Application of Micro-Western Array for Identifying Different Serum Protein Expression Profile among Healthy Control, Alzheimer’s Disease Patients and Patients’ Adult Children
Abstract
1. Introduction
2. Results
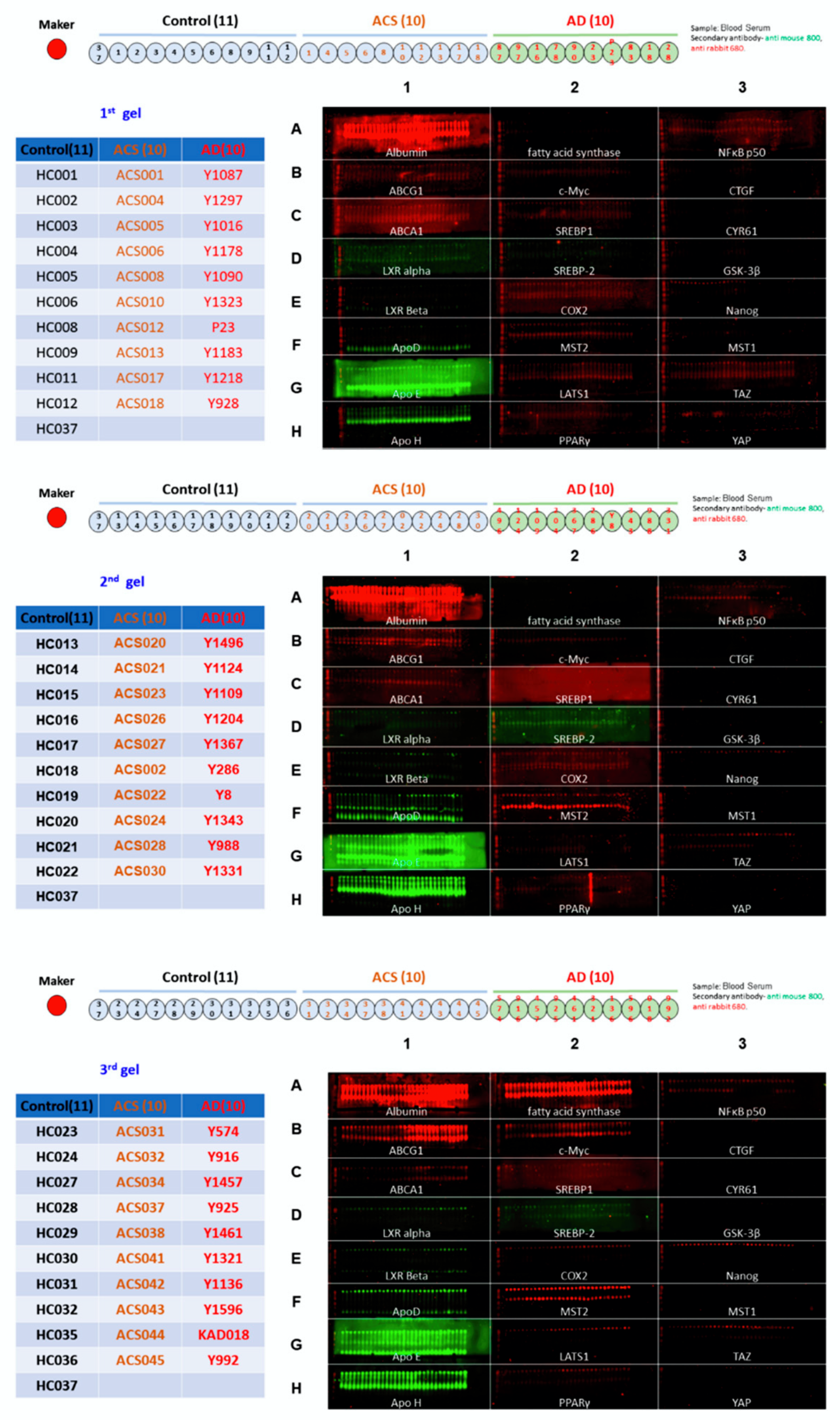
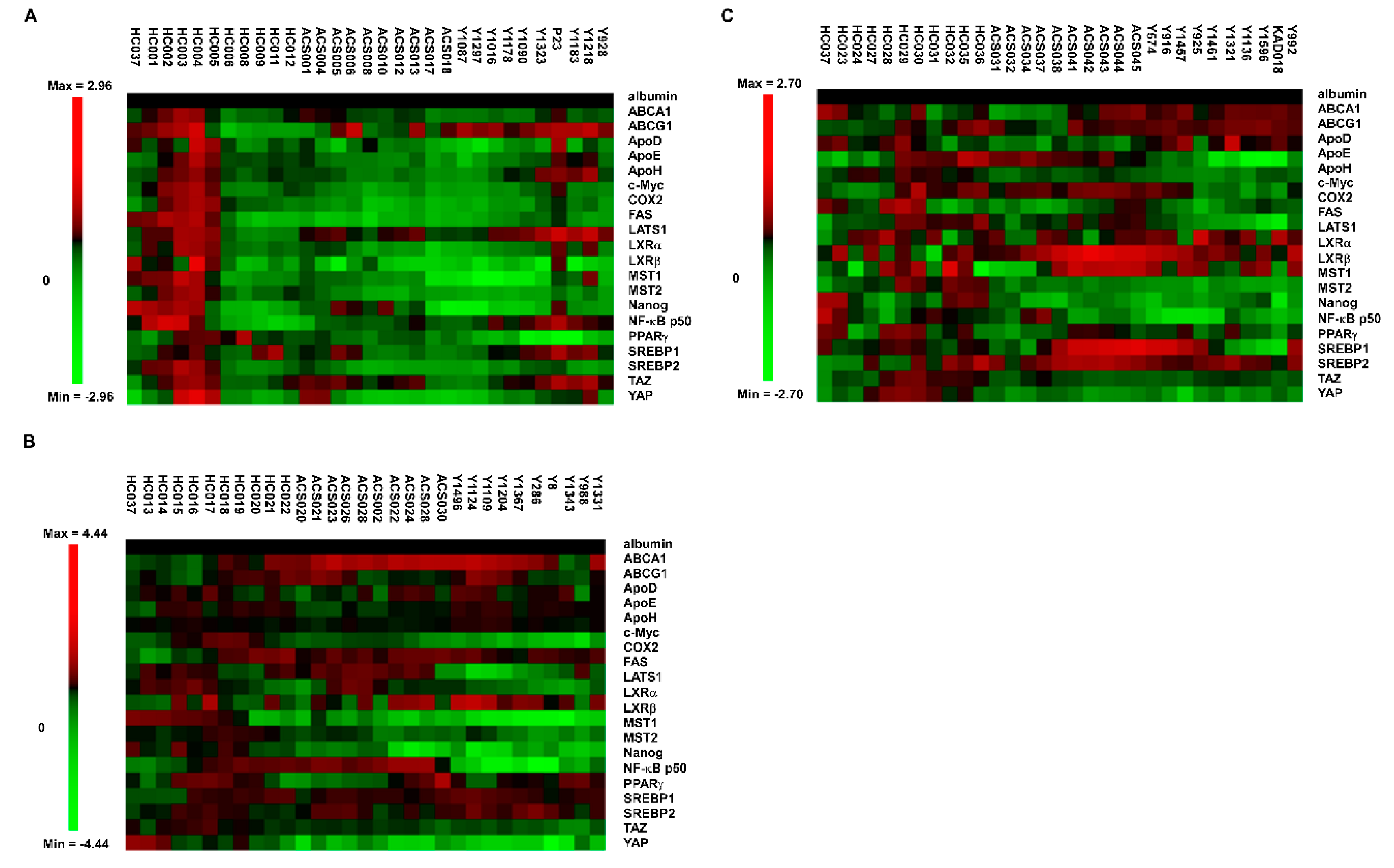
2.1. Micro-Western Array Reveals the Difference in Serum Proteins Profile between Healthy Control versus ACS and AD
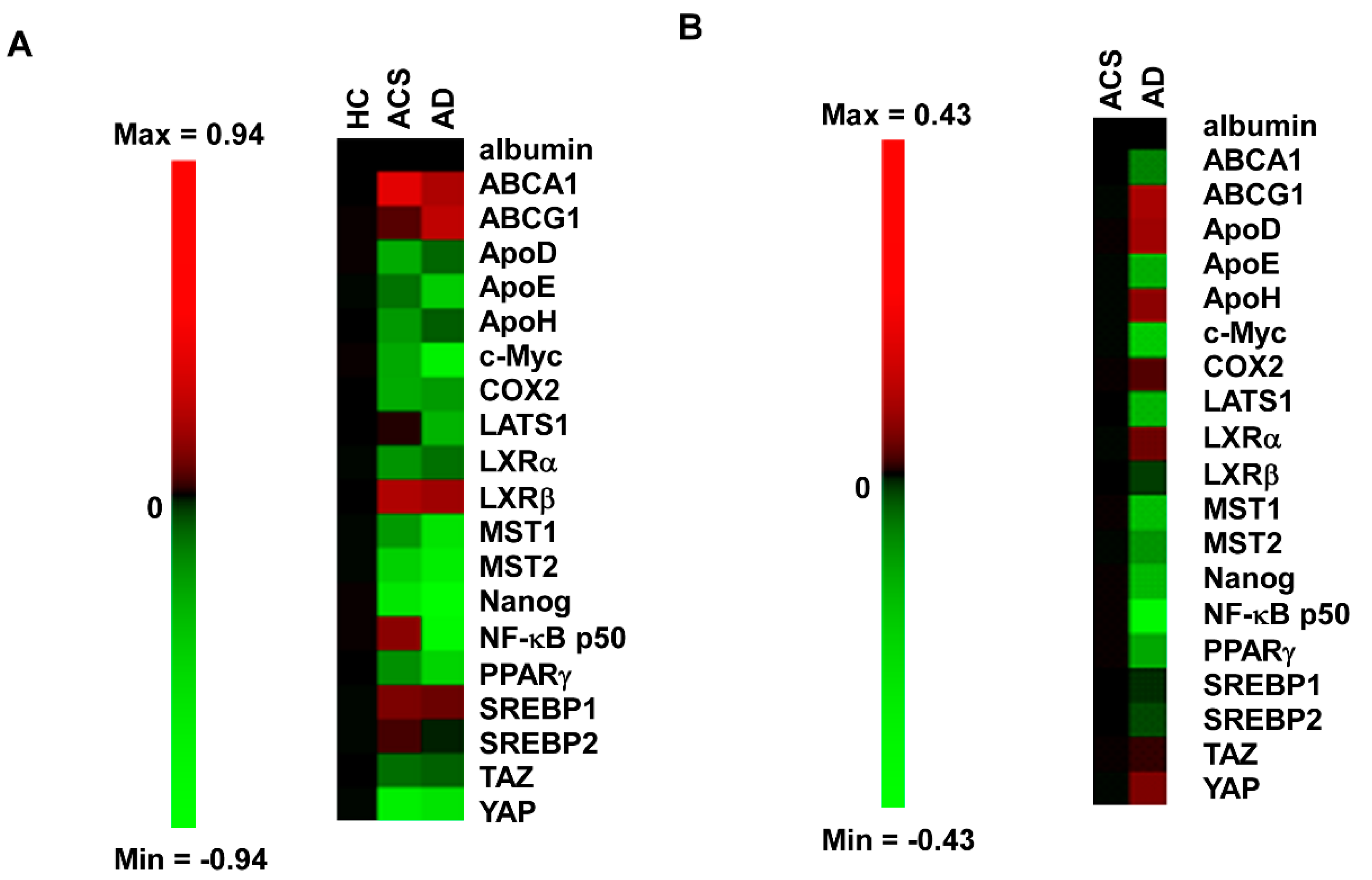
2.2. Profiles of Serum Proteins in ACS and AD Are Different as Determined by MWA
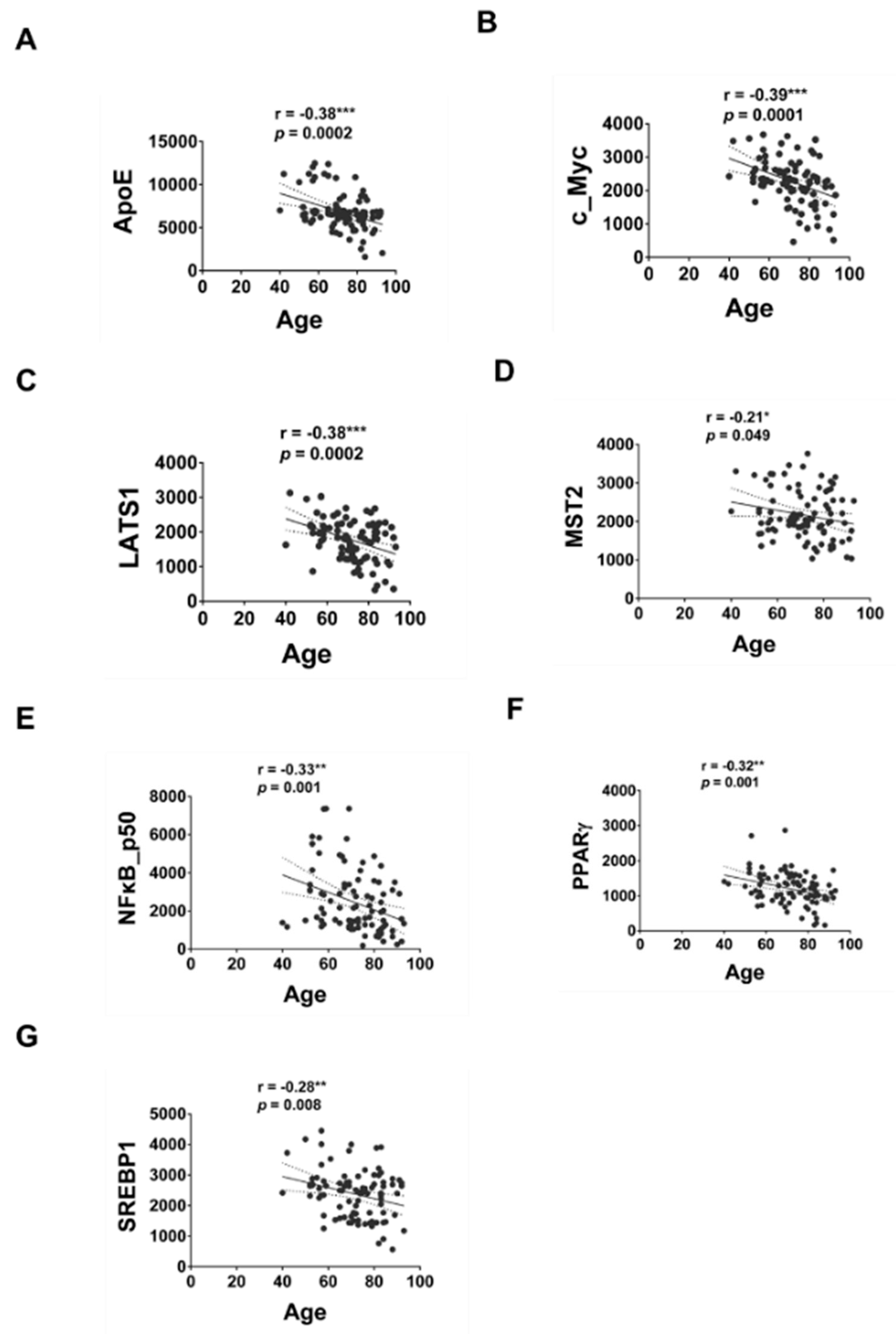
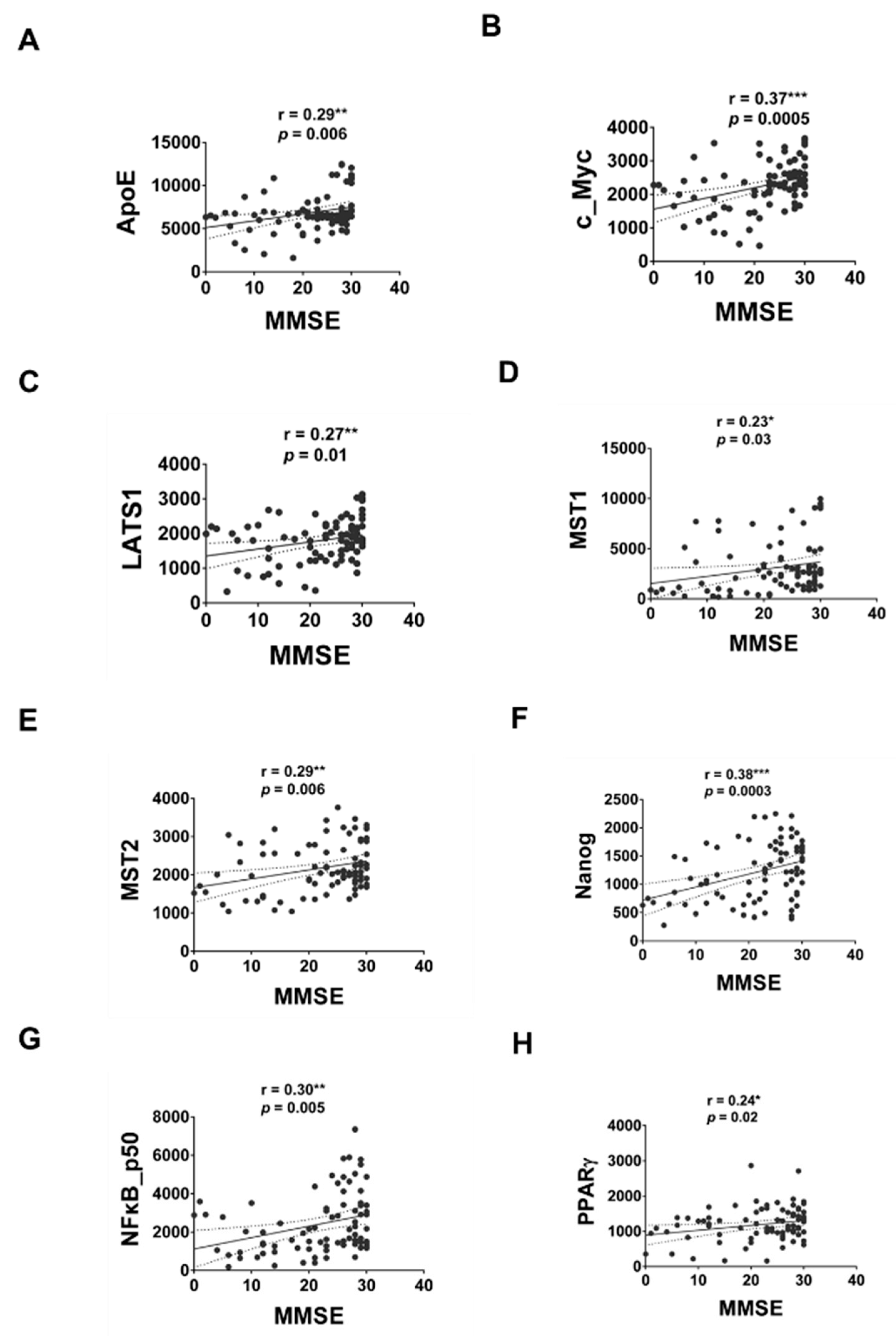
2.3. Correlation of Serum Proteins with MMSE or Age
3. Discussion
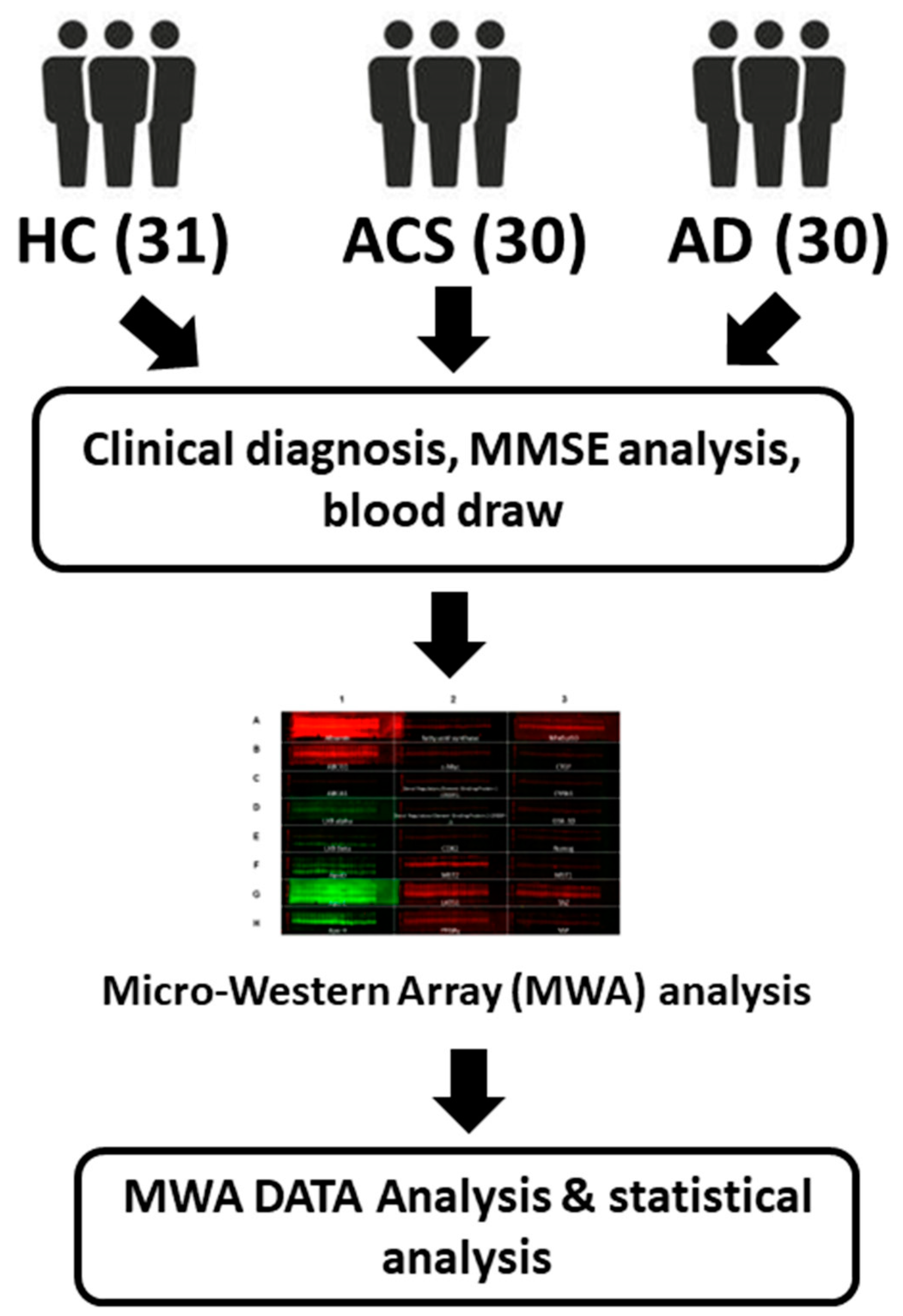
4. Materials and Methods
4.1. Recruitment of Participants
4.2. Clinical and Cognitive Assessments
4.3. Micro-Western Array
4.4. Statistics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murphy, M.P.; LeVine, H., 3rd. Alzheimer’s disease and the amyloid-beta peptide. J. Alzheimers Dis 2010, 19, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.G.; Li, Y.; Ng, C.T.; Song, Y.Q. Inflammation in Alzheimer’s Disease and Molecular Genetics: Recent Update. Arch. Immunol. Ther. Exp. 2015, 63, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Swanson, K.V.; Deng, M.; Ting, J.P. The NLRP3 inflammasome: Molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 2019, 19, 477–489. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Dong, Z.; Song, W. NLRP3 inflammasome as a novel therapeutic target for Alzheimer’s disease. Signal. Transduct. Target. Ther. 2020, 5, 37. [Google Scholar] [CrossRef] [PubMed]
- Heneka, M.T.; Kummer, M.P.; Stutz, A.; Delekate, A.; Schwartz, S.; Vieira-Saecker, A.; Griep, A.; Axt, D.; Remus, A.; Tzeng, T.C.; et al. NLRP3 is activated in Alzheimer’s disease and contributes to pathology in APP/PS1 mice. Nature 2013, 493, 674–678. [Google Scholar] [CrossRef]
- Ising, C.; Venegas, C.; Zhang, S.; Scheiblich, H.; Schmidt, S.V.; Vieira-Saecker, A.; Schwartz, S.; Albasset, S.; McManus, R.M.; Tejera, D.; et al. NLRP3 inflammasome activation drives tau pathology. Nature 2019, 575, 669–673. [Google Scholar] [CrossRef] [PubMed]
