Wen, P.Y.; Vogelbaum, M.A. Meningiomas: Knowledge base, treatment outcomes, and uncertainties. A RANO review. J. Neurosurg. 2015, 122, 4–23. [Google Scholar] [CrossRef]
- Tiwari, A.; Srivastava, S.; Pant, M. Brain tumor segmentation and classification from magnetic resonance images: Review of selected methods from 2014 to 2019. Pattern Recognit. Lett. 2019, 131, 244–260. [Google Scholar] [CrossRef]
- Iwendi, C.; Khan, S.; Anajemba, J.H.; Mittal, M.; Alenezi, M.; Alazab, M. The Use of Ensemble Models for Multiple Class and Binary Class Classification for Improving Intrusion Detection Systems. Sensors 2020, 20, 2559. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Ullah, T.; Ahmad, I.; Al-Sharabi, A.; Ullah, K.; Khan, R.A.; Rasheed, S.; Ullah, I.; Uddin, N.; Ali, S. A Novel Hybrid Deep Learning Model for Metastatic Cancer Detection. Comput. Intell. Neurosci. 2022, 2022, 8141530. [Google Scholar] [CrossRef]
- Yousafzai, B.K.; Khan, S.A.; Rahman, T.; Khan, I.; Ullah, I.; Rehman, A.U.; Baz, M.; Hamam, H.; Cheikhrouhou, O. Student-Performulator: Student Academic Performance Using Hybrid Deep Neural Network. Sustainability 2021, 13, 9775. [Google Scholar] [CrossRef]
- Hassan, H.; Ren, Z.; Zhou, C.; Khan, M.A.; Pan, Y.; Zhao, J.; Huang, B. Supervised and weakly supervised deep learning models for COVID-19 CT diagnosis: A systematic review. Comput. Methods Programs Biomed. 2022, 218, 106731. [Google Scholar] [CrossRef]
- Hassan, H.; Ren, Z.; Zhao, H.; Huang, S.; Li, D.; Xiang, S.; Kang, Y.; Chen, S.; Huang, B. Review and classification of AI-enabled COVID-19 CT imaging models based on computer vision tasks. Comput. Biol. Med. 2021, 141, 105123. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, H.; Yan, Y.; Hassan, H. 3D Cross-Pseudo Supervision (3D-CPS): A Semi-supervised nnU-Net Architecture for Abdominal Organ Segmentation. In Proceedings of the Fast and Low-Resource Semi-Supervised Abdominal Organ Segmentation: MICCAI 2022 Challenge, FLARE 2022, Held in Conjunction with MICCAI, Singapore, 22 September 2022; pp. 87–100. [Google Scholar] [CrossRef]
- Iwendi, C.; Khan, S.; Anajemba, J.H.; Bashir, A.K.; Noor, F. Realizing an Efficient IoMT-Assisted Patient Diet Recommendation System Through Machine Learning Model. IEEE Access 2020, 8, 28462–28474. [Google Scholar] [CrossRef]
- Kaplan, K.; Kaya, Y.; Kuncan, M.; Ertunç, H.M. Brain tumor classification using modified local binary patterns (LBP) feature extraction methods. Med. Hypotheses 2020, 139, 109696. [Google Scholar] [CrossRef]
- Rathi, V.P.G.P.; Palani, S. Brain Tumor Detection and Classification Using Deep Learning Classifier on MRI Images. Appl. Sci. Eng. Technol. 2015, 10, 177–187. [Google Scholar]
- Ho, R.; Sharma, V.; Tan, B.; Ng, A.; Lui, Y.-S.; Husain, S.; Ho, C.; Tran, B.; Pham, Q.-H.; McIntyre, R.; et al. Comparison of Brain Activation Patterns during Olfactory Stimuli between Recovered COVID-19 Patients and Healthy Controls: A Functional Near-Infrared Spectroscopy (fNIRS) Study. Brain Sci. 2021, 11, 968. [Google Scholar] [CrossRef] [PubMed]
- McGrowder, D.; Miller, F.; Vaz, K.; Nwokocha, C.; Wilson-Clarke, C.; Anderson-Cross, M.; Brown, J.; Anderson-Jackson, L.; Williams, L.; Latore, L.; et al. Cerebrospinal Fluid Biomarkers of Alzheimer’s Disease: Current Evidence and Future Perspectives. Brain Sci. 2021, 11, 215. [Google Scholar] [CrossRef] [PubMed]
