Characterization of Ex Vivo and In Vitro Wnt Transcriptome Induced by Spinal Cord Injury in Rat Microglial Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Surgical Procedures
2.2. Primary Microglia Cell Cultures and Stimulation with SCE
2.2.1. Preparation of the SCE
2.2.2. Microglial Cell Cultures
2.2.3. Assessment of Microglia Cell Culture Purity
2.3. Microglia Isolation from Spinal Cord Tissue
2.4. mRNA Isolation and Quantitative qRT-PCR
2.5. Statistical Analysis
3. Results
3.1. Differential Expression of Wnt Family Genes in Intact and Injured Rat Spinal Cord Isolated Microglia/MDM Cells
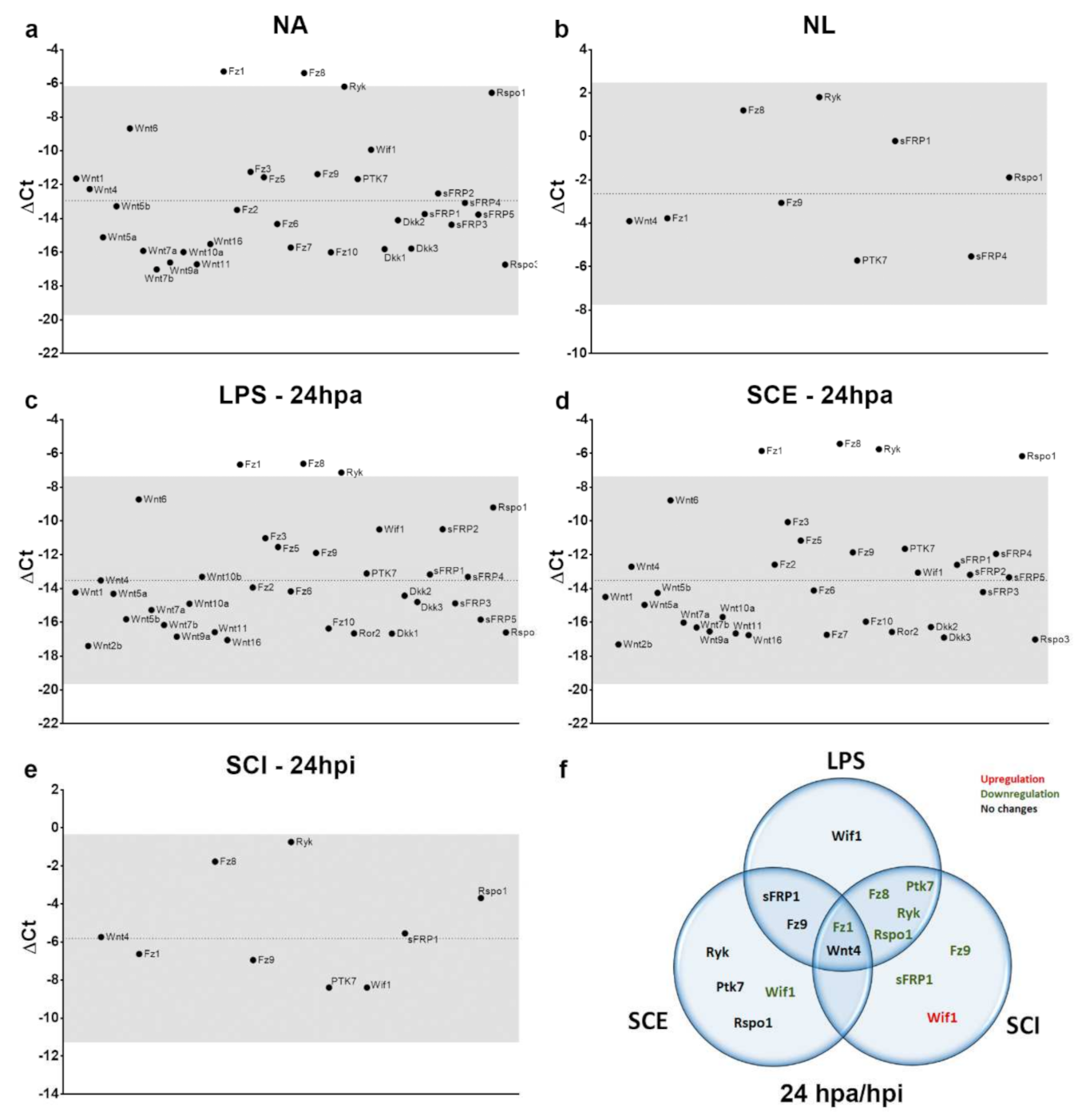
3.2. Differential Expression of Wnt Family Genes in Rat Non-Activated and Activated Microglial Cultured Cells When Exposed to a 24 hpi-SCE Versus a Standard LPS Protocol of Stimulation
3.2.1. Wnt Ligands
3.2.2. Wnt Receptors
3.2.3. Wnt Modulators
3.3. Differential Expression of Wnt Family Genes by Microglia/MDM Cells Depending on the Experimental Approach (Ex Vivo or In Vitro) Used
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Kroner, A.; Almanza, J.R. Role of microglia in spinal cord injury. Neurosci. Lett. 2019, 709, 134370. [Google Scholar] [CrossRef] [PubMed]
- Kabba, J.A.; Xu, Y.; Christian, H.; Ruan, W.; Chenai, K.; Xiang, Y.; Zhang, L.; Saavedra, J.M.; Pang, T. Microglia: Housekeeper of the Central Nervous System. Cell. Mol. Neurobiol. 2018, 38, 53–71. [Google Scholar] [CrossRef]
- Masuda, T.; Sankowski, R.; Staszewski, O.; Prinz, M. Microglia Heterogeneity in the Single-Cell Era. Cell Rep. 2020, 30, 1271–1281. [Google Scholar] [CrossRef] [PubMed]
- Brockie, S.; Hong, J.; Fehlings, M.G. The Role of Microglia in Modulating Neuroinflammation after Spinal Cord Injury. Int. J. Mol. Sci. 2021, 22, 9706. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Tan, Y.H.; Light, A.R.; Fu, K.Y. Different peripheral tissue injury induces differential phenotypic changes of spinal activated microglia. Clin. Dev. Immunol. 2013, 2013, 901420. [Google Scholar] [CrossRef] [PubMed]