- Engelhart, M.J.; Geerlings, M.I.; Meijer, J.; Kiliaan, A.; Ruitenberg, A.; van Swieten, J.C.; Stijnen, T.; Hofman, A.; Witteman, J.C.; Breteler, M.M. Inflammatory proteins in plasma and the risk of dementia: The rotterdam study. Arch. Neurol. 2004, 61, 668–672. [Google Scholar] [CrossRef]
- Leung, R.; Proitsi, P.; Simmons, A.; Lunnon, K.; Guntert, A.; Kronenberg, D.; Pritchard, M.; Tsolaki, M.; Mecocci, P.; Kloszewska, I.; et al. Inflammatory proteins in plasma are associated with severity of Alzheimer’s disease. PLoS ONE 2013, 8, e64971. [Google Scholar] [CrossRef] [PubMed]
- Kinney, J.W.; Bemiller, S.M.; Murtishaw, A.S.; Leisgang, A.M.; Salazar, A.M.; Lamb, B.T. Inflammation as a central mechanism in Alzheimer’s disease. Alzheimers Dement. 2018, 4, 575–590. [Google Scholar] [CrossRef]
- Ciaccio, M.F.; Wagner, J.P.; Chuu, C.P.; Lauffenburger, D.A.; Jones, R.B. Systems analysis of EGF receptor signaling dynamics with microwestern arrays. Nat. Methods 2010, 7, 148–155. [Google Scholar] [CrossRef]
- Serrano-Pozo, A.; Das, S.; Hyman, B.T. APOE and Alzheimer’s disease: Advances in genetics, pathophysiology, and therapeutic approaches. Lancet Neurol. 2021, 20, 68–80. [Google Scholar] [CrossRef]
- Guo, L.; LaDu, M.J.; Van Eldik, L.J. A dual role for apolipoprotein e in neuroinflammation: Anti- and pro-inflammatory activity. J. Mol. Neurosci. 2004, 23, 205–212. [Google Scholar] [CrossRef]
- Bell, R.D.; Winkler, E.A.; Singh, I.; Sagare, A.P.; Deane, R.; Wu, Z.; Holtzman, D.M.; Betsholtz, C.; Armulik, A.; Sallstrom, J.; et al. Apolipoprotein E controls cerebrovascular integrity via cyclophilin A. Nature 2012, 485, 512–516. [Google Scholar] [CrossRef] [PubMed]