- Perri, R.L.; Castelli, P.; La Rosa, C.; Zucchi, T.; Onofri, A. COVID-19, Isolation, Quarantine: On the Efficacy of for Ongoing Trauma. Brain Sci. 2021, 11, 579. [Google Scholar] [CrossRef] [PubMed]
- Gębska, M.; Dalewski, B.; Pałka, Ł.; Kołodziej, Ł.; Sobolewska, E. The Importance of Type D Personality in the Development of Temporomandibular Disorders (TMDs) and Depression in Students during the COVID-19 Pandemic. Brain Sci. 2021, 12, 28. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Huang, W.; Cao, S.; Yang, R.; Yang, W.; Yun, Z.; Wang, Z.; Feng, Q. Enhanced Performance of Brain Tumor Classification via Tumor Region Augmentation and Partition. PLoS ONE 2015, 10, e0140381. [Google Scholar] [CrossRef] [PubMed]
- Sasikala, M.; Kumaravel, N. A wavelet-based optimal texture feature set for classification of brain tumours. J. Med. Eng. Technol. 2008, 32, 198–205. [Google Scholar] [CrossRef]
- El-Dahshan, E.-S.A.; Hosny, T.; Salem, A.-B.M. Hybrid intelligent techniques for MRI brain images classification. Digit. Signal Process. 2010, 20, 433–441. [Google Scholar] [CrossRef]
- Mohsen, H.; El-Dahshan, E.-S.A.; El-Horbaty, E.-S.M.; Salem, A.-B.M. Classification using deep learning neural networks for brain tumors. Future Comput. Inform. J. 2018, 3, 68–71. [Google Scholar] [CrossRef]
- Cheng, J. Figshare Brain Tumor Dataset. 2017. Available online: https://figshare.com/articles/dataset/brain_tumor_dataset/1512427/5 (accessed on 13 May 2022).
- Ismael, M.R.; Abdel-Qader, I. Brain Tumor Classification via Statistical Features and Back-Propagation Neural Network. In Proceedings of the 2018 IEEE International Conference on Electro/Information Technology (EIT), Rochester, MI, USA, 3–5 May 2018; pp. 252–257. [Google Scholar] [CrossRef]
- Abiwinanda, N.; Hanif, M.; Hesaputra, S.T.; Handayani, A.; Mengko, T.R. Brain Tumor Classification Using Convolutional Neural Network. In Proceedings of the World Congress on Medical Physics and Biomedical Engineering 2018, Prague, Czech Republic, 3–8 June 2018; Springer: Singapore, 2019; Volume 68, pp. 183–189. [Google Scholar] [CrossRef]
- Pashaei, A.; Sajedi, H.; Jazayeri, N. Brain Tumor Classification via Convolutional Neural Network and Extreme Learning Machines. In Proceedings of the 2018 8th International Conference on Computer and Knowledge Engineering (ICCKE), Mashhad, Iran, 25–26 October 2018; pp. 314–319. [Google Scholar]
- Phaye, S.S.R.; Sikka, A.; Dhall, A.; Bathula, D. Dense and Diverse Capsule Networks: Making the Capsules Learn Better. arXiv 2018, arXiv:1805.04001. [Google Scholar]
- Avsar, E.; Salçin, K. Detection and classification of brain tumours from MRI images using faster R-CNN. Teh. Glas. 2019, 13, 337–342. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, Z.; Zhu, H.; Chen, C.; Gao, M.; Xu, K.; Xu, J. Holistic Brain Tumor Screening and Classification Based on DenseNet and Recurrent Neural Network. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Springer: Cham, Switzerland, 2019; Volume 11383, pp. 208–217. [Google Scholar] [CrossRef]
