Identification of Dysregulated microRNAs in Glioblastoma Stem-like Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. GSC Differentiation
2.3. miR-425-5p Mimic Transfection
2.4. RNA and miRNA Isolation
2.5. RNA Reverse Transcription (RT) and Quantitive Real-Time Polymerase Chain Reaction (qPCR)
2.6. miRNA Reverse Transcription and miRNA PCR Array
2.7. miRNA Reverse Transcription and qPCR
2.8. Protein Extraction and Western Blot Analysis
2.9. Kyoto Encyclopedia of Genes and Genomes (KEGG) Analyses
2.10. Statistical Analysis
3. Results
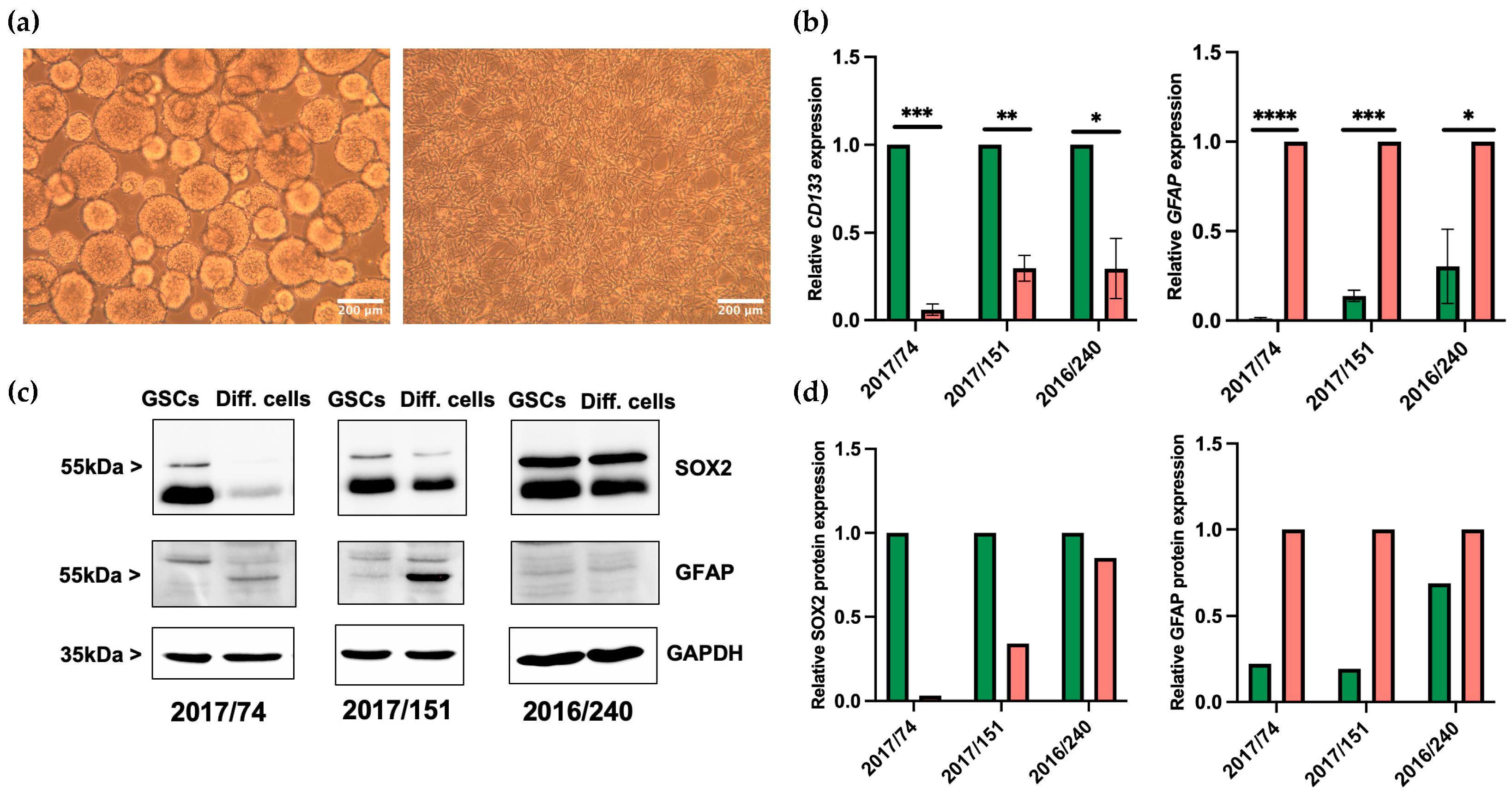
3.1. Differentiation of GSCs
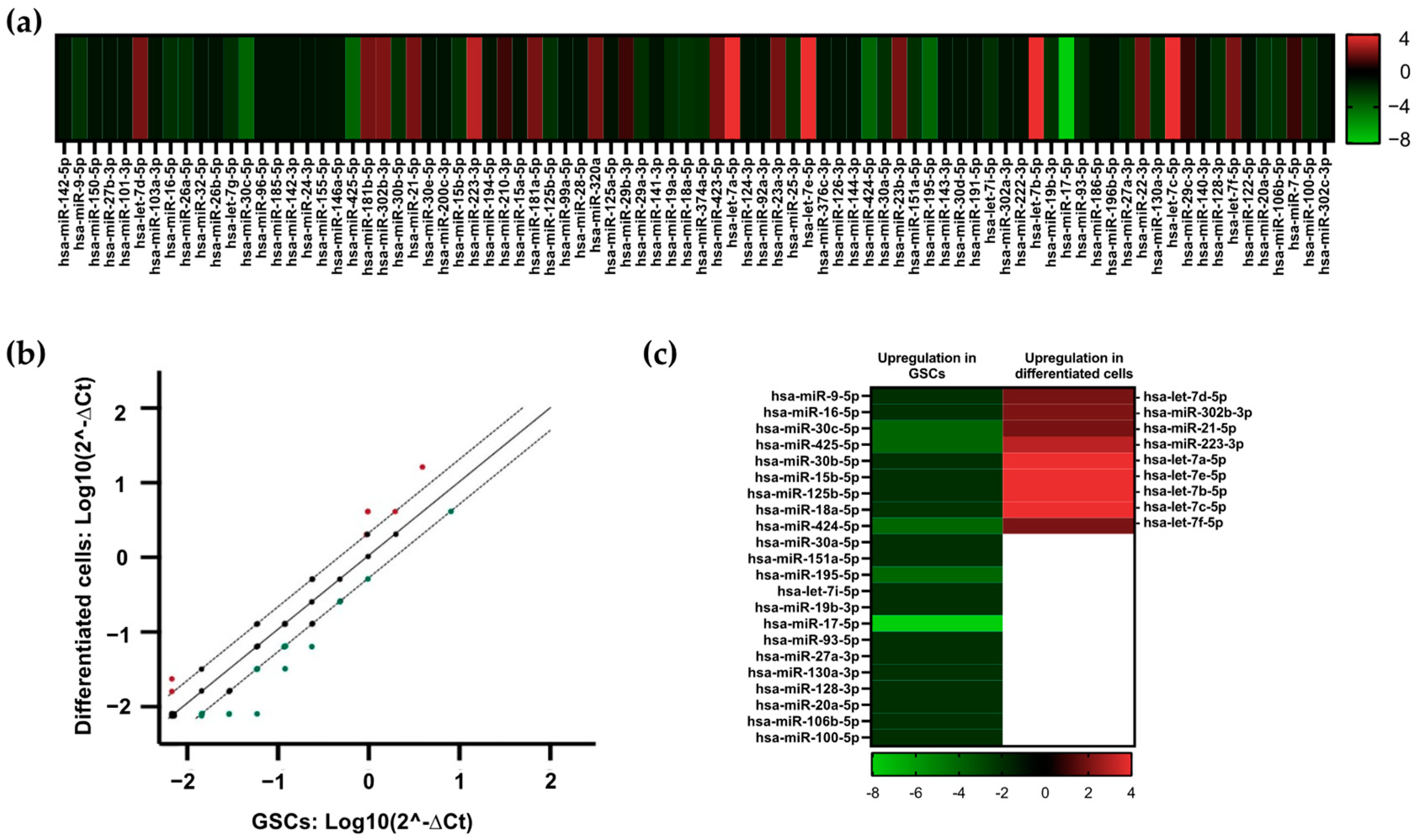
3.2. Identification of Dysregulated miRNAs in GSCs and Differentiated Cells
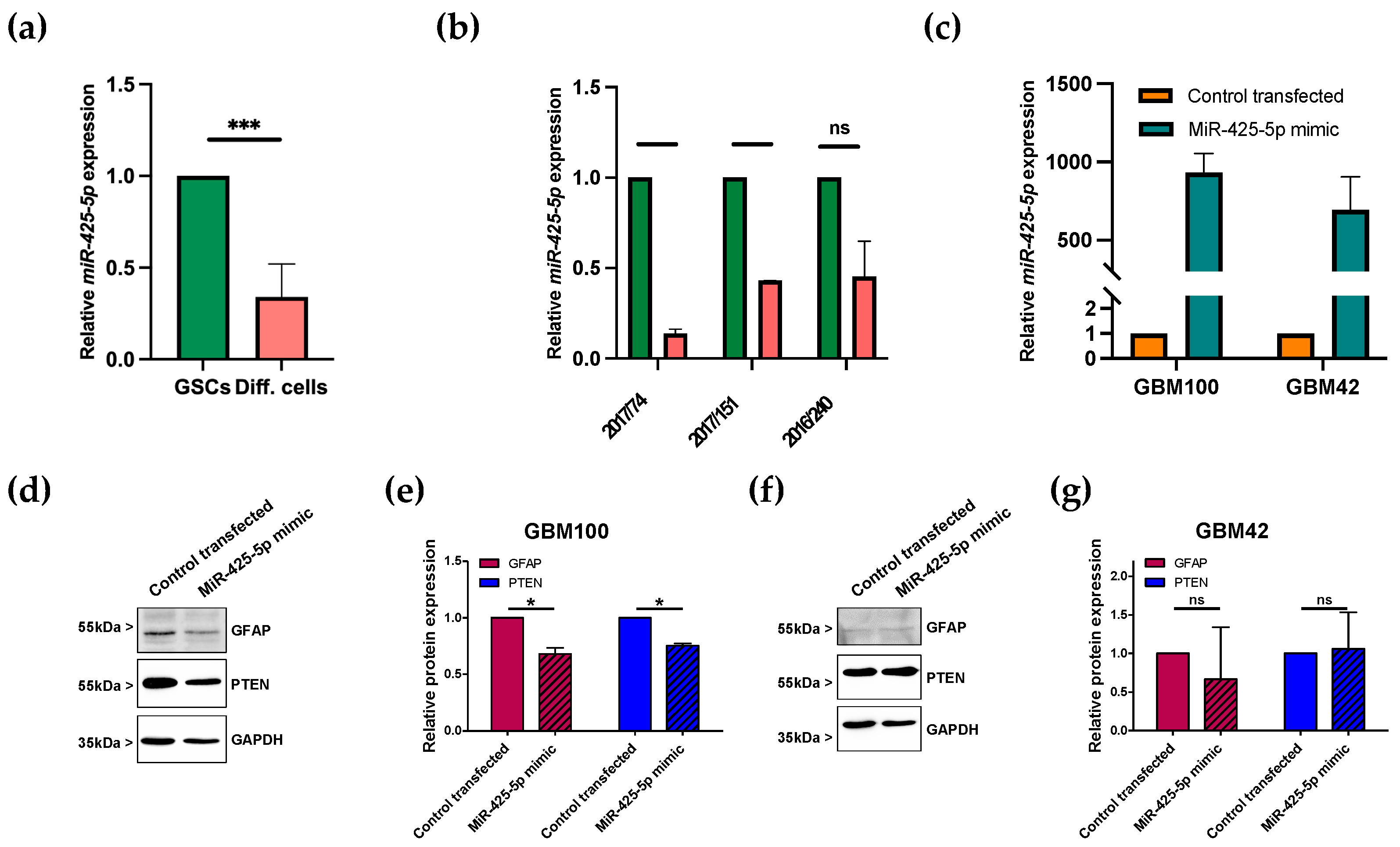
3.3. miR-425-5p Is Downregulated in Differentiated GSCs
3.4. Transfection of miRNA Mimic Affects Protein Levels of GFAP in Patient-Derived GBM Cells
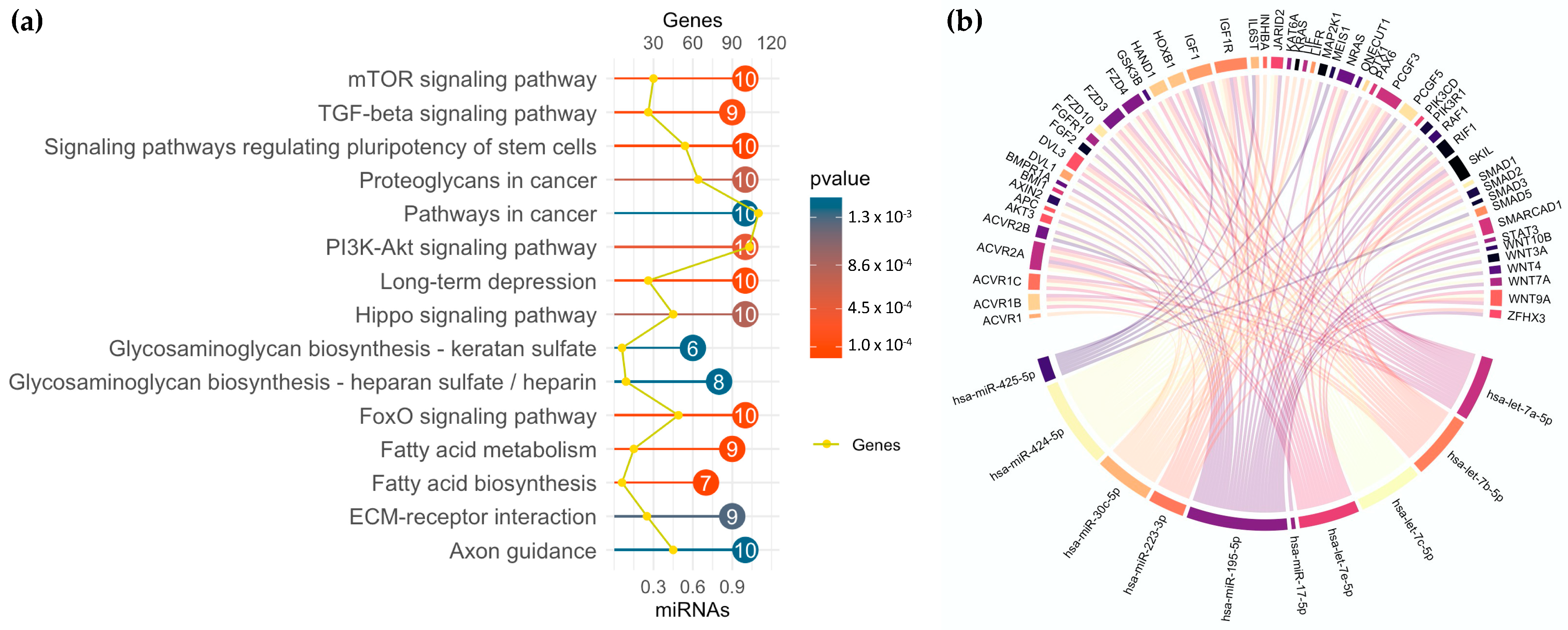
3.5. miRNA Profiling in GSC Maintenance and Differentiation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vollmann-Zwerenz, A.; Leidgens, V.; Feliciello, G.; Klein, C.A.; Hau, P. Tumor Cell Invasion in Glioblastoma. Int. J. Mol. Sci. 2020, 21, 1932. [Google Scholar] [CrossRef] [PubMed]
- Anjum, K.; Shagufta, B.I.; Abbas, S.Q.; Patel, S.; Khan, I.; Shah, S.A.A.; Akhter, N.; Hassan, S.S.U. Current status and future therapeutic perspectives of glioblastoma multiforme (GBM) therapy: A review. Biomed. Pharmacother. 2017, 92, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Alifieris, C.; Trafalis, D.T. Glioblastoma multiforme: Pathogenesis and treatment. Pharmacol. Ther. 2015, 152, 63–82. [Google Scholar] [CrossRef] [PubMed]
- Garnier, D.; Renoult, O.; Alves-Guerra, M.-C.; Paris, F.; Pecqueur, C. Glioblastoma Stem-Like Cells, Metabolic Strategy to Kill a Challenging Target. Front. Oncol. 2019, 9, 118. [Google Scholar] [CrossRef]
