Identifying the Hub Genes of Glioma Peritumoral Brain Edema Using Bioinformatical Methods
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Sources
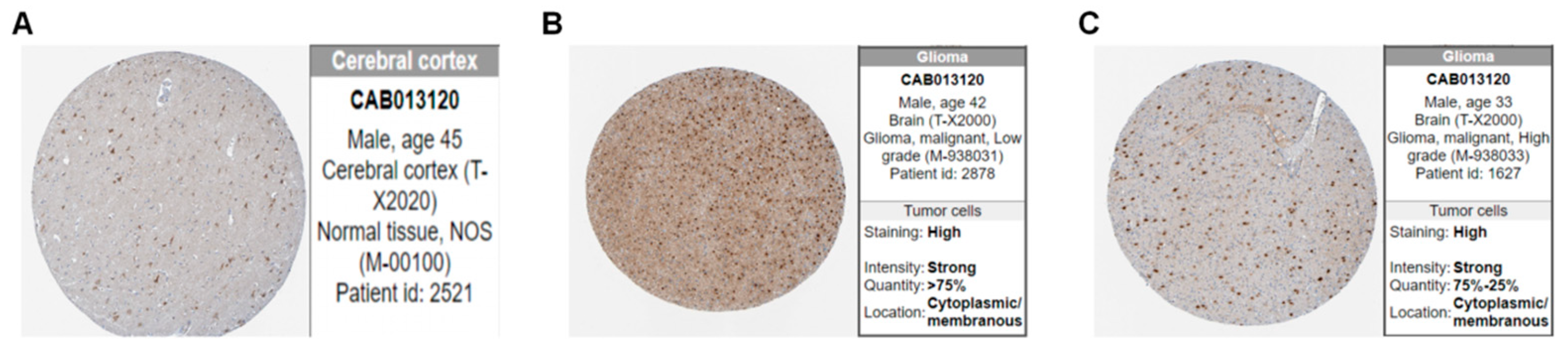
2.2. Assessments of Image Features
2.3. Differentially Expressed Gene (DEGs) Analysis
2.4. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis
2.5. Correlation Analysis and PPI Network Construction
2.6. Survival Analysis of Hub Genes
2.7. Immune infiltration analysis
2.8. Statistical Analysis
3. Results
3.1. Clinical Features
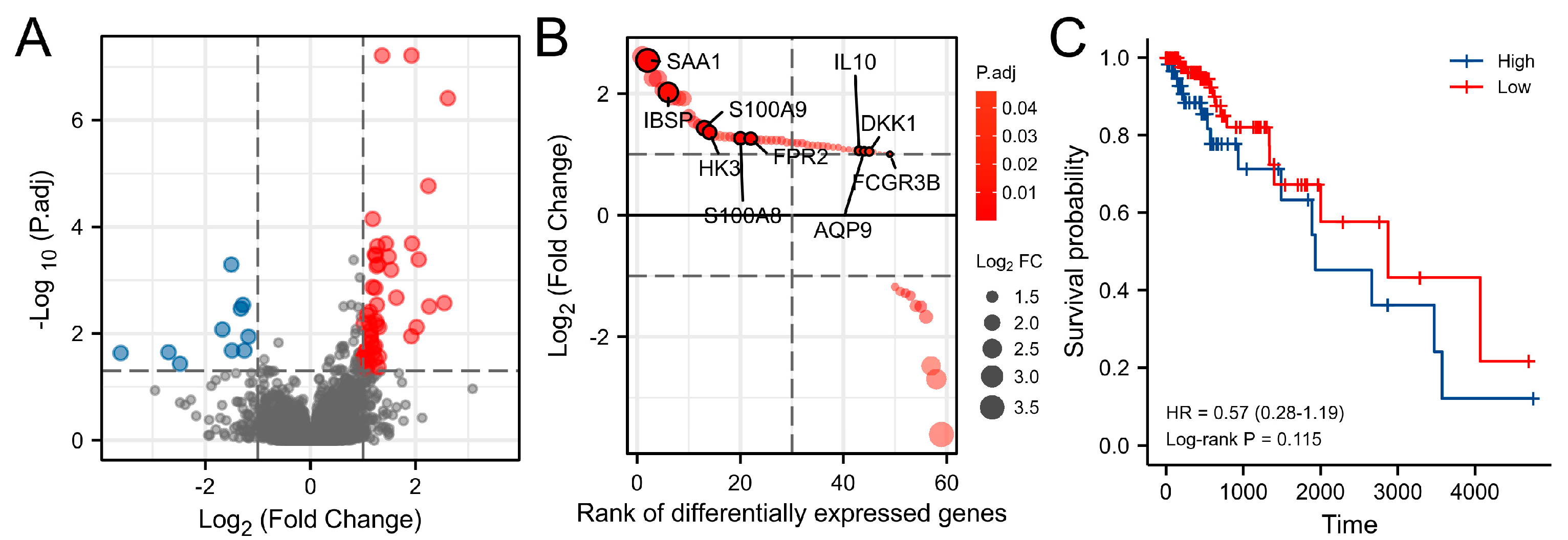
3.2. Differentially Expressed Genes (DEGs) Analysis
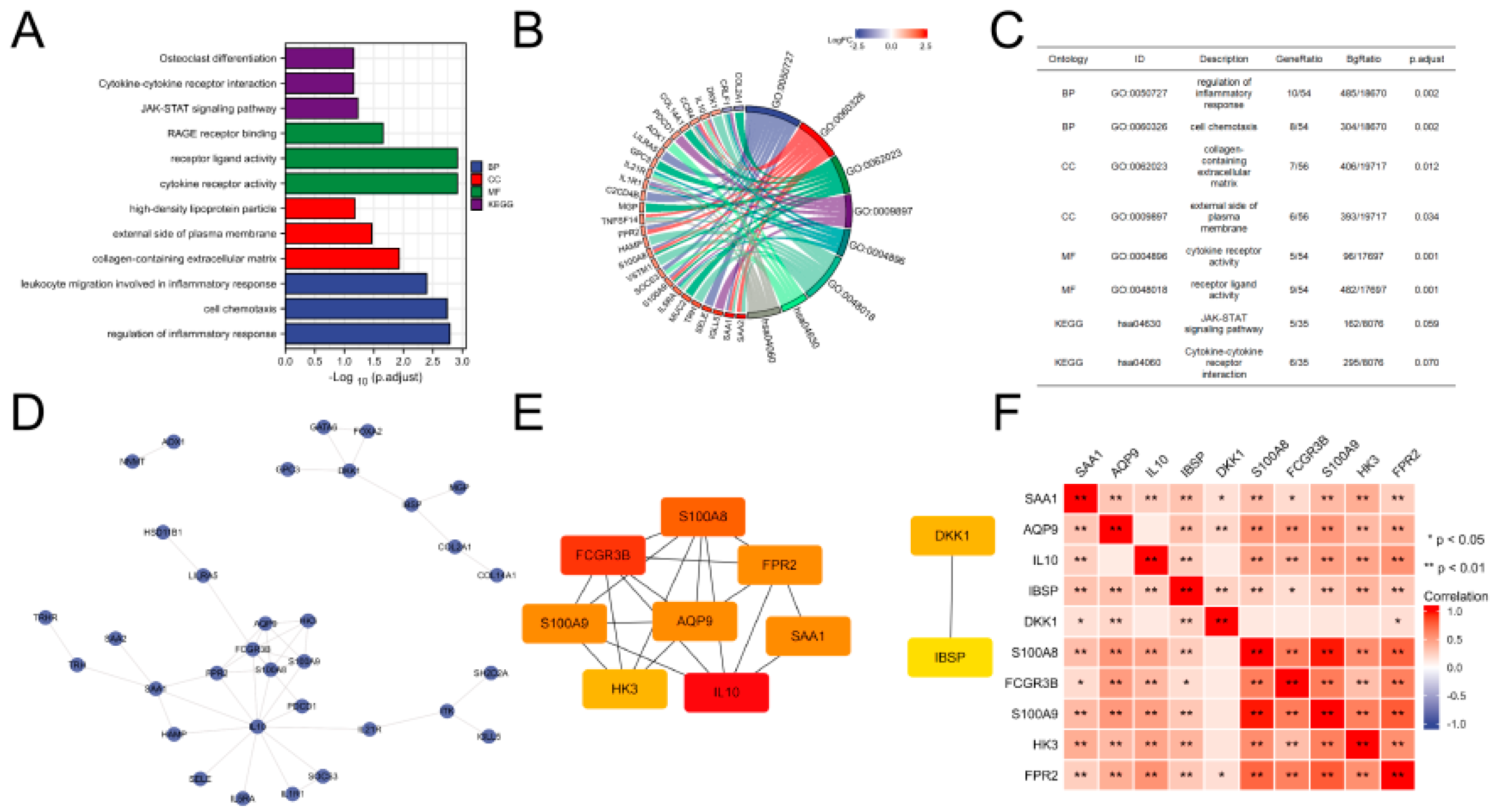
3.3. Functional Analysis of DEGs
3.4. PPI Network Construction and Key Gene Identification
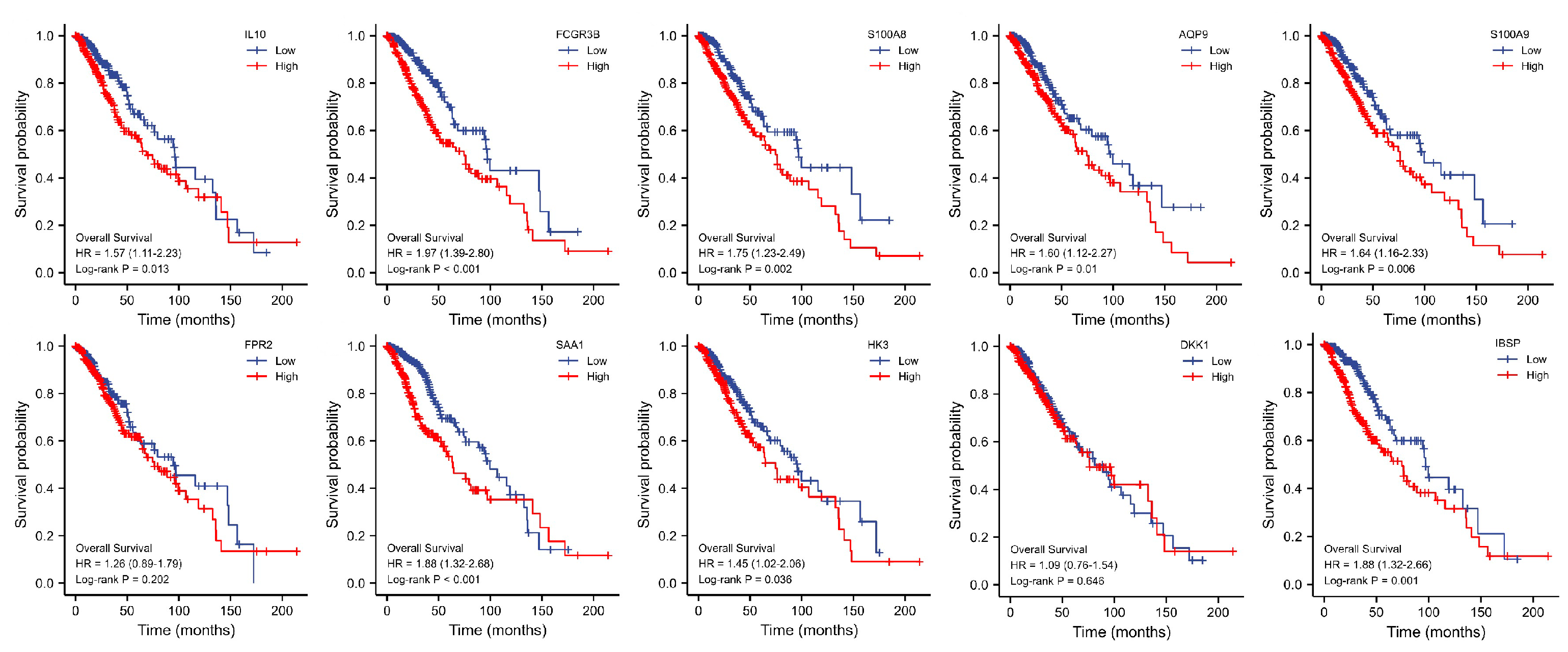
3.5. Key Genes Prognostic Analysis
3.6. Immune Infiltration Analysis of Gene Sets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AQPs | Aquaporins |
| BBB | Blood–brain barrier |
| BP | Biological processes |
| CC | Cellular components |
| CTLs | Cytotoxic T cells |
| DEGs | Differential genes |
| DKK1 | Dickkopf Wnt signaling pathway inhibitor 1 |
| FCGR3B | Fc gamma receptor IIIb |
| FDR | False discovery rate |
| FPR2 | Formyl peptide receptor 2 |
| GBM | Glioblastoma |
| GPTBE | Glioma peritumoral brain edema |
| GSCA | Gene Set Caner Analysis |
| GSVA | Gene set variation analysis |
| GO | Gene Ontology |
| HK3 | Hexokinase 3 |
| HPA | Human protein atlas |
| IBSP | Integrin binding sialoprotein |
| IDH1 | Lactate dehydrogenase 1 |
| IL10 | Interleukin 10 |
| JAK | Janus kinase |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KM | Kaplan–Meier |
| LGG | Low-grade gliomas |
| MF | Molecular function |
| MMP | Metalloproteinase |
| MR | Magnetic resonance |
| OS | Overall survival |
| PPI | Protein–protein interaction |
| PTBE | Peritumoral brain edema |
| RAGE | Receptor of advanced glycation end-products |
| SAA1 | Serum amyloid A1 |
| STAT | Signal transducer and activator of transcription |