- Anaraki, A.K.; Ayati, M.; Kazemi, F. Magnetic resonance imaging-based brain tumor grades classification and grading via convolutional neural networks and genetic algorithms. Biocybern. Biomed. Eng. 2018, 39, 63–74. [Google Scholar] [CrossRef]
- Gumaei, A.; Hassan, M.M.; Hassan, R.; Alelaiwi, A.; Fortino, G. A Hybrid Feature Extraction Method with Regularized Extreme Learning Machine for Brain Tumor Classification. IEEE Access 2019, 7, 36266–36273. [Google Scholar] [CrossRef]
- Ghassemi, N.; Shoeibi, A.; Rouhani, M. Deep neural network with generative adversarial networks pre-training for brain tumor classification based on MR images. Biomed. Signal Process. Control. 2019, 57, 101678. [Google Scholar] [CrossRef]
- Swati, Z.N.K.; Zhao, Q.; Kabir, M.; Ali, F.; Ali, Z.; Ahmed, S.; Lu, J. Brain tumor classification for MR images using transfer learning and fine-tuning. Comput. Med. Imaging Graph. 2019, 75, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Noreen, N.; Palaniappan, S.; Qayyum, A.; Ahmad, I.; Alassafi, M.O. Brain Tumor Classification Based on Fine-Tuned Models and the Ensemble Method. Comput. Mater. Contin. 2021, 67, 3967–3982. [Google Scholar] [CrossRef]
- Cheng, J. Brain Tumor Image Dataset. Kaggle. Available online: https://www.kaggle.com/datasets/denizkavi1/brain-tumor (accessed on 6 August 2022).
- Goodfellow, I. Deep Learning; MIT Press: Cambridge, MA, USA, 2016; pp. 1–10. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift . In Proceedings of the 32nd International Conference on Machine Learning (ICML 2015), Lille, France, 6–11 July 2015; Volume 1, pp. 448–456. [Google Scholar]
- Koffas, S.; Picek, S.; Conti, M. Dynamic Backdoors with Global Average Pooling. In Proceedings of the 2022 IEEE 4th International Conference on Artificial Intelligence Circuits and Systems (AICAS), Incheon, Republic of Korea, 13–15 June 2022; pp. 320–323. [Google Scholar] [CrossRef]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. J. Big Data 2021, 8, 53. [Google Scholar] [CrossRef]
- Bin Tufail, A.; Ullah, I.; Rehman, A.U.; Khan, R.A.; Khan, M.A.; Ma, Y.-K.; Khokhar, N.H.; Sadiq, M.T.; Khan, R.; Shafiq, M.; et al. On Disharmony in Batch Normalization and Dropout Methods for Early Categorization of Alzheimer’s Disease. Sustainability 2022, 14, 14695. [Google Scholar] [CrossRef]
- Nair, V.; Hinton, G.E. Rectified linear units improve Restricted Boltzmann machines. In Proceedings of the 27th International Conference on Machine Learning (ICML-10), Haifa, Israel, 21–24 June 2010. [Google Scholar]
- Kingma, D.P.; Ba, J.; Bengio, Y.; LeCun, Y. Adam: A method for stochastic optimization. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015—Conference Track Proceedings, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Robbins, H.; Monro, S. A Stochastic Approximation Method. Ann. Math. Stat. 1951, 22, 400–407. [Google Scholar] [CrossRef]
- Moradi, R.; Berangi, R.; Minaei, B. A survey of regularization strategies for deep models. Artif. Intell. Rev. 2019, 53, 3947–3986. [Google Scholar] [CrossRef]
- Mele, B.; Altarelli, G. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 299, 345–350. [Google Scholar] [CrossRef]
- Keras. ReduceLROnPlateau. Available online: https://keras.io/api/callbacks/reduce_lr_on_plateau/ (accessed on 21 October 2022).