- Lathia, J.D.; Mack, S.C.; Mulkearns-Hubert, E.E.; Valentim, C.L.L.; Rich, J.N. Cancer stem cells in glioblastoma. Genes Dev. 2015, 29, 1203–1217. [Google Scholar] [CrossRef]
- Gimple, R.C.; Bhargava, S.; Dixit, D.; Rich, J.N. Glioblastoma stem cells: Lessons from the tumor hierarchy in a lethal cancer. Genes Dev. 2019, 33, 591–609. [Google Scholar] [CrossRef] [PubMed]
- Aghajani, M.; Mansoori, B.; Mohammadi, A.; Asadzadeh, Z.; Baradaran, B. New emerging roles of CD133 in cancer stem cell: Signaling pathway and miRNA regulation. J. Cell. Physiol. 2019, 234, 21642–21661. [Google Scholar] [CrossRef]
- Barzegar Behrooz, A.; Syahir, A.; Ahmad, S. CD133: Beyond a cancer stem cell biomarker. J. Drug Target. 2019, 27, 257–269. [Google Scholar] [CrossRef]
- Ricci-Vitiani, L.; Pallini, R.; Larocca, L.M.; Lombardi, D.G.; Signore, M.; Pierconti, F.; Petrucci, G.; Montano, N.; Maira, G.; de Maria, R. Mesenchymal differentiation of glioblastoma stem cells. Cell Death Differ. 2008, 15, 1491–1498. [Google Scholar] [CrossRef]
- Aldaz, B.; Sagardoy, A.; Nogueira, L.; Guruceaga, E.; Grande, L.; Huse, J.T.; Aznar, M.A.; Díez-Valle, R.; Tejada-Solís, S.; Alonso, M.M.; et al. Involvement of miRNAs in the differentiation of human glioblastoma multiforme stem-like cells. PLoS ONE 2013, 8, e77098. [Google Scholar] [CrossRef]
- Shea, A.; Harish, V.; Afzal, Z.; Chijioke, J.; Kedir, H.; Dusmatova, S.; Roy, A.; Ramalinga, M.; Harris, B.; Blancato, J.; et al. MicroRNAs in glioblastoma multiforme pathogenesis and therapeutics. Cancer Med. 2016, 5, 1917–1946. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Buruiană, A.; Florian, Ș.I.; Florian, A.I.; Timiș, T.-L.; Mihu, C.M.; Miclăuș, M.; Oșan, S.; Hrapșa, I.; Cataniciu, R.C.; Farcaș, M.; et al. The Roles of miRNA in Glioblastoma Tumor Cell Communication: Diplomatic and Aggressive Negotiations. Int. J. Mol. Sci. 2020, 21, 1950. [Google Scholar] [CrossRef] [PubMed]
