Axial Attention Convolutional Neural Network for Brain Tumor Segmentation with Multi-Modality MRI Scans
Abstract
1. Introduction
2. Related Work
2.1. DL-Based Methods for Brain Tumor Segmentation
2.2. The Attention-Based Module for Brain Tumor Segmentation
3. Methodology
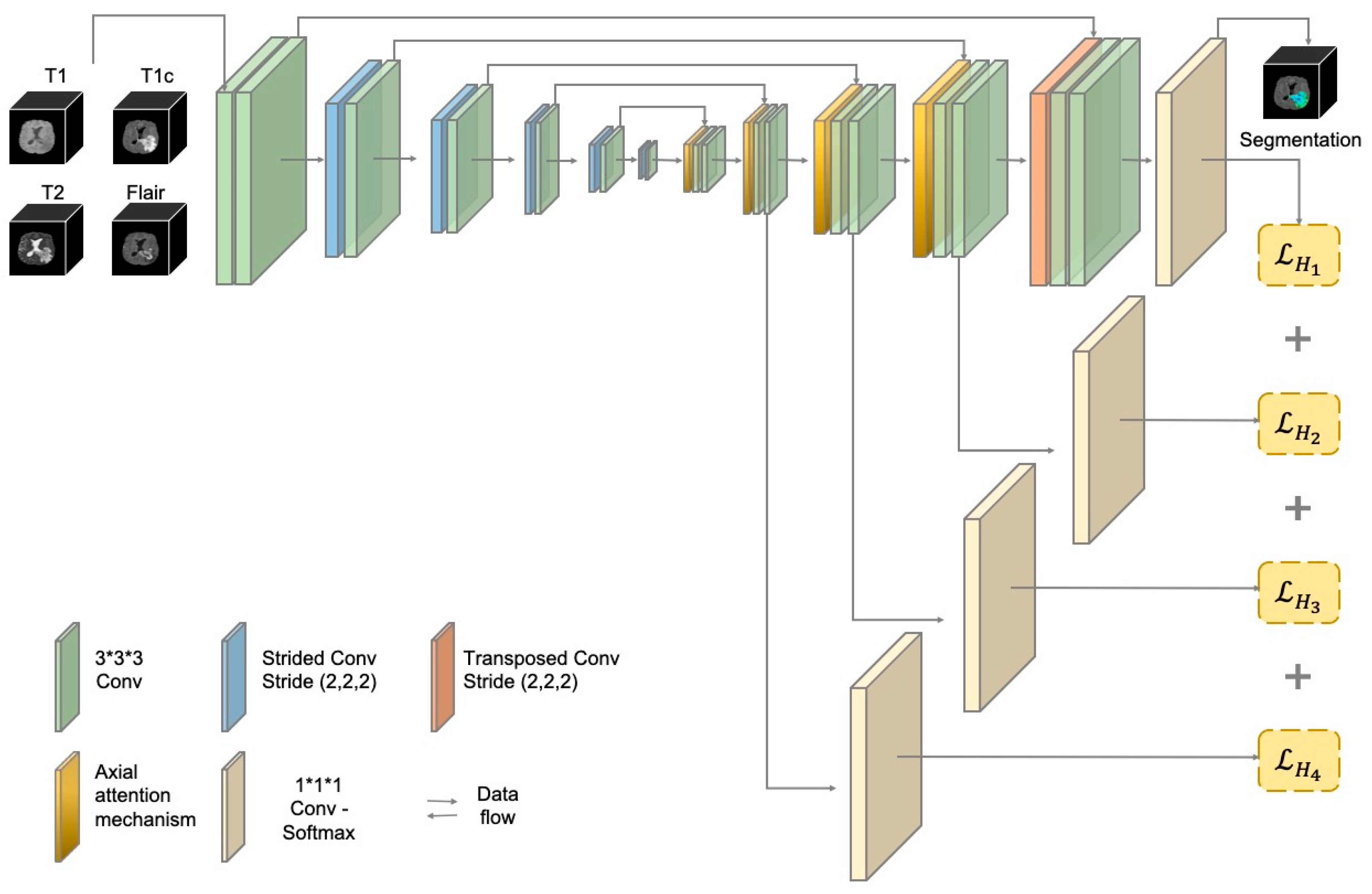
3.1. The Structure of the AABTS-Net
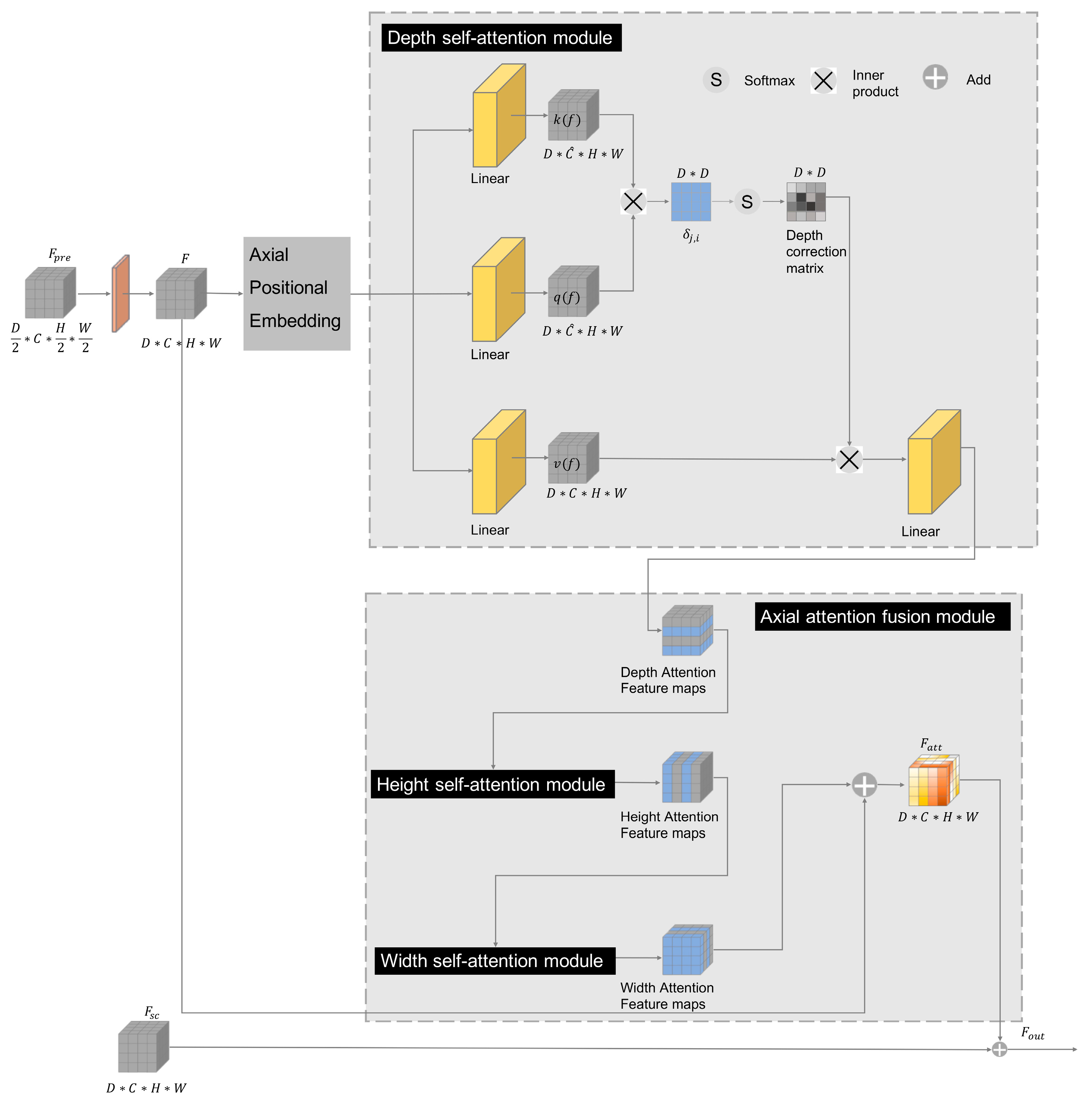
3.2. The Axial Attention Mechanism
3.3. The Deep Supervision (DS) Mechanism
3.4. The Hybrid Loss Function
3.5. Experiments
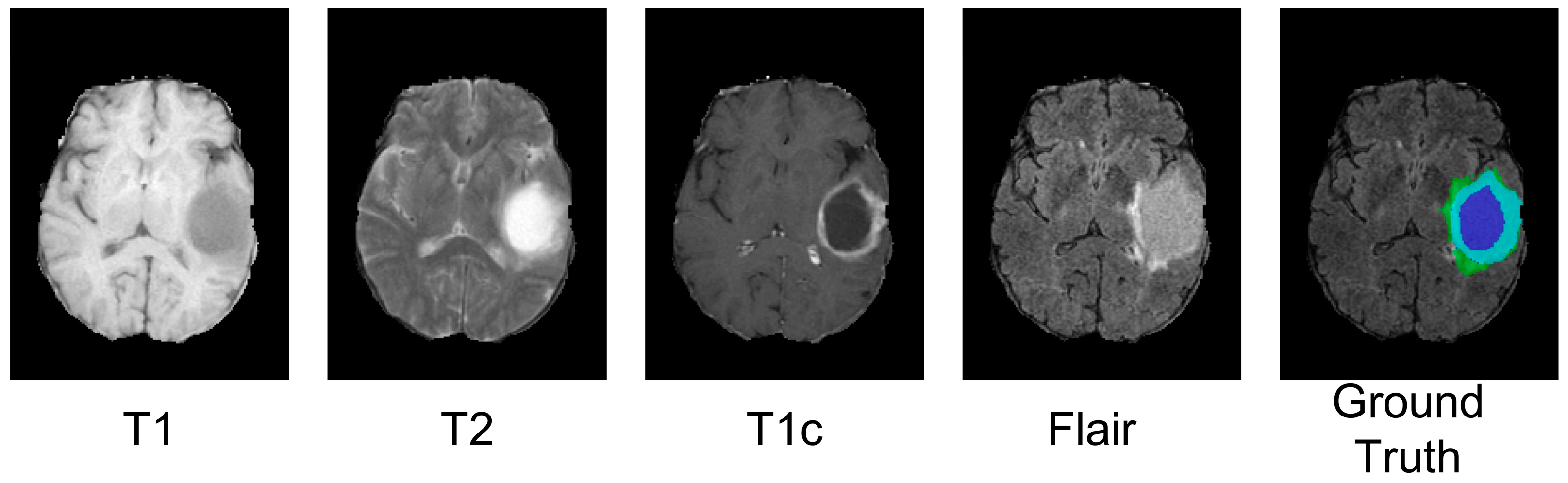
3.5.1. Datasets and Pre-Processing
3.5.2. Evaluation Metrics
3.5.3. The Training Details
4. Results
4.1. Comparison with State-of-the-Art Methods
4.1.1. The Experiment Results on the BraTS 2019 Dataset
4.1.2. The Experiment Results on the Brats 2021 Dataset
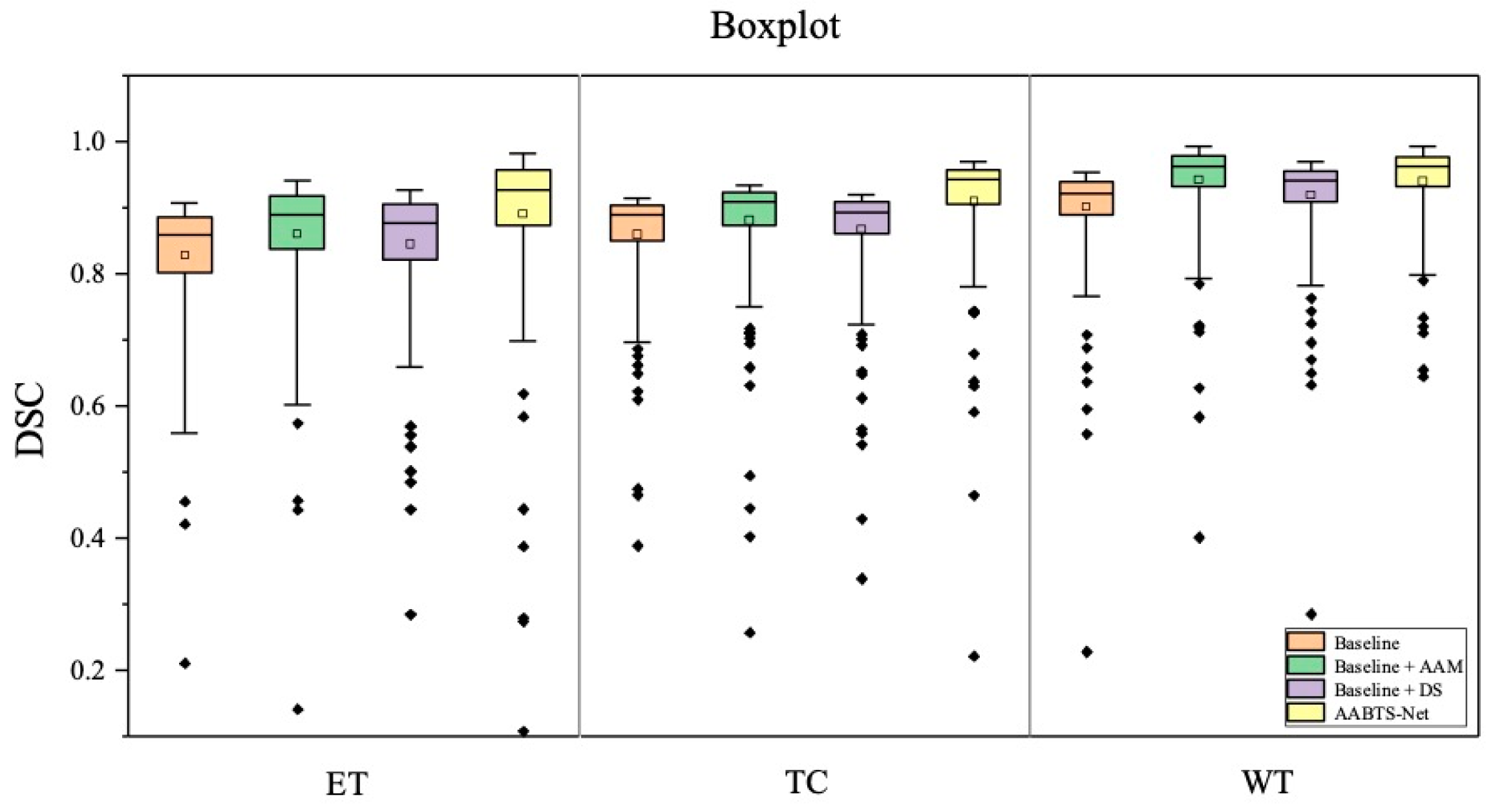
4.2. The Analysis of Key Components
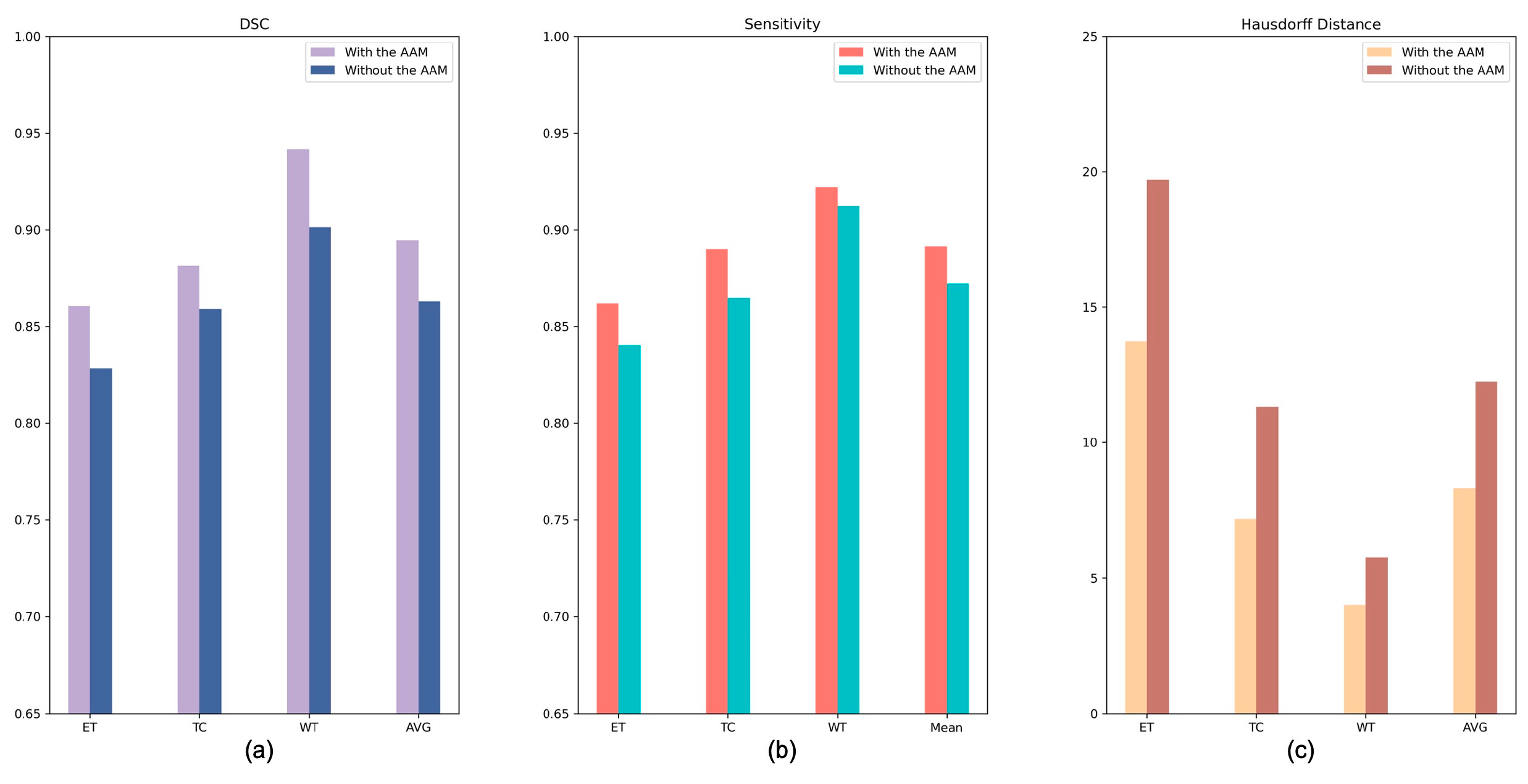
4.2.1. Analysis of the Axial Attention Mechanism
4.2.2. Analysis of the Deep Supervision Mechanism
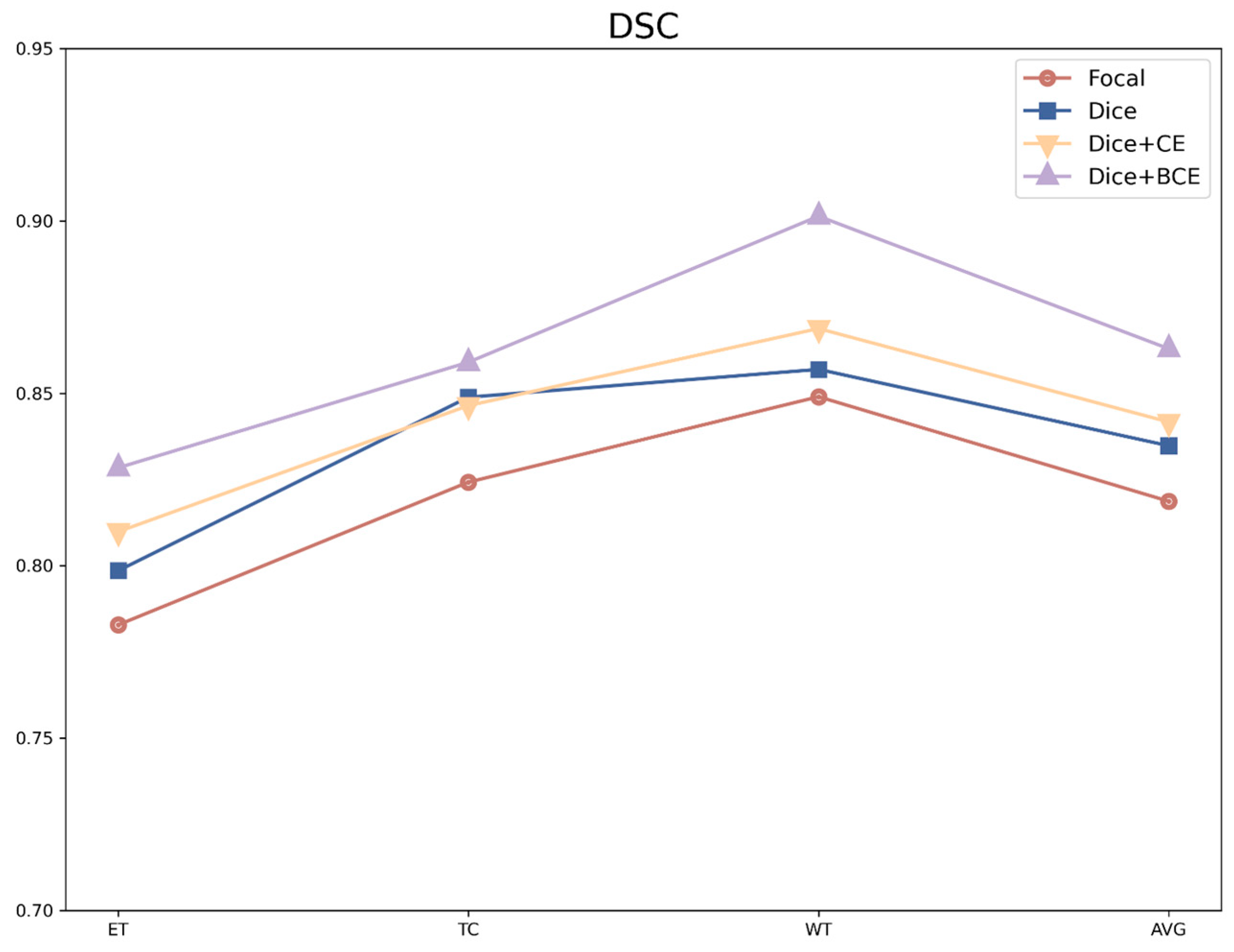
4.2.3. Analysis of the Different Loss Functions
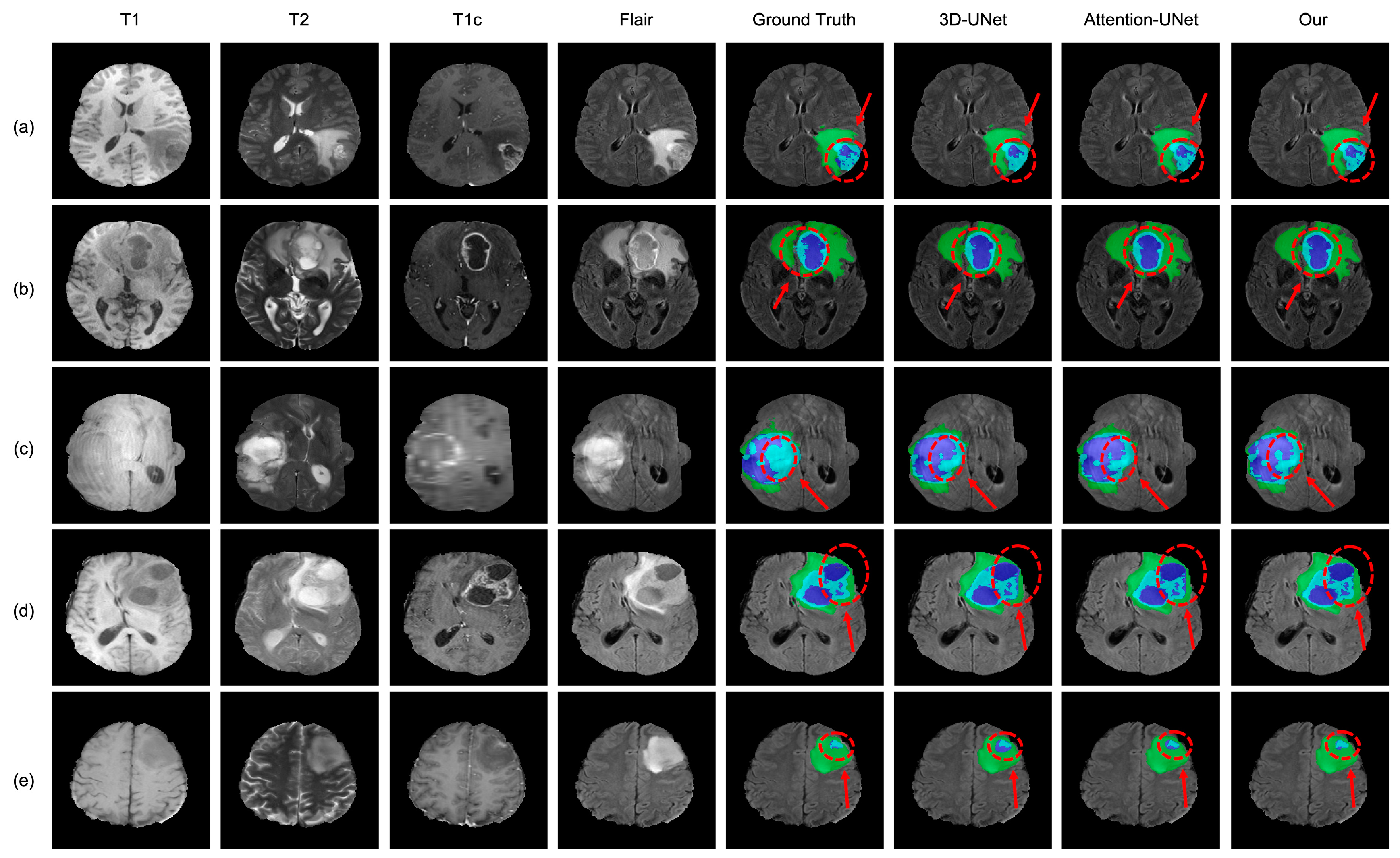
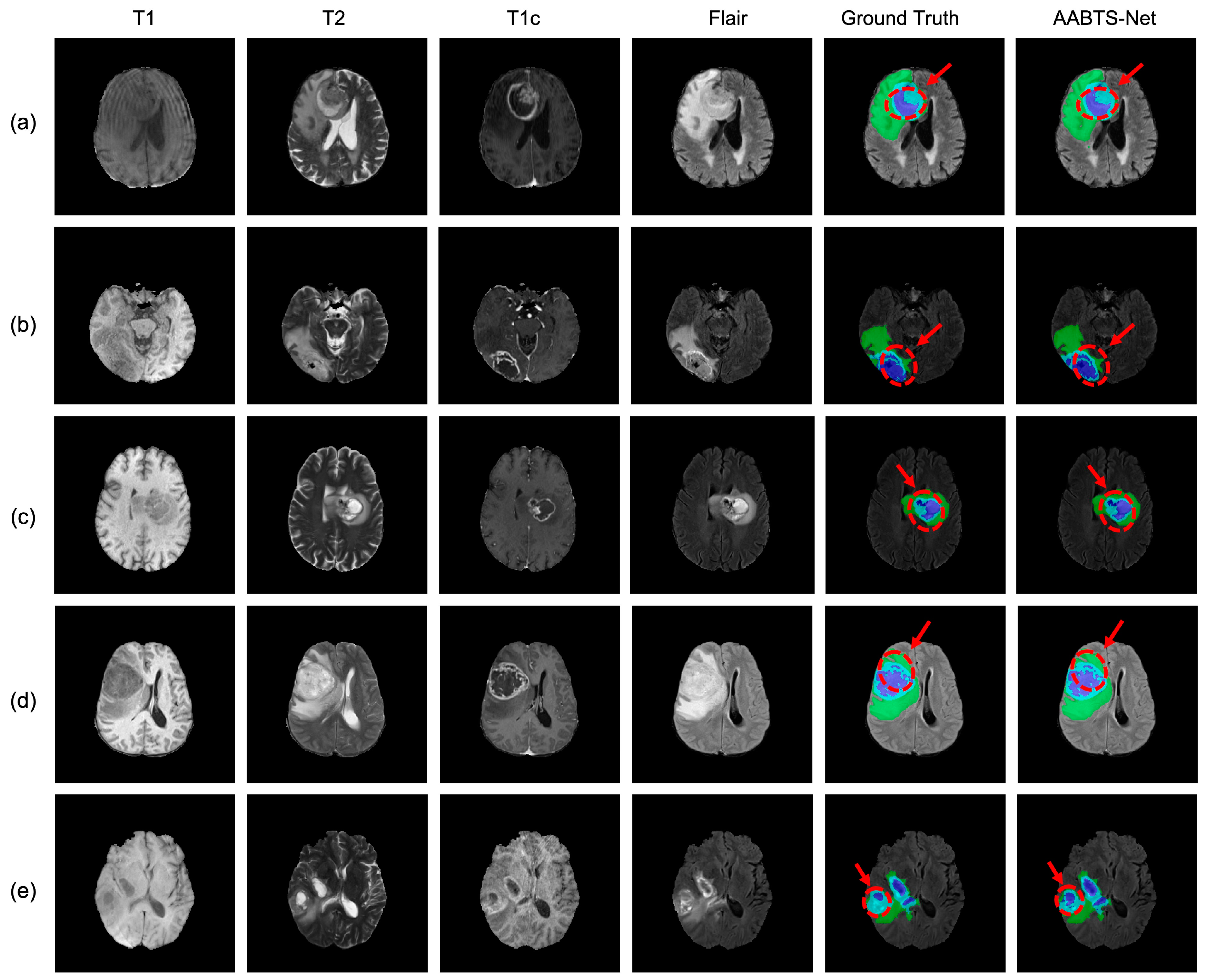
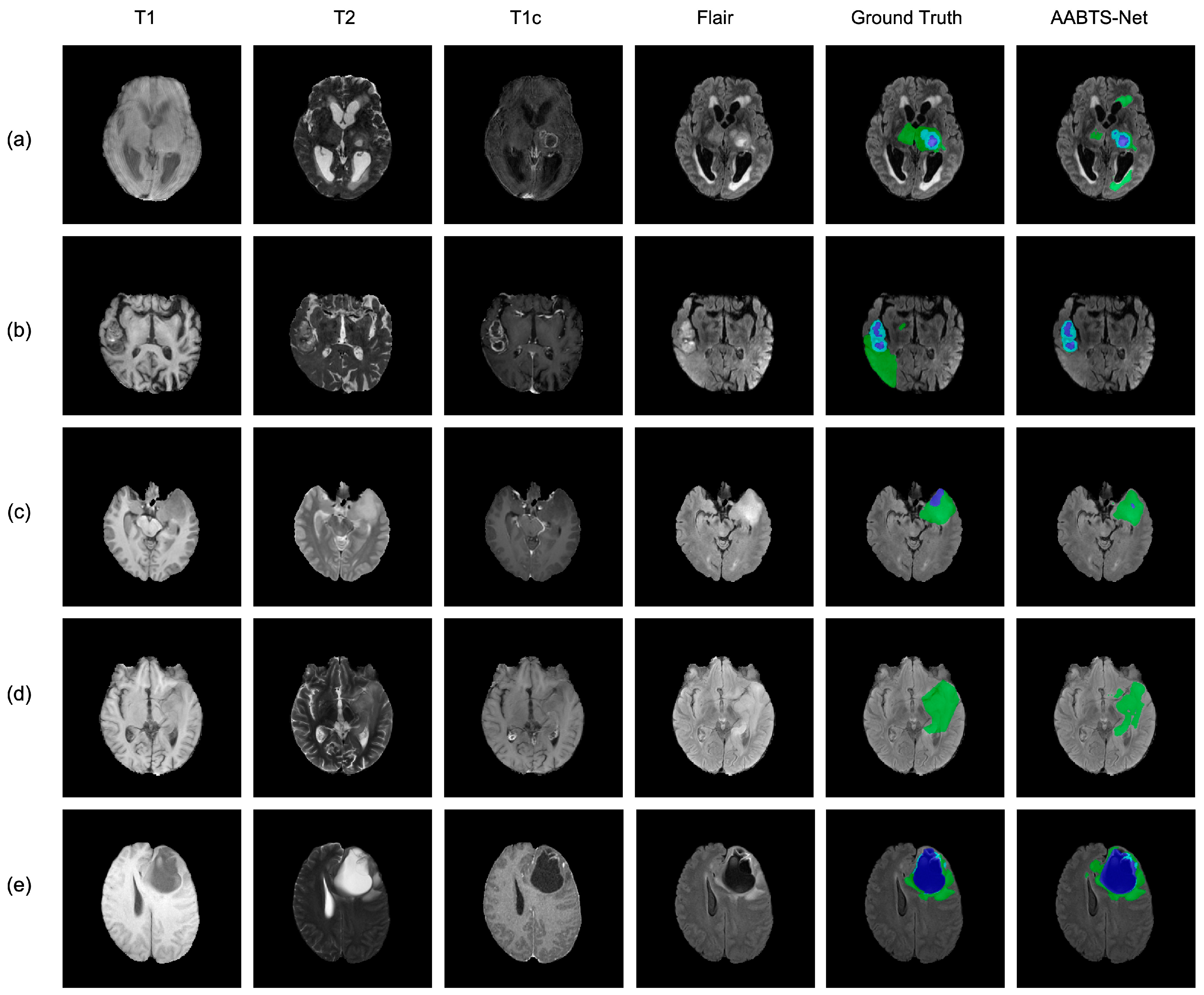
4.3. Analysis of the Bad Tumor Segmentation Results
5. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Holland, E.C. Progenitor Cells and Glioma Formation. Curr. Opin. Neurol. 2001, 14, 683–688. [Google Scholar] [CrossRef] [PubMed]
- Kleihues, P.; Burger, P.C.; Scheithauer, B.W. The New Who Classification of Brain Tumours. Brain Pathol. 1993, 3, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Bauer, S.; Wiest, R.; Nolte, L.-P.; Reyes, M. A Survey of Mri-Based Medical Image Analysis for Brain Tumor Studies. Phys. Med. Biol. 2013, 58, R97. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.-P.; Lauterbur, P.C. Principles of Magnetic Resonance Imaging: A Signal Processing Perspective; The Institute of Electrical and Electronics Engineers Press: Manhattan, NY, USA, 2000. [Google Scholar]