- Dietschy, J.M.; Turley, S.D. Cholesterol metabolism in the brain. Curr. Opin. Lipidol. 2001, 12, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Liao, F.; Yoon, H.; Kim, J. Apolipoprotein E metabolism and functions in brain and its role in Alzheimer’s disease. Curr. Opin. Lipidol. 2017, 28, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Grubman, A.; Chew, G.; Ouyang, J.F.; Sun, G.; Choo, X.Y.; McLean, C.; Simmons, R.K.; Buckberry, S.; Vargas-Landin, D.B.; Poppe, D.; et al. A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat. Neurosci. 2019, 22, 2087–2097. [Google Scholar] [CrossRef]
- Pasinetti, G.M.; Aisen, P.S. Cyclooxygenase-2 expression is increased in frontal cortex of Alzheimer’s disease brain. Neuroscience 1998, 87, 319–324. [Google Scholar] [CrossRef]
- Han, S.H.; Hulette, C.; Saunders, A.M.; Einstein, G.; Pericak-Vance, M.; Strittmatter, W.J.; Roses, A.D.; Schmechel, D.E. Apolipoprotein E is present in hippocampal neurons without neurofibrillary tangles in Alzheimer’s disease and in age-matched controls. Exp. Neurol 1994, 128, 13–26. [Google Scholar] [CrossRef]
- Flowers, S.A.; Rebeck, G.W. APOE in the normal brain. Neurobiol Dis 2020, 136, 104724. [Google Scholar] [CrossRef]
- Lefterov, I.; Fitz, N.F.; Cronican, A.; Lefterov, P.; Staufenbiel, M.; Koldamova, R. Memory deficits in APP23/Abca1+/− mice correlate with the level of Abeta oligomers. ASN Neuro 2009, 1, e00006. [Google Scholar] [CrossRef]
- Wahrle, S.E.; Jiang, H.; Parsadanian, M.; Legleiter, J.; Han, X.; Fryer, J.D.; Kowalewski, T.; Holtzman, D.M. ABCA1 is required for normal central nervous system ApoE levels and for lipidation of astrocyte-secreted apoE. J. Biol. Chem. 2004, 279, 40987–40993. [Google Scholar] [CrossRef] [PubMed]
- Do, T.M.; Ouellet, M.; Calon, F.; Chimini, G.; Chacun, H.; Farinotti, R.; Bourasset, F. Direct evidence of abca1-mediated efflux of cholesterol at the mouse blood-brain barrier. Mol. Cell Biochem. 2011, 357, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Fitz, N.F.; Carter, A.Y.; Tapias, V.; Castranio, E.L.; Kodali, R.; Lefterov, I.; Koldamova, R. ABCA1 Deficiency Affects Basal Cognitive Deficits and Dendritic Density in Mice. J. Alzheimers Dis. 2017, 56, 1075–1085. [Google Scholar] [CrossRef] [PubMed]