- Hannen, R.; Selmansberger, M.; Hauswald, M.; Pagenstecher, A.; Nist, A.; Stiewe, T.; Acker, T.; Carl, B.; Nimsky, C.; Bartsch, J.W. Comparative Transcriptomic Analysis of Temozolomide Resistant Primary GBM Stem-Like Cells and Recurrent GBM Identifies Up-Regulation of the Carbonic Anhydrase CA2 Gene as Resistance Factor. Cancers 2019, 11, 921. [Google Scholar] [CrossRef]
- Dong, F.; Eibach, M.; Bartsch, J.W.; Dolga, A.M.; Schlomann, U.; Conrad, C.; Schieber, S.; Schilling, O.; Biniossek, M.L.; Culmsee, C.; et al. The metalloprotease-disintegrin ADAM8 contributes to temozolomide chemoresistance and enhanced invasiveness of human glioblastoma cells. Neuro Oncol. 2015, 17, 1474–1485. [Google Scholar] [CrossRef] [PubMed]
- Schlomann, U.; Koller, G.; Conrad, C.; Ferdous, T.; Golfi, P.; Garcia, A.M.; Höfling, S.; Parsons, M.; Costa, P.; Soper, R.; et al. ADAM8 as a drug target in pancreatic cancer. Nat. Commun. 2015, 6, 6175. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer International Publishing: New York, NY, USA, 2016; ISBN 978-3-319-24275-0. [Google Scholar]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. circlize Implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef] [PubMed]
- Gruszka, R.; Zakrzewska, M. The Oncogenic Relevance of miR-17-92 Cluster and Its Paralogous miR-106b-25 and miR-106a-363 Clusters in Brain Tumors. Int. J. Mol. Sci. 2018, 19, 879. [Google Scholar] [CrossRef]
- Ernst, A.; Campos, B.; Meier, J.; Devens, F.; Liesenberg, F.; Wolter, M.; Reifenberger, G.; Herold-Mende, C.; Lichter, P.; Radlwimmer, B. De-repression of CTGF via the miR-17-92 cluster upon differentiation of human glioblastoma spheroid cultures. Oncogene 2010, 29, 3411–3422. [Google Scholar] [CrossRef]
- Schraivogel, D.; Weinmann, L.; Beier, D.; Tabatabai, G.; Eichner, A.; Zhu, J.Y.; Anton, M.; Sixt, M.; Weller, M.; Beier, C.P.; et al. CAMTA1 is a novel tumour suppressor regulated by miR-9/9* in glioblastoma stem cells. EMBO J. 2011, 30, 4309–4322. [Google Scholar] [CrossRef]