- Jayadevappa, D.; Srinivas Kumar, S.; Murty, D. Medical Image Segmentation Algorithms Using Deformable Models: A Review. IETE Tech. Rev. 2011, 28, 248–255. [Google Scholar] [CrossRef]
- Mansourvar, M.; Ismail, M.A.; Herawan, T.; Gopal Raj, R.; Abdul Kareem, S.; Nasaruddin, F.H. Automated Bone Age Assessment: Motivation, Taxonomies, and Challenges. Comput. Math Methods Med. 2013, 2013, 391626. [Google Scholar] [CrossRef]
- Kamnitsas, K.; Ledig, C.; Newcombe, V.F.; Simpson, J.P.; Kane, A.D.; Menon, D.K.; Rueckert, D.; Glocker, B. Efficient Multi-Scale 3d Cnn with Fully Connected Crf for Accurate Brain Lesion Segmentation. Med. Image Anal. 2017, 36, 61–78. [Google Scholar] [CrossRef]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.M.; Larochelle, H. Brain Tumor Segmentation with Deep Neural Networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar] [CrossRef]
- Pereira, S.; Pinto, A.; Alves, V.; Silva, C.A. Brain Tumor Segmentation Using Convolutional Neural Networks in Mri Images. IEEE Trans. Med. Imaging 2016, 35, 1240–1251. [Google Scholar] [CrossRef]
- Zhao, X.; Wu, Y.; Song, G.; Li, Z.; Zhang, Y.; Fan, Y. A Deep Learning Model Integrating Fcnns and Crfs for Brain Tumor Segmentation. Med. Image Anal. 2018, 43, 98–111. [Google Scholar] [CrossRef]
- Chen, L.; Wu, Y.; DSouza, A.M.; Abidin, A.Z.; Wismüller, A.; Xu, C. (Eds.) Mri Tumor Segmentation with Densely Connected 3d Cnn. In Medical Imaging 2018: Image Processing; SPIE: Bellingham, WA, USA, 2018. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. (Eds.) U-Net: Convolutional Networks for Biomedical Image Segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Manhattan, NY, USA, 2015. [Google Scholar]