- Schwab, J.M.; Zhang, Y.; Kopp, M.A.; Brommer, B.; Popovich, P.G. The paradox of chronic neuroinflammation, systemic immune suppression, autoimmunity after traumatic chronic spinal cord injury. Exp. Neurol. 2014, 258, 121–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heneka, M.T.; Kummer, M.P.; Latz, E. Innate immune activation in neurodegenerative disease. Nat. Rev. Immunol. 2014, 14, 463–477. [Google Scholar] [CrossRef]
- Stephenson, J.; Nutma, E.; van der Valk, P.; Amor, S. Inflammation in CNS neurodegenerative diseases. Immunology 2018, 154, 204–219. [Google Scholar] [CrossRef] [Green Version]
- Donnelly, D.J.; Popovich, P.G. Inflammation and its role in neuroprotection, axonal regeneration and functional recovery after spinal cord injury. Exp. Neurol. 2008, 209, 378–388. [Google Scholar] [CrossRef] [Green Version]
- Bradbury, E.J.; Burnside, E.R. Moving beyond the glial scar for spinal cord repair. Nat. Commun. 2019, 10, 3879. [Google Scholar] [CrossRef]
- Alizadeh, A.; Dyck, S.M.; Karimi-Abdolrezaee, S. Traumatic Spinal Cord Injury: An Overview of Pathophysiology, Models and Acute Injury Mechanisms. Front. Neurol. 2019, 10, 282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Profyris, C.; Cheema, S.S.; Zang, D.; Azari, M.F.; Boyle, K.; Petratos, S. Degenerative and regenerative mechanisms governing spinal cord injury. Neurobiol. Dis. 2004, 15, 415–436. [Google Scholar] [CrossRef] [PubMed]
- Akhmetzyanova, E.; Kletenkov, K.; Mukhamedshina, Y.; Rizvanov, A. Different Approaches to Modulation of Microglia Phenotypes After Spinal Cord Injury. Front. Syst. Neurosci. 2019, 13, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dokalis, N.; Prinz, M. Resolution of neuroinflammation: Mechanisms and potential therapeutic option. Semin. Immunopathol. 2019, 41, 699–709. [Google Scholar] [CrossRef]
- David, S.; Kroner, A.; Greenhalgh, A.D.; Zarruk, J.G.; Lopez-Vales, R. Myeloid cell responses after spinal cord injury. J. Neuroimmunol. 2018, 321, 97–108. [Google Scholar] [CrossRef] [Green Version]
- Bellver-Landete, V.; Bretheau, F.; Mailhot, B.; Vallieres, N.; Lessard, M.; Janelle, M.E.; Vernoux, N.; Tremblay, M.E.; Fuehrmann, T.; Shoichet, M.S.; et al. Microglia are an essential component of the neuroprotective scar that forms after spinal cord injury. Nat. Commun. 2019, 10, 518. [Google Scholar] [CrossRef] [Green Version]