- Li, H.; Yang, B.B. Stress response of glioblastoma cells mediated by miR-17-5p targeting PTEN and the passenger strand miR-17-3p targeting MDM2. Oncotarget 2012, 3, 1653–1668. [Google Scholar] [CrossRef]
- Kehl, T.; Kern, F.; Backes, C.; Fehlmann, T.; Stöckel, D.; Meese, E.; Lenhof, H.-P.; Keller, A. miRPathDB 2.0: A novel release of the miRNA Pathway Dictionary Database. Nucleic Acids Res. 2020, 48, D142–D147. [Google Scholar] [CrossRef]
- De La Rocha, A.M.A.; González-Huarriz, M.; Guruceaga, E.; Mihelson, N.; Tejada-Solís, S.; Díez-Valle, R.; Martínez-Vélez, N.; Fueyo, J.; Gomez-Manzano, C.; Alonso, M.M.; et al. miR-425-5p, a SOX2 target, regulates the expression of FOXJ3 and RAB31 and promotes the survival of GSCs. Arch. Clin. Biomed. Res. 2020, 4, 221–238. [Google Scholar] [CrossRef]
- Zhou, J.-S.; Yang, Z.-S.; Cheng, S.-Y.; Yu, J.-H.; Huang, C.-J.; Feng, Q. miRNA-425-5p enhances lung cancer growth via the PTEN/PI3K/AKT signaling axis. BMC Pulm. Med. 2020, 20, 223. [Google Scholar] [CrossRef] [PubMed]
- Quintavalle, C.; Donnarumma, E.; Iaboni, M.; Roscigno, G.; Garofalo, M.; Romano, G.; Fiore, D.; de Marinis, P.; Croce, C.M.; Condorelli, G. Effect of miR-21 and miR-30b/c on TRAIL-induced apoptosis in glioma cells. Oncogene 2013, 32, 4001–4008. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, X.; Zhuang, S. miR-30c Impedes Glioblastoma Cell Proliferation and Migration by Targeting SOX9. Oncol. Res. 2019, 27, 165–171. [Google Scholar] [CrossRef]
- Gheidari, F.; Arefian, E.; Adegani, F.J.; Kalhori, M.R.; Seyedjafari, E.; Kabiri, M.; Teimoori-Toolabi, L.; Soleimani, M. miR-424 induces apoptosis in glioblastoma cells and targets AKT1 and RAF1 oncogenes from the ERBB signaling pathway. Eur. J. Pharmacol. 2021, 906, 174273. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhang, H.; Tan, L.; Yang, Y.; Wang, H.; Zhao, Q.; Lu, J. FAM87A as a Competing Endogenous RNA of miR-424-5p Suppresses Glioma Progression by Regulating PPM1H. Comput. Math. Methods Med. 2021, 2021, 7952922. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Wang, X.-B.; Zhang, Y.-H.; Zhou, Y.-M.; Yin, Q.; Yao, W.-C. MicroRNA-424 inhibits cell migration, invasion and epithelial-mesenchymal transition in human glioma by targeting KIF23 and functions as a novel prognostic predictor. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 6369–6378. [Google Scholar] [CrossRef]
- Wang, H.; Ren, S.; Xu, Y.; Miao, W.; Huang, X.; Qu, Z.; Li, J.; Liu, X.; Kong, P. MicroRNA-195 reverses the resistance to temozolomide through targeting cyclin E1 in glioma cells. Anticancer. Drugs 2019, 30, 81–88. [Google Scholar] [CrossRef]
- Xu, N.; Liu, B.; Lian, C.; Doycheva, D.M.; Fu, Z.; Liu, Y.; Zhou, J.; He, Z.; Yang, Z.; Huang, Q.; et al. Long noncoding RNA AC003092.1 promotes temozolomide chemosensitivity through miR-195/TFPI-2 signaling modulation in glioblastoma. Cell Death Dis. 2018, 9, 1139. [Google Scholar] [CrossRef]
- Liu, K.; Deng, Y.; Yang, Y.; Wang, H.; Zhou, P. MicorRNA-195 links long non-coding RNA SEMA3B antisense RNA 1 (head to head) and cyclin D1 to regulate the proliferation of glioblastoma cells. Bioengineered 2022, 13, 8798–8805. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Liang, X.; Li, X.; Zhang, Y.; Sun, Z.; Liu, Y.; Wang, J. MicroRNA-195: A review of its role in cancers. OncoTargets Ther. 2018, 11, 7109–7123. [Google Scholar] [CrossRef]
- Wang, X.-R.; Luo, H.; Li, H.-L.; Cao, L.; Wang, X.-F.; Yan, W.; Wang, Y.-Y.; Zhang, J.-X.; Jiang, T.; Kang, C.-S.; et al. Overexpressed let-7a inhibits glioma cell malignancy by directly targeting K-ras, independently of PTEN. Neuro Oncol. 2013, 15, 1491–1501. [Google Scholar] [CrossRef]
- Song, H.; Zhang, Y.; Liu, N.; Zhang, D.; Wan, C.; Zhao, S.; Kong, Y.; Yuan, L. Let-7b inhibits the malignant behavior of glioma cells and glioma stem-like cells via downregulation of E2F2. J. Physiol. Biochem. 2016, 72, 733–744. [Google Scholar] [CrossRef]