- Wang, F.; Jiang, R.; Zheng, L.; Meng, C.; Biswal, B. (Eds.) 3d U-Net Based Brain Tumor Segmentation and Survival Days Prediction. In International MICCAI Brainlesion Workshop; Springer: Manhattan, NY, USA, 2019. [Google Scholar]
- Cheng, X.; Jiang, Z.; Sun, Q.; Zhang, J. (Eds.) Memory-Efficient Cascade 3d U-Net for Brain Tumor Segmentation. In International Miccai Brainlesion Workshop; Springer: Manhattan, NY, USA, 2019. [Google Scholar]
- Chen, W.; Liu, B.; Peng, S.; Sun, J.; Qiao, X. (Eds.) S3d-Unet: Separable 3d U-Net for Brain Tumor Segmentation. In International MICCAI Brainlesion Workshop; Springer: Manhattan, NY, USA, 2018. [Google Scholar]
- Jiang, Z.; Ding, C.; Liu, M.; Tao, D. (Eds.) Two-Stage Cascaded U-Net: 1st Place Solution to Brats Challenge 2019 Segmentation Task. In International MICCAI Brainlesion Workshop; Springer: Manhattan, NY, USA, 2019. [Google Scholar]
- Zhou, T.; Canu, S.; Vera, P.; Ruan, S. Latent Correlation Representation Learning for Brain Tumor Segmentation with Missing Mri Modalities. IEEE Trans. Image Processing 2021, 30, 4263–4274. [Google Scholar] [CrossRef]
- Xu, W.; Yang, H.; Zhang, M.; Cao, Z.; Pan, X.; Liu, W. Brain Tumor Segmentation with Corner Attention and High-Dimensional Perceptual Loss. Biomed. Signal Processing Control. 2022, 73, 103438. [Google Scholar] [CrossRef]
- Zhang, J.; Jiang, Z.; Dong, J.; Hou, Y.; Liu, B. Attention Gate Resu-Net for Automatic Mri Brain Tumor Segmentation. IEEE Access 2020, 8, 58533–58545. [Google Scholar] [CrossRef]
- Mazumdar, I.; Mukherjee, J. Fully Automatic Mri Brain Tumor Segmentation Using Efficient Spatial Attention Convolutional Networks with Composite Loss. Neurocomputing 2022, 500, 243–254. [Google Scholar] [CrossRef]
- Kong, D.; Liu, X.; Wang, Y.; Li, D.; Xue, J. 3d Hierarchical Dual-Attention Fully Convolutional Networks with Hybrid Losses for Diverse Glioma Segmentation. Knowl.-Based Syst. 2022, 237, 107692. [Google Scholar] [CrossRef]