- Fitz, N.F.; Cronican, A.A.; Saleem, M.; Fauq, A.H.; Chapman, R.; Lefterov, I.; Koldamova, R. Abca1 deficiency affects Alzheimer’s disease-like phenotype in human ApoE4 but not in ApoE3-targeted replacement mice. J. Neurosci. 2012, 32, 13125–13136. [Google Scholar] [CrossRef]
- Wahrle, S.E.; Jiang, H.; Parsadanian, M.; Kim, J.; Li, A.; Knoten, A.; Jain, S.; Hirsch-Reinshagen, V.; Wellington, C.L.; Bales, K.R.; et al. Overexpression of ABCA1 reduces amyloid deposition in the PDAPP mouse model of Alzheimer disease. J. Clin. Invest. 2008, 118, 671–682. [Google Scholar] [CrossRef]
- Boehm-Cagan, A.; Bar, R.; Liraz, O.; Bielicki, J.K.; Johansson, J.O.; Michaelson, D.M. ABCA1 Agonist Reverses the ApoE4-Driven Cognitive and Brain Pathologies. J. Alzheimers Dis. 2016, 54, 1219–1233. [Google Scholar] [CrossRef]
- Fitz, N.F.; Cronican, A.; Pham, T.; Fogg, A.; Fauq, A.H.; Chapman, R.; Lefterov, I.; Koldamova, R. Liver X receptor agonist treatment ameliorates amyloid pathology and memory deficits caused by high-fat diet in APP23 mice. J. Neurosci. 2010, 30, 6862–6872. [Google Scholar] [CrossRef]
- Donkin, J.J.; Stukas, S.; Hirsch-Reinshagen, V.; Namjoshi, D.; Wilkinson, A.; May, S.; Chan, J.; Fan, J.; Collins, J.; Wellington, C.L. ATP-binding cassette transporter A1 mediates the beneficial effects of the liver X receptor agonist GW3965 on object recognition memory and amyloid burden in amyloid precursor protein/presenilin 1 mice. J. Biol. Chem. 2010, 285, 34144–34154. [Google Scholar] [CrossRef]
- Nordestgaard, L.T.; Tybjaerg-Hansen, A.; Nordestgaard, B.G.; Frikke-Schmidt, R. Loss-of-function mutation in ABCA1 and risk of Alzheimer’s disease and cerebrovascular disease. Alzheimers Dement. 2015, 11, 1430–1438. [Google Scholar] [CrossRef]
- Marchi, C.; Adorni, M.P.; Caffarra, P.; Ronda, N.; Spallazzi, M.; Barocco, F.; Galimberti, D.; Bernini, F.; Zimetti, F. ABCA1- and ABCG1-mediated cholesterol efflux capacity of cerebrospinal fluid is impaired in Alzheimer’s disease. J. Lipid Res. 2019, 60, 1449–1456. [Google Scholar] [CrossRef]