- Degrauwe, N.; Schlumpf, T.B.; Janiszewska, M.; Martin, P.; Cauderay, A.; Provero, P.; Riggi, N.; Suvà, M.-L.; Paro, R.; Stamenkovic, I. The RNA Binding Protein IMP2 Preserves Glioblastoma Stem Cells by Preventing let-7 Target Gene Silencing. Cell Rep. 2016, 15, 1634–1647. [Google Scholar] [CrossRef] [PubMed]
- Ventayol, M.; Viñas, J.L.; Sola, A.; Jung, M.; Brüne, B.; Pi, F.; Mastora, C.; Hotter, G. miRNA let-7e targeting MMP9 is involved in adipose-derived stem cell differentiation toward epithelia. Cell Death Dis. 2014, 5, e1048. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Zhu, J.; Zaorsky, N.G.; Deng, Y.; Wu, X.; Liu, Y.; Liu, F.; Cai, G.; Gu, W.; Shen, L.; et al. MicroRNA-223 enhances radiation sensitivity of U87MG cells in vitro and in vivo by targeting ataxia telangiectasia mutated. Int. J. Radiat. Oncol. Biol. Phys. 2014, 88, 955–960. [Google Scholar] [CrossRef]
- Boyd, N.H.; Tran, A.N.; Bernstock, J.D.; Etminan, T.; Jones, A.B.; Gillespie, G.Y.; Friedman, G.K.; Hjelmeland, A.B. Glioma stem cells and their roles within the hypoxic tumor microenvironment. Theranostics 2021, 11, 665–683. [Google Scholar] [CrossRef]
- Karsy, M.; Arslan, E.; Moy, F. Current Progress on Understanding MicroRNAs in Glioblastoma Multiforme. Genes Cancer 2012, 3, 3–15. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Zhao, Z.; Cai, Q.; Zhang, Y.; Zhang, P.; Shi, S.; Xie, H.; Peng, X.; Yin, W.; Tao, Y.; et al. miRNA-based biomarkers, therapies, and resistance in Cancer. Int. J. Biol. Sci. 2020, 16, 2628–2647. [Google Scholar] [CrossRef]
- Da Ros, M.; de Gregorio, V.; Iorio, A.; Giunti, L.; Guidi, M.; de Martino, M.; Genitori, L.; Sardi, I. Glioblastoma Chemoresistance: The Double Play by Microenvironment and Blood-Brain Barrier. Int. J. Mol. Sci. 2018, 19, 2879. [Google Scholar] [CrossRef]
- Olive, V.; Jiang, I.; He, L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int. J. Biochem. Cell Biol. 2010, 42, 1348–1354. [Google Scholar] [CrossRef] [PubMed]