| TAMs | Tumor associated macrophages |
| TCGA | The cancer genome atlas |
| TCIA | The Cancer Imaging Archive |
| VASARI | Visually accessible Rembrandt images |
| VEGF | Vascular endothelial growth factor |
References
- Ostrom, Q.T.; Francis, S.S.; Barnholtz-Sloan, J.S. Epidemiology of Brain and Other CNS Tumors. Curr. Neurol. Neurosci. Rep. 2021, 21, 68. [Google Scholar] [CrossRef] [PubMed]
- Montemurro, N.; Fanelli, G.N.; Scatena, C.; Ortenzi, V.; Pasqualetti, F.; Mazzanti, C.M.; Morganti, R.; Paiar, F.; Naccarato, A.G.; Perrini, P. Surgical outcome and molecular pattern characterization of recurrent glioblastoma multiforme: A single-center retrospective series. Clin. Neurol. Neurosurg. 2021, 207, 106735. [Google Scholar] [CrossRef] [PubMed]
- Niedbała, M.; Malarz, K.; Sharma, G.; Kramer-Marek, G.; Kaspera, W. Glioblastoma: Pitfalls and Opportunities of Immunotherapeutic Combinations. OncoTargets Ther. 2022, 15, 437–468. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-Y.; Mei, W.-Z.; Lin, Z.-X. Pre-Operative Peritumoral Edema and Survival Rate in Glioblastoma Multiforme. Onkologie 2013, 36, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Seidel, C.; Dörner, N.; Osswald, M.; Wick, A.; Platten, M.; Bendszus, M.; Wick, W. Does age matter? A MRI study on peritumoral edema in newly diagnosed primary glioblastoma. BMC Cancer 2011, 11, 127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Vallières, M.; Bai, H.X.; Su, C.; Tang, H.; Oldridge, D.; Zhang, Z.; Xiao, B.; Liao, W.; Tao, Y.; et al. MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro-Oncology 2017, 19, 862–870. [Google Scholar] [CrossRef]
- Xingfu, W.; Xueyong, L.; Yupeng, C.; Guoshi, L.; Wenzhong, M.; Jianwu, C.; Ying, L.; Zhixiong, L.; Sheng, Z. Histopathological findings in the peritumoral edema area of human glioma. Histol. Histopathol. 2015, 30, 1101–1109. [Google Scholar] [CrossRef]