- AboElenein, N.M.; Piao, S.; Noor, A.; Ahmed, P.N. Mirau-Net: An Improved Neural Network Based on U-Net for Gliomas Segmentation. Signal Processing: Image Commun. 2022, 101, 116553. [Google Scholar] [CrossRef]
- Wang, W.; Chen, C.; Ding, M.; Yu, H.; Zha, S.; Li, J. (Eds.) Transbts: Multimodal Brain Tumor Segmentation Using Transformer. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Manhattan, NY, USA, 2021. [Google Scholar]
- Jia, Q.; Shu, H. (Eds.) Bitr-Unet: A Cnn-Transformer Combined Network for Mri Brain Tumor Segmentation. In International MICCAI Brainlesion Workshop; Springer: Manhattan, NY, USA, 2022. [Google Scholar]
- Zhou, Z.; Zhou, Y.; Wang, D.; Mu, J.; Zhou, H. Self-Attention Feature Fusion Network for Semantic Segmentation. Neurocomputing 2021, 453, 50–59. [Google Scholar] [CrossRef]
- Wang, H.; Zhu, Y.; Green, B.; Adam, H.; Yuille, A.; Chen, L.-C. (Eds.) Axial-Deeplab: Stand-Alone Axial-Attention for Panoptic Segmentation. In European Conference on Computer Vision; Springer: Manhattan, NY, USA, 2020. [Google Scholar]
- Liu, P.; Dou, Q.; Wang, Q.; Heng, P.-A. An Encoder-Decoder Neural Network with 3d Squeeze-and-Excitation and Deep Supervision for Brain Tumor Segmentation. IEEE Access 2020, 8, 34029–34037. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, Y.; Chen, W.; Chang, Y.; Gu, H.; Yu, B. Msmanet: A Multi-Scale Mesh Aggregation Network for Brain Tumor Segmentation. Appl. Soft Comput. 2021, 110, 107733. [Google Scholar] [CrossRef]
- Zhou, Z.; He, Z.; Shi, M.; Du, J.; Chen, D. 3d Dense Connectivity Network with Atrous Convolutional Feature Pyramid for Brain Tumor Segmentation in Magnetic Resonance Imaging of Human Heads. Comput Biol. Med. 2020, 121, 103766. [Google Scholar] [CrossRef]
- Chen, H.; Qin, Z.; Ding, Y.; Tian, L.; Qin, Z. Brain Tumor Segmentation with Deep Convolutional Symmetric Neural Network. Neurocomputing 2020, 392, 305–313. [Google Scholar] [CrossRef]