- Sano, O.; Tsujita, M.; Shimizu, Y.; Kato, R.; Kobayashi, A.; Kioka, N.; Remaley, A.T.; Michikawa, M.; Ueda, K.; Matsuo, M. ABCG1 and ABCG4 Suppress gamma-Secretase Activity and Amyloid beta Production. PLoS ONE 2016, 11, e0155400. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.; Kim, W.S.; Shepherd, C.E.; Halliday, G.M. Apolipoprotein D Upregulation in Alzheimer’s Disease but Not Frontotemporal Dementia. J. Mol. Neurosci. 2019, 67, 125–132. [Google Scholar] [CrossRef]
- Kuiperij, H.B.; Hondius, D.C.; Kersten, I.; Versleijen, A.A.M.; Rozemuller, A.J.M.; Greenberg, S.M.; Schreuder, F.; Klijn, C.J.M.; Verbeek, M.M. Apolipoprotein D: A potential biomarker for cerebral amyloid angiopathy. Neuropathol. Appl. Neurobiol. 2020, 46, 431–440. [Google Scholar] [CrossRef]
- Li, H.; Ruberu, K.; Munoz, S.S.; Jenner, A.M.; Spiro, A.; Zhao, H.; Rassart, E.; Sanchez, D.; Ganfornina, M.D.; Karl, T.; et al. Apolipoprotein D modulates amyloid pathology in APP/PS1 Alzheimer’s disease mice. Neurobiol. Aging 2015, 36, 1820–1833. [Google Scholar] [CrossRef] [PubMed]
- Nagano, S.; Huang, X.; Moir, R.D.; Payton, S.M.; Tanzi, R.E.; Bush, A.I. Peroxidase activity of cyclooxygenase-2 (COX-2) cross-links beta-amyloid (Abeta) and generates Abeta-COX-2 hetero-oligomers that are increased in Alzheimer’s disease. J. Biol. Chem. 2004, 279, 14673–14678. [Google Scholar] [CrossRef]
- Lee, J.Y.; Han, S.H.; Park, M.H.; Baek, B.; Song, I.S.; Choi, M.K.; Takuwa, Y.; Ryu, H.; Kim, S.H.; He, X.; et al. Neuronal SphK1 acetylates COX2 and contributes to pathogenesis in a model of Alzheimer’s Disease. Nat. Commun. 2018, 9, 1479. [Google Scholar] [CrossRef] [PubMed]
- Spell, C.; Kolsch, H.; Lutjohann, D.; Kerksiek, A.; Hentschel, F.; Damian, M.; von Bergmann, K.; Rao, M.L.; Maier, W.; Heun, R. SREBP1a polymorphism influences the risk of Alzheimer’s disease in carriers of the ApoE4 allele. Dement. Geriatr. Cogn. Disord. 2004, 18, 245–249. [Google Scholar] [CrossRef]
- Barbero-Camps, E.; Fernandez, A.; Martinez, L.; Fernandez-Checa, J.C.; Colell, A. APP/PS1 mice overexpressing SREBP2 exhibit combined Abeta accumulation and tau pathology underlying Alzheimer’s disease. Hum. Mol. Genet. 2013, 22, 3460–3476. [Google Scholar] [CrossRef]