- Wang, X.F.; Lin, G.S.; Lin, Z.X.; Chen, Y.P.; Chen, Y.; Zhang, J.D.; Tan, W.L. Association of pSTAT3-VEGF signaling pathway with peritumoral edema in newly diagnosed glioblastoma: An immunohistochemical study. Int. J. Clin. Exp. Pathol. 2014, 7, 6133–6140. [Google Scholar]
- Mou, K.; Chen, M.; Mao, Q.; Wang, P.; Ni, R.; Xia, X.; Liu, Y. AQP-4 in peritumoral edematous tissue is correlated with the degree of glioma and with expression of VEGF and HIF-alpha. J. Neuro-Oncol. 2010, 100, 375–383. [Google Scholar] [CrossRef]
- Yang, W.-C.; Zhou, L.-J.; Zhang, R.; Yue, Z.-Y.; Dong, H.; Song, C.-Y.; Qian, H.; Lu, S.-J.; Chang, F.-F. Effects of propofol and sevoflurane on aquaporin-4 and aquaporin-9 expression in patients performed gliomas resection. Brain Res. 2015, 1622, 1–6. [Google Scholar] [CrossRef]
- Beltran, E.; Matiasek, K.; De Risio, L.; De Stefani, A.; Feliu-Pascual, A.L.; Matiasek, L.A. Expression of MMP-2 and MMP-9 in Benign Canine Rostrotentorial Meningiomas Is Not Correlated to the Extent of Peritumoral Edema. Vet. Pathol. 2013, 50, 1091–1098. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-J.; Hu, F.-F.; Xia, M.-X.; Han, L.; Zhang, Q.; Guo, A.-Y. GSCALite: A web server for gene set cancer analysis. Bioinformatics 2018, 34, 3771–3772. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Chow, P.H.; Bowen, J.; Yool, A.J. Combined Systematic Review and Transcriptomic Analyses of Mammalian Aquaporin Classes 1 to 10 as Biomarkers and Prognostic Indicators in Diverse Cancers. Cancers 2020, 12, 1911. [Google Scholar] [CrossRef]
- Walcott, B.P.; Kahle, K.T.; Simard, J.M. Novel Treatment Targets for Cerebral Edema. Neurotherapeutics 2012, 9, 65–72. [Google Scholar] [CrossRef] [Green Version]
- Schoenegger, K.; Oberndorfer, S.; Wuschitz, B.; Struhal, W.; Hainfellner, J.; Prayer, D.; Heinzl, H.; Lahrmann, H.; Marosi, C.; Grisold, W. Peritumoral edema on MRI at initial diagnosis: An independent prognostic factor for glioblastoma? Eur. J. Neurol. 2009, 16, 874–878. [Google Scholar] [CrossRef]