- Yang, Q.; Guo, X.; Chen, Z.; Woo, P.Y.; Yuan, Y. D2-Net: Dual Disentanglement Network for Brain Tumor Segmentation with Missing Modalities. IEEE Trans. Med. Imaging 2022, 41, 2953–2964. [Google Scholar] [CrossRef] [PubMed]
- Baid, U.; Ghodasara, S.; Mohan, S.; Bilello, M.; Calabrese, E.; Colak, E.; Farahani, K.; Kalpathy-Cramer, J.; Kitamura, F.C.; Pati, S.; et al. The Rsna-Asnr-Miccai Brats 2021 Benchmark on Brain Tumor Segmentation and Radiogenomic Classification. arXiv preprint 2021, arXiv:210702314. [Google Scholar]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (Brats). IEEE Trans. Med Imaging 2014, 34, 1993–2024. [Google Scholar] [CrossRef]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.S.; Freymann, J.B.; Farahani, K.; Davatzikos, C. Advancing the Cancer Genome Atlas Glioma Mri Collections with Expert Segmentation Labels and Radiomic Features. Sci. Data 2017, 4, 1–13. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXivpreprint 2014, arXiv:14126980. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. (Eds.) 3d U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Manhattan, NY, USA, 2016. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning Where to Look for the Pancreas. arXiv preprint 2018, arXiv:180403999. [Google Scholar]
- Shi, W.; Pang, E.; Wu, Q.; Lin, F. (Eds.) Brain Tumor Segmentation Using Dense Channels 2d U-Net and Multiple Feature Extraction Network. In International MICCAI Brainlesion Workshop; Springer: Manhattan, NY, USA, 2019. [Google Scholar]
- Zhao, Y.-X.; Zhang, Y.-M.; Liu, C.-L. (Eds.) Bag of Tricks for 3d Mri Brain Tumor Segmentation. In International MICCAI Brainlesion Workshop; Springer: Manhattan, NY, USA, 2019. [Google Scholar]