- Montano, N. Biomarkers in Glioblastoma multiforme. Evidences from a literature review. J. Clin. Transl. Res. 2016, 2, 3–10. [Google Scholar] [CrossRef]
- Kudinov, A.E.; Karanicolas, J.; Golemis, E.A.; Boumber, Y. Musashi RNA-Binding Proteins as Cancer Drivers and Novel Therapeutic Targets. Clin. Cancer Res. 2017, 23, 2143–2153. [Google Scholar] [CrossRef]
- Ferri, A.; Stagni, V.; Barilà, D. Targeting the DNA Damage Response to Overcome Cancer Drug Resistance in Glioblastoma. Int. J. Mol. Sci. 2020, 21, 4910. [Google Scholar] [CrossRef]
| GSC Line | Age at Diagnosis (in Years) | Sex | Survival in Days | Localization | MGMT Promotor Methylation Status | Ki67-Li | p53 Accumulation |
|---|---|---|---|---|---|---|---|
| 2016/240 | 48 | female | 641 | right frontal lobe | methylated | up to 10% | moderately accumulated |
| 2017/151 | 66 | male | 126 | right temporal lobe and right insula | methylated | up to 20% | accumulated |
| 2017/74 | 61 | male | 398 | right temporal lobe | not methylated | up to 50% | moderately accumulated |
| miRNA | Upregulated in | Fold Regulation | Role in GBM | Role in GSCs | mRNA Target |
|---|---|---|---|---|---|
| miR-17-5p | GSCs | −8.05 | Onco-miRNA [20] Highly expressed in GBM, correlated with poor prognosis [20] | Highly expressed in GSCs [21,22,23] Increases GSC proliferation [21] | PTEN [23] GFAP (predicted) [24] |
| miR-425-5p | GSCs | −4.00 | Onco-miRNA [25] Associated with poor prognosis [25] | Highly expressed in GSCs [25] Promotes neurosphere formation and GSC survival [25] | PTEN [26] GFAP (predicted) [24] |
| miR-30c-5p | GSCs | −4.00 | Conflicting data Promotes chemoresistance [27] Inhibition of proliferation, migration, and invasion [28] Downregulation in GBM tissue [28] | Unexplored | SOX-9 [28] |
| miR-424-5p | GSCs | −4.04 | Conflicting data Effects on migration and proliferation, induction of apoptosis [29,30,31] Inhibition of epithelial-to-mesenchymal transition (EMT) and tumor growth [31] Enhances chemoresistance [30] | Unexplored | Akt-1, RAF1 [29] GFAP (predicted) [24] |