- Wang, W.; DA, R.; Wang, M.; Wang, T.; Qi, L.; Jiang, H.; Chen, W.; Li, Q. Expression of brain-specific angiogenesis inhibitor 1 is inversely correlated with pathological grade, angiogenesis and peritumoral brain edema in human astrocytomas. Oncol. Lett. 2013, 5, 1513–1518. [Google Scholar] [CrossRef] [Green Version]
- Easton, A.S. Regulation of Permeability Across the Blood-Brain Barrier. In Biology and Regulation of Blood-Tissue Barriers; Springer: Cham, Switzerland, 2012; Volume 763, pp. 1–19. [Google Scholar] [CrossRef]
- Dickinson, P.J.; Sturges, B.K.; Higgins, R.J.; Roberts, B.N.; Leutenegger, C.M.; Bollen, A.W.; LeCouteur, R.A. Vascular Endothelial Growth Factor mRNA Expression and Peritumoral Edema in Canine Primary Central Nervous System Tumors. Vet. Pathol. 2008, 45, 131–139. [Google Scholar] [CrossRef] [Green Version]
- Hou, J.; Kshettry, V.R.; Selman, W.R.; Bambakidis, N.C. Peritumoral brain edema in intracranial meningiomas: The emergence of vascular endothelial growth factor–directed therapy. Neurosurg. Focus 2013, 35, E2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fanelli, G.; Grassini, D.; Ortenzi, V.; Pasqualetti, F.; Montemurro, N.; Perrini, P.; Naccarato, A.; Scatena, C. Decipher the Glioblastoma Microenvironment: The First Milestone for New Groundbreaking Therapeutic Strategies. Genes 2021, 12, 445. [Google Scholar] [CrossRef]
- Gadina, M.; Johnson, C.; Schwartz, D.; Bonelli, M.; Hasni, S.; Kanno, Y.; Changelian, P.; Laurence, A.; O’Shea, J.J. Translational and clinical advances in JAK-STAT biology: The present and future of jakinibs. J. Leukoc. Biol. 2018, 104, 499–514. [Google Scholar] [CrossRef] [PubMed]
- Engelhorn, T.; Savaskan, N.; Schwarz, M.A.; Kreutzer, J.; Meyer, E.P.; Hahnen, E.; Ganslandt, O.; Dörfler, A.; Nimsky, C.; Buchfelder, M.; et al. Cellular characterization of the peritumoral edema zone in malignant brain tumors. Cancer Sci. 2009, 100, 1856–1862. [Google Scholar] [CrossRef] [PubMed]