- Ribalta Lorenzo, P.; Marcinkiewicz, M.; Nalepa, J. (Eds.) Multi-Modal U-Nets with Boundary Loss and Pre-Training for Brain Tumor Segmentation. In International MICCAI Brainlesion Workshop; Springer: Manhattan, NY, USA, 2019. [Google Scholar]
- Ahmad, P.; Qamar, S.; Hashemi, S.R.; Shen, L. (Eds.) Hybrid Labels for Brain Tumor Segmentation. In International MICCAI Brainlesion Workshop; Springer: Manhattan, NY, USA, 2019. [Google Scholar]
- Choi, Y.; Al-masni, M.A.; Kim, D.-H. (Eds.) 3d Cmm-Net with Deeper Encoder for Semantic Segmentation of Brain Tumors in Brats 2021 Challenge; Springer International Publishing: Cham, Switzerland, 2022. [Google Scholar]
- Jiang, Y.; Zhang, Y.; Lin, X.; Dong, J.; Cheng, T.; Liang, J. Swinbts: A Method for 3d Multimodal Brain Tumor Segmentation Using Swin Transformer. Brain Sci. 2022, 12, 797. [Google Scholar] [CrossRef]
- Singh, G.; Phophalia, A. (Eds.) Multimodal Brain Tumor Segmentation Using Modified Unet Architecture; Springer International Publishing: Cham, Switzerland, 2022. [Google Scholar]
- Akbar, A.S.; Fatichah, C.; Suciati, N. (Eds.) Unet3d with multiple Atrous Convolutions Attention Block for Brain Tumor Segmentation; Springer International Publishing: Cham, Switzerland, 2022. [Google Scholar]
- Singh, H.S. (Ed.) Brain Tumor Segmentation Using Attention Activated U-Net with Positive Mining; Springer International Publishing: Cham, Switzerland, 2022. [Google Scholar]
- Luu, H.M.; Park, S.-H. (Eds.) Extending Nn-Unet for Brain Tumor Segmentation. In International MICCAI Brainlesion Workshop; Springer: Manhattan, NY, USA, 2022. [Google Scholar]
- Bukhari, S.T.; Mohy-Ud-Din, H. E1d3 U-Net for Brain Tumor Segmentation: Submission to the Rsna-Asnr-Miccai Brats 2021 Challenge. In International MICCAI Brainlesion Workshop 2022; Springer: Cham, Switzerland, 2021. [Google Scholar]



| Dataset | Training Dataset | Validation Dataset * |
|---|---|---|
| BraTS 2019 | 335 cases | 125 cases |
| BraTS 2021 | 1251 cases | 219 cases |