- Xu, X.; Shen, X.; Wang, J.; Feng, W.; Wang, M.; Miao, X.; Wu, Q.; Wu, L.; Wang, X.; Ma, Y.; et al. YAP prevents premature senescence of astrocytes and cognitive decline of Alzheimer’s disease through regulating CDK6 signaling. Aging Cell 2021, 20, e13465. [Google Scholar] [CrossRef]
- Tanaka, H.; Homma, H.; Fujita, K.; Kondo, K.; Yamada, S.; Jin, X.; Waragai, M.; Ohtomo, G.; Iwata, A.; Tagawa, K.; et al. YAP-dependent necrosis occurs in early stages of Alzheimer’s disease and regulates mouse model pathology. Nat. Commun. 2020, 11, 507. [Google Scholar] [CrossRef]
- Haramizu, S.; Ota, N.; Hase, T.; Murase, T. Aging-associated changes in physical performance and energy metabolism in the senescence-accelerated mouse. J. Gerontol. A Biol. Sci. Med. Sci. 2011, 66, 646–655. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ulrich-Lai, Y.M.; Ryan, K.K. PPARgamma and stress: Implications for aging. Exp. Gerontol. 2013, 48, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Heneka, M.; Landreth, G.E. The role of peroxisome proliferator-activated receptor-gamma (PPARgamma) in Alzheimer’s disease: Therapeutic implications. CNS Drugs 2008, 22, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, J.W.; Zhao, X.; De Cecco, M.; Peterson, A.L.; Pagliaroli, L.; Manivannan, J.; Hubbard, G.B.; Ikeno, Y.; Zhang, Y.; Feng, B.; et al. Reduced expression of MYC increases longevity and enhances healthspan. Cell 2015, 160, 477–488. [Google Scholar] [CrossRef]
- Garcia-Garcia, V.A.; Alameda, J.P.; Page, A.; Casanova, M.L. Role of NF-kappaB in Ageing and Age-Related Diseases: Lessons from Genetically Modified Mouse Models. Cells 2021, 10, 1906. [Google Scholar] [CrossRef]
- Lojkowska, W.; Witkowski, G.; Bednarska-Makaruk, M.; Wehr, H.; Sienkiewicz-Jarosz, H.; Graban, A.; Bochynska, A.; Wisniewska, A.; Gugala, M.; Slawinska, K.; et al. Correlations between cerebellar and brain volumes, cognitive impairments, ApoE levels, and APOE genotypes in patients with AD and MCI. Curr. Alzheimer Res. 2013, 10, 964–972. [Google Scholar] [CrossRef]