- Klemm, F.; Maas, R.R.; Bowman, R.L.; Kornete, M.; Soukup, K.; Nassiri, S.; Brouland, J.-P.; Iacobuzio-Donahue, C.A.; Brennan, C.; Tabar, V.; et al. Interrogation of the Microenvironmental Landscape in Brain Tumors Reveals Disease-Specific Alterations of Immune Cells. Cell 2020, 181, 1643–1660. [Google Scholar] [CrossRef]
- Reszec, J.; Hermanowicz, A.; Rutkowski, R.; Bernaczyk, P.; Mariak, Z.; Chyczewski, L. Evaluation of mast cells and hypoxia inducible factor-1 expression in meningiomas of various grades in correlation with peritumoral brain edema. J. Neuro-Oncol. 2013, 115, 119–125. [Google Scholar] [CrossRef] [Green Version]
- Qi, L.; Yu, H.; Zhang, Y.; Zhao, D.; Lv, P.; Zhong, Y.; Xu, Y. IL-10 secreted by M2 macrophage promoted tumorigenesis through interaction with JAK2 in glioma. Oncotarget 2016, 7, 71673–71685. [Google Scholar] [CrossRef] [Green Version]
- Zahir, S.T.; Mortaz, M.; Yazdi, M.B.; Sharahjin, N.S.; Shabani, M. Calvarium mass as the first presentation of glioblastoma multiforme: A very rare manifestation of high-grade glioma. Neurochirurgie 2018, 64, 76–78. [Google Scholar] [CrossRef]
- Knebel, F.; Uno, M.; Galatro, T.; Bellé, L.P.; Shinjo, S.; Marie, S.; Campa, A. Serum amyloid A1 is upregulated in human glioblastoma. J. Neuro-Oncol. 2017, 132, 383–391. [Google Scholar] [CrossRef]
- Liu, L.; Li, X.; Shi, J.; Li, L.; Wang, J.; Luo, Z.Z. Effects of FPR2 gene silencing on the proliferation, migration and invasion of human glioma U87 cells. Zhonghua Zhong Liu Za Zhi (Chin. J. Oncol.) 2018, 40, 659–666. [Google Scholar] [CrossRef]
- Albertini, R.; Bianchi, R. Aquaporins and Glia. Curr. Neuropharmacol. 2010, 8, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Manley, G.; Binder, D.; Papadopoulos, M.; Verkman, A. New insights into water transport and edema in the central nervous system from phenotype analysis of aquaporin-4 null mice. Neuroscience 2004, 129, 981–989. [Google Scholar] [CrossRef] [PubMed]
- Badaut, J. Aquaglyceroporin 9 in brain pathologies. Neuroscience 2010, 168, 1047–1057. [Google Scholar] [CrossRef] [PubMed]