| Name | Parameter |
|---|---|
| Batch_size | 2 |
| Epoch | 300 |
| Learning rate | 0.0001 |
| GPU memory | 16 GB |
| Patch size | 128 × 128 × 128 |
| Optimizer | Adam |
| Framework | Pytorch |
| Methods | DSC | HD | ||||||
|---|---|---|---|---|---|---|---|---|
| ET | TC | WT | AVG | ET | TC | WT | AVG | |
| 3D UNet [36] | 0.709 | 0.725 | 0.874 | 0.769 | 5.062 | 8.719 | 9.432 | 7.738 |
| Attention UNet [37] | 0.760 | 0.772 | 0.888 | 0.807 | 5.202 | 7.756 | 8.258 | 7.072 |
| Shi et al. [38] | 0.691 | 0.770 | 0.887 | 0.783 | 5.888 | 12.192 | 21.190 | 13.090 |
| Zhao et al. [39] | 0.702 | 0.800 | 0.893 | 0.798 | 4.766 | 6.472 | 5.078 | 5.439 |
| Lorenzo et al. [40] | 0.663 | 0.751 | 0.890 | 0.768 | / | / | / | / |
| Ahmad et al. [41] | 0.623 | 0.758 | 0.852 | 0.744 | 8.468 | 10.674 | 9.008 | 9.383 |
| Our Method | 0.777 | 0.838 | 0.911 | 0.842 | 3.246 | 6.028 | 3.988 | 4.421 |
| Methods | DSC | HD | ||||||
|---|---|---|---|---|---|---|---|---|
| ET | TC | WT | AVG | ET | TC | WT | AVG | |
| ESA-Net [20] | 0.812 | 0.852 | 0.907 | 0.857 | 26.470 | 13.830 | 5.940 | 15.413 |
| 3D CMM-Net [42] | 0.732 | 0.751 | 0.874 | 0.786 | 35.001 | 24.638 | 10.161 | 23.267 |
| SwinBTS [43] | 0.832 | 0.848 | 0.918 | 0.866 | 16.030 | 14.510 | 3.650 | 11.397 |
| Singh et al. [44] | 0.730 | 0.760 | 0.870 | 0.787 | 30.500 | 14.700 | 6.290 | 17.163 |
| Akbar et al. [45] | 0.780 | 0.807 | 0.891 | 0.826 | 25.820 | 21.170 | 11.780 | 19.590 |
| Singh et al. [46] | 0.753 | 0.808 | 0.899 | 0.820 | 21.800 | 12.500 | 6.450 | 13.583 |
| Extending nn-UNet [47] | 0.845 | 0.878 | 0.928 | 0.884 | 20.730 | 7.623 | 3.470 | 10.608 |
| E1D3-UNet [48] | 0.818 | 0.863 | 0.923 | 0.868 | 18.240 | 9.620 | 4.340 | 10.733 |
| Bitr-UNet [24] | 0.819 | 0.843 | 0.910 | 0.857 | 17.847 | 16.689 | 4.508 | 13.015 |
| Our Method | 0.830 | 0.861 | 0.922 | 0.871 | 17.728 | 11.178 | 3.996 | 10.967 |