| miR-195-5p | GSCs | −4.05 | Conflicting data Affects response to TMZ [32,33] Inhibits proliferation [34] Upregulated in recurrent GBM samples [35] | Unexplored | Cyclin E1 [32] Cyclin D1 [34] GFAP (predicted) [24] |
| let-7a-5p | Differentiated cells | 4.01 | Tumor-suppressor miRNA family [36,37] Inhibition of tumor cells’ migration, proliferation, and invasion [36,37] Promotes cell cycle arrest and apoptosis [36] | Low expression in GSCs [38] Inhibition of neurosphere growth [37] | K-Ras [36] Musashi-2 (predicted) [24] |
| let-7e-5p | 3.97 | MMP9 [39] Musashi-2 (predicted) [24] | |||
| let-7b-5p | 3.97 | E2F2 [37] Musashi-2, Musashi-1 (predicted) [24] | |||
| let-7c-5p | 4.00 | Musashi-2 (predicted) [24] | |||
| miR-233-3p | Differentiated cells | 3.09 | Tumor-suppressor miRNA [40] Enhances radiation sensitivity of GBM cells [40] | Unexplored | ATM [40] Musashi-2 (predicted) [24] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Evers, L.; Schäfer, A.; Pini, R.; Zhao, K.; Stei, S.; Nimsky, C.; Bartsch, J.W. Identification of Dysregulated microRNAs in Glioblastoma Stem-like Cells. Brain Sci. 2023, 13, 350. https://doi.org/10.3390/brainsci13020350
Evers L, Schäfer A, Pini R, Zhao K, Stei S, Nimsky C, Bartsch JW. Identification of Dysregulated microRNAs in Glioblastoma Stem-like Cells. Brain Sciences. 2023; 13(2):350. https://doi.org/10.3390/brainsci13020350
Chicago/Turabian StyleEvers, Lara, Agnes Schäfer, Raffaella Pini, Kai Zhao, Susanne Stei, Christopher Nimsky, and Jörg W. Bartsch. 2023. "Identification of Dysregulated microRNAs in Glioblastoma Stem-like Cells" Brain Sciences 13, no. 2: 350. https://doi.org/10.3390/brainsci13020350
APA StyleEvers, L., Schäfer, A., Pini, R., Zhao, K., Stei, S., Nimsky, C., & Bartsch, J. W. (2023). Identification of Dysregulated microRNAs in Glioblastoma Stem-like Cells. Brain Sciences, 13(2), 350. https://doi.org/10.3390/brainsci13020350