- Jelen, S.; Ulhøi, B.P.; Larsen, A.; Frøkiær, J.; Nielsen, S.; Rützler, M. AQP9 Expression in Glioblastoma Multiforme Tumors Is Limited to a Small Population of Astrocytic Cells and CD15+/CalB+ Leukocytes. PLoS ONE 2013, 8, e75764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, K.-T.; Fu, P.; Juerchott, K.; Motaln, H.; Selbig, J.; Lah, T.; Tonn, J.-C.; Schichor, C. The expression of Wnt-inhibitor DKK1 (Dickkopf 1) is determined by intercellular crosstalk and hypoxia in human malignant gliomas. J. Cancer Res. Clin. Oncol. 2014, 140, 1261–1270. [Google Scholar] [CrossRef]
- Chen, Y.; Qin, Y.; Dai, M.; Liu, L.; Ni, Y.; Sun, Q.; Li, L.; Zhou, Y.; Qiu, C.; Jiang, Y. IBSP, a potential recurrence biomarker, promotes the progression of colorectal cancer via Fyn/β-catenin signaling pathway. Cancer Med. 2021, 10, 4030–4045. [Google Scholar] [CrossRef]
- Mueller, W.; Lass, U.; Wellmann, S.; Kunitz, F.; Von Deimling, A. Mutation analysis of DKK1 and in vivo evidence of predominant p53-independent DKK1 function in gliomas. Acta Neuropathol. 2005, 109, 314–320. [Google Scholar] [CrossRef]
- Ghinda, D.C.; Yang, Y.; Wu, S.; Lu, J.; Su, L.; Damiani, S.; Tumati, S.; Jansen, G.; Duffau, H.; Wu, J.-S.; et al. Personalized Multimodal Demarcation of Peritumoral Tissue in Glioma. JCO Precis. Oncol. 2020, 4, 1128–1140. [Google Scholar] [CrossRef] [PubMed]
- Darmanis, S.; Sloan, S.A.; Croote, D.; Mignardi, M.; Chernikova, S.; Samghababi, P.; Zhang, Y.; Neff, N.; Kowarsky, M.; Caneda, C.; et al. Single-Cell RNA-Seq Analysis of Infiltrating Neoplastic Cells at the Migrating Front of Human Glioblastoma. Cell Rep. 2017, 21, 1399–1410. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Characteristic | Low Edema Group | High Edema Group | p |
|---|---|---|---|
| n | 116 | 62 | |
| Gender, n (%) | 0.354 | ||
| Female | 50 (28.7%) | 33 (19%) | |
| Male | 62 (35.6%) | 29 (16.7%) | |
| Laterality, n (%) | 0.787 | ||
| Left | 55 (32%) | 27 (15.7%) | |
| Midline | 2 (1.2%) | 1 (0.6%) | |
| Right | 54 (31.4%) | 33 (19.2%) | |