| Methods | DSC | HD | Sensitivity | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ET | TC | WT | AVG | ET | TC | WT | AVG | ET | TC | WT | AVG | |
| Baseline | 0.828 | 0.859 | 0.901 | 0.863 | 19.703 | 11.310 | 5.743 | 12.252 | 0.840 | 0.865 | 0.912 | 0.872 |
| Baseline + AAM | 0.860 | 0.881 | 0.942 | 0.894 | 13.728 | 7.178 | 3.996 | 8.301 | 0.862 | 0.890 | 0.922 | 0.891 |
| Baseline + DS | 0.845 | 0.867 | 0.919 | 0.877 | 14.583 | 8.042 | 3.723 | 8.783 | 0.854 | 0.882 | 0.927 | 0.887 |
| AABTS-Net | 0.890 | 0.909 | 0.940 | 0.913 | 10.132 | 7.132 | 3.343 | 6.869 | 0.905 | 0.915 | 0.952 | 0.924 |
| Cases | DSC | |||
|---|---|---|---|---|
| ET | TC | WT | AVG | |
| (a) BraTS2021_00493 | 0.885 | 0.924 | 0.270 | 0.693 |
| (b) BraTS2021_00494 | 0.964 | 0.990 | 0.730 | 0.895 |
| (c) BraTS2021_01666 | 0 | 0.738 | 0.916 | 0.551 |
| (d) BraTS2021_01179 | 1.000 | 0 | 0.769 | 0.590 |
| (e) BraTS2021_01293 | 0.541 | 0.929 | 0.874 | 0.781 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, W.; Li, D.; Lv, M.; Huang, P. Axial Attention Convolutional Neural Network for Brain Tumor Segmentation with Multi-Modality MRI Scans. Brain Sci. 2023, 13, 12. https://doi.org/10.3390/brainsci13010012
Tian W, Li D, Lv M, Huang P. Axial Attention Convolutional Neural Network for Brain Tumor Segmentation with Multi-Modality MRI Scans. Brain Sciences. 2023; 13(1):12. https://doi.org/10.3390/brainsci13010012
Chicago/Turabian StyleTian, Weiwei, Dengwang Li, Mengyu Lv, and Pu Huang. 2023. "Axial Attention Convolutional Neural Network for Brain Tumor Segmentation with Multi-Modality MRI Scans" Brain Sciences 13, no. 1: 12. https://doi.org/10.3390/brainsci13010012
APA StyleTian, W., Li, D., Lv, M., & Huang, P. (2023). Axial Attention Convolutional Neural Network for Brain Tumor Segmentation with Multi-Modality MRI Scans. Brain Sciences, 13(1), 12. https://doi.org/10.3390/brainsci13010012