- Coats, M.; Morris, J.C. Antecedent biomarkers of Alzheimer’s disease: The adult children study. J. Geriatr. Psychiatry Neurol. 2005, 18, 242–244. [Google Scholar] [CrossRef]
- Wang, W.F.; Chiu, P.Y.; Lin, Y.T.; Hu, C.J.; Fuh, J.L.; Yang, Y.H. Registration of Alzheimer’s disease in Taiwan: Patient and informant. Am. J. Alzheimers Dis. Other Demen. 2014, 29, 18–22. [Google Scholar] [CrossRef]
- McKhann, G.; Drachman, D.; Folstein, M.; Katzman, R.; Price, D.; Stadlan, E.M. Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 1984, 34, 939–944. [Google Scholar] [CrossRef]
- Morris, J.C. The Clinical Dementia Rating (CDR): Current version and scoring rules. Neurology 1993, 43, 2412–2414. [Google Scholar] [CrossRef]
- Galvin, J.E.; Roe, C.M.; Powlishta, K.K.; Coats, M.A.; Muich, S.J.; Grant, E.; Miller, J.P.; Storandt, M.; Morris, J.C. The AD8: A brief informant interview to detect dementia. Neurology 2005, 65, 559–564. [Google Scholar] [CrossRef] [PubMed]
- Folstein, M.F.; Folstein, S.E.; McHugh, P.R. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res. 1975, 12, 189–198. [Google Scholar] [CrossRef]
| Characteristics | AD (n = 30) | AC (n = 30) | NC (n = 31) | p Value | |||
|---|---|---|---|---|---|---|---|
| Three Groups | AD vs. AC | AD vs. NC | AC vs. NC | ||||
| Gender, female (%) | 80.0 | 90.0 | 64.5 | <0.001 | <0.001 | <0.001 | <0.001 |
| APOE ε4 positive (%) | 46.7 | 46.7 | 16.1 | <0.001 | <0.001 | <0.001 | <0.001 |
| Age (years) | 82.6 ± 6.2 | 57.5 ± 7.1 | 74.2 ± 6.0 | <0.001 | <0.001 | <0.01 | <0.001 |
| Education (years) † | 5.3 ± 5.3 | 13.7 ± 4.0 | 10.5 ± 3.8 | <0.001 | <0.001 | <0.01 | 0.055 |
| MMSE ‡ | 13.6 ± 7.2 | 28.4 ± 1.6 | 24.5 ± 3.7 | <0.001 | <0.001 | <0.001 | <0.01 |
| Proteins | Pearson Correlation Coefficient (r) with Age | p Value | Pearson Correlation Coefficient (r) with MMSE | p Value |
|---|---|---|---|---|
| ABCA1 | −0.2 | 0.06 | 0.014 | 0.90 |
| ABCG1 | 0.02 | 0.81 | −0.13 | 0.23 |
| ApoD | 0.04 | 0.72 | −0.003 | 0.97 |
| ApoE | −0.38 *** | 0.0002 | 0.29 ** | 0.006 |
| ApoH | −0.15 | 0.16 | −0.15 | 0.17 |