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. In Proceedings of the 3rd International Conference on Learning Representations, ICLR, San Diego, CA, USA, 7–9 May 2015; pp. 1–14. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A.; Liu, W.; et al. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar] [CrossRef]
- Glorot, X.; Bengio, Y. Understanding the difficulty of training deep feedforward neural networks. J. Mach. Learn. Res. 2010, 9, 249–256. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L. MobileNetV2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar] [CrossRef]
- Kuraparthi, S.; Reddy, M.K.; Sujatha, C.; Valiveti, H.; Duggineni, C.; Kollati, M.; Kora, P.; Sravan, V. Brain Tumor Classification of MRI Images Using Deep Convolutional Neural Network. Trait. Signal 2021, 38, 1171–1179. [Google Scholar] [CrossRef]
- Ting, K.M. Confusion Matrix. In Encyclopedia of Machine Learning and Data Mining; Sammut, C., Webb, G.I., Eds.; Springer: Boston, MA, USA, 2017; p. 206. [Google Scholar] [CrossRef]
- Hajian-Tilaki, K. Receiver Operating Characteristic (ROC) Curve Analysis for Medical Diagnostic Test Evaluation. Casp. J. Intern. Med. 2013, 4, 627. [Google Scholar]
- Ding, T.; Li, D.; Sun, R. Sub-Optimal Local Minima Exist for Neural Networks with Almost All Non-Linear Activations. arXiv 2019, arXiv:1911.01413. [Google Scholar]
- Yadav, S.; Shukla, S. Analysis of k-fold cross-validation over hold-out validation on colossal datasets for quality classification. In Proceedings of the 2016 IEEE 6th International Conference on Advanced Computing (IACC), Bhimavaram, India, 27–28 February 2016; pp. 78–83. [Google Scholar]

| Authors | Methods | Average Precision | Average Recall | Average F1-Score | Accuracy |
|---|---|---|---|---|---|
| Ismael and Abdel-Qader [35] | DWT-Gabor-NN | X | X | X | 91.9 |
| Afshar [14] | CapsNet | X | X | X | 90.89 |
| Pashaei [37] | CNN + KELM | 94.6 | 58.43 | 93 | 93.68 |
| Avşar and Salçin [39] | R-CNN | 97 | X | 95 | 91.66 |
| Zhou [40] | LSTM + DenseNet | X | X | X | 92.13 |
| Anaraki [41] | CCN + GA | X | X | X | 94.20 |
| Gumaei [42] | Hybrid PCA-NGIST + RELM | X | X | X | 94.23 |
| Ghassemi [43] | CNN based GAN | 95.29 | X | 95.10 | 95.60 |
| Swati [44] | VGG16 Finetune | 89.17 | X | 91.50 | 94.65 |
| Swati [44] | VGG19 Finetune | 89.52 | X | 91.73 | 94.82 |
| Swati [44] | AlexNet | 84.56 | X | 86.83 | 89.95 |
| Noreen [45] | InceptionV3 Ensemble | 93 | 92 | 92 | 94.34 |
| Our studies | Proposed CNN | 98 | 98 | 98 | 98.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rasheed, Z.; Ma, Y.-K.; Ullah, I.; Al Shloul, T.; Tufail, A.B.; Ghadi, Y.Y.; Khan, M.Z.; Mohamed, H.G. Automated Classification of Brain Tumors from Magnetic Resonance Imaging Using Deep Learning. Brain Sci. 2023, 13, 602. https://doi.org/10.3390/brainsci13040602
Rasheed Z, Ma Y-K, Ullah I, Al Shloul T, Tufail AB, Ghadi YY, Khan MZ, Mohamed HG. Automated Classification of Brain Tumors from Magnetic Resonance Imaging Using Deep Learning. Brain Sciences. 2023; 13(4):602. https://doi.org/10.3390/brainsci13040602
Chicago/Turabian StyleRasheed, Zahid, Yong-Kui Ma, Inam Ullah, Tamara Al Shloul, Ahsan Bin Tufail, Yazeed Yasin Ghadi, Muhammad Zubair Khan, and Heba G. Mohamed. 2023. "Automated Classification of Brain Tumors from Magnetic Resonance Imaging Using Deep Learning" Brain Sciences 13, no. 4: 602. https://doi.org/10.3390/brainsci13040602
APA StyleRasheed, Z., Ma, Y.-K., Ullah, I., Al Shloul, T., Tufail, A. B., Ghadi, Y. Y., Khan, M. Z., & Mohamed, H. G. (2023). Automated Classification of Brain Tumors from Magnetic Resonance Imaging Using Deep Learning. Brain Sciences, 13(4), 602. https://doi.org/10.3390/brainsci13040602