| Tumor location, n (%) | 0.721 | ||
| Posterior Fossa, Cerebellum | 1 (0.6%) | 0 (0%) | |
| Supratentorial, Frontal Lobe | 64 (36.8%) | 38 (21.8%) | |
| Supratentorial, Not Otherwise | 3 (1.7%) | 1 (0.6%) | |
| Supratentorial, Occipital Lobe | 0 (0%) | 1 (0.6%) | |
| Supratentorial, Parietal Lobe | 13 (7.5%) | 6 (3.4%) | |
| Supratentorial, Temporal Lobe | 31 (17.8%) | 16 (9.2%) | |
| Histologic grade, n (%) | 0.094 | ||
| G2 | 65 (37.4%) | 27 (15.5%) | |
| G3 | 47 (27%) | 35 (20.1%) | |
| IDH1, n (%) | 1.000 | ||
| Wild | 28 (15.8%) | 15 (8.5%) | |
| Mutant | 88 (49.7%) | 46 (26%) | |
| 1p/19q co-del status, n (%) | 0.377 | ||
| Wild | 85 (48.6%) | 42 (24%) | |
| Mutant | 28 (16%) | 20 (11.4%) | |
| TP53, n (%) | 0.870 | ||
| Mutant | 60 (33.9%) | 30 (16.9%) | |
| Wild | 56 (31.6%) | 31 (17.5%) | |
| Age, median (IQR) | 39 (30, 52.25) | 48.5 (37, 57) | 0.022 |
| Rank | Name | Score |
|---|---|---|
| 1 | IL10 | 12 |
| 2 | FCGR3B | 8 |
| 3 | S100A8 | 6 |
| 4 | AQP9 | 5 |
| 4 | S100A9 | 5 |
| 4 | FPR2 | 5 |
| 4 | SAA1 | 5 |
| 8 | HK3 | 4 |
| 8 | DKK1 | 4 |
| 10 | IBSP | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Peng, Z.; Wang, H.; Xiang, W. Identifying the Hub Genes of Glioma Peritumoral Brain Edema Using Bioinformatical Methods. Brain Sci. 2022, 12, 805. https://doi.org/10.3390/brainsci12060805
Wu Y, Peng Z, Wang H, Xiang W. Identifying the Hub Genes of Glioma Peritumoral Brain Edema Using Bioinformatical Methods. Brain Sciences. 2022; 12(6):805. https://doi.org/10.3390/brainsci12060805
Chicago/Turabian StyleWu, Yuxi, Zesheng Peng, Haofei Wang, and Wei Xiang. 2022. "Identifying the Hub Genes of Glioma Peritumoral Brain Edema Using Bioinformatical Methods" Brain Sciences 12, no. 6: 805. https://doi.org/10.3390/brainsci12060805
APA StyleWu, Y., Peng, Z., Wang, H., & Xiang, W. (2022). Identifying the Hub Genes of Glioma Peritumoral Brain Edema Using Bioinformatical Methods. Brain Sciences, 12(6), 805. https://doi.org/10.3390/brainsci12060805