| c_Myc | −0.39 *** | 0.0001 | 0.37 *** | 0.0005 |
| COX2 | −0.11 | 0.31 | 0.03 | 0.80 |
| LATS1 | −0.38 *** | 0.0002 | 0.27 ** | 0.01 |
| LXRα | 0.04 | 0.70 | 0.01 | 0.93 |
| LXRβ | −0.10 | 0.34 | 0.03 | 0.74 |
| MST1 | −0.12 | 0.24 | 0.23 * | 0.03 |
| MST2 | −0.21 * | 0.049 | 0.29 ** | 0.006 |
| Nanog | −0.16 | 0.12 | 0.38 *** | 0.0003 |
| NFκB p50 | −0.33 ** | 0.001 | 0.30 ** | 0.005 |
| PPARγ | −0.32 ** | 0.001 | 0.24 * | 0.02 |
| SREBP1 | −0.28 ** | 0.008 | 0.04 | 0.67 |
| SREBP2 | −0.12 | 0.26 | 0.05 | 0.65 |
| TAZ | −0.16 | 0.12 | 0.05 | 0.66 |
| YAP | −0.06 | 0.55 | 0.03 | 0.81 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huo, C.; Chen, M.-H.; Hour, T.-C.; Huang, L.-C.; Fong, Y.-O.; Kuo, Y.-Y.; Yang, Y.-H.; Chuu, C.-P. Application of Micro-Western Array for Identifying Different Serum Protein Expression Profile among Healthy Control, Alzheimer’s Disease Patients and Patients’ Adult Children. Brain Sci. 2022, 12, 1134. https://doi.org/10.3390/brainsci12091134
Huo C, Chen M-H, Hour T-C, Huang L-C, Fong Y-O, Kuo Y-Y, Yang Y-H, Chuu C-P. Application of Micro-Western Array for Identifying Different Serum Protein Expression Profile among Healthy Control, Alzheimer’s Disease Patients and Patients’ Adult Children. Brain Sciences. 2022; 12(9):1134. https://doi.org/10.3390/brainsci12091134
Chicago/Turabian StyleHuo, Chieh, Ming-Hui Chen, Tzyh-Chyuan Hour, Ling-Chun Huang, Yi-On Fong, Ying-Yu Kuo, Yuan-Han Yang, and Chih-Pin Chuu. 2022. "Application of Micro-Western Array for Identifying Different Serum Protein Expression Profile among Healthy Control, Alzheimer’s Disease Patients and Patients’ Adult Children" Brain Sciences 12, no. 9: 1134. https://doi.org/10.3390/brainsci12091134
APA StyleHuo, C., Chen, M.-H., Hour, T.-C., Huang, L.-C., Fong, Y.-O., Kuo, Y.-Y., Yang, Y.-H., & Chuu, C.-P. (2022). Application of Micro-Western Array for Identifying Different Serum Protein Expression Profile among Healthy Control, Alzheimer’s Disease Patients and Patients’ Adult Children. Brain Sciences, 12(9), 1134. https://doi.org/10.3390/brainsci12091134