- Marchetti, B.; Pluchino, S. Wnt your brain be inflamed? Yes, it Wnt! Trends Mol. Med. 2013, 19, 144–156. [Google Scholar] [CrossRef] [Green Version]
- Lambert, C.; Cisternas, P.; Inestrosa, N.C. Role of Wnt Signaling in Central Nervous System Injury. Mol. Neurobiol. 2015, 53, 2297–2311. [Google Scholar] [CrossRef]
- Libro, R.; Bramanti, P.; Mazzon, E. The role of the Wnt canonical signaling in neurodegenerative diseases. Life Sci. 2016, 158, 78–88. [Google Scholar] [CrossRef]
- L’Episcopo, F.; Tirolo, C.; Serapide, M.F.; Caniglia, S.; Testa, N.; Leggio, L.; Vivarelli, S.; Iraci, N.; Pluchino, S.; Marchetti, B. Microglia Polarization, Gene-Environment Interactions and Wnt/beta-Catenin Signaling: Emerging Roles of Glia-Neuron and Glia-Stem/Neuroprogenitor Crosstalk for Dopaminergic Neurorestoration in Aged Parkinsonian Brain. Front. Aging Neurosci. 2018, 10, 12. [Google Scholar] [CrossRef] [Green Version]
- Lu, P.; Han, D.; Zhu, K.; Jin, M.; Mei, X.; Lu, H. Effects of Sirtuin 1 on microglia in spinal cord injury: Involvement of Wnt/beta-catenin signaling pathway. Neuroreport 2019, 30, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Itokazu, T.; Hayano, Y.; Takahashi, R.; Yamashita, T. Involvement of Wnt/beta-catenin signaling in the development of neuropathic pain. Neurosci. Res. 2014, 79, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Sonn, I.; Nakamura, M.; Renault-Mihara, F.; Okano, H. Polarization of Reactive Astrocytes in Response to Spinal Cord Injury is Enhanced by M2 Macrophage-Mediated Activation of Wnt/beta-Catenin Pathway. Mol. Neurobiol. 2019, 57, 1847–1862. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, X.; Lu, C.C.; Kerman, R.; Steward, O.; Xu, X.M.; Zou, Y. Repulsive Wnt signaling inhibits axon regeneration after CNS injury. J. Neurosci. 2008, 28, 8376–8382. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, T.; Koda, M.; Kitajo, K.; Yamazaki, M.; Takahashi, K.; Kikuchi, A.; Yamashita, T. Wnt-Ryk signaling mediates axon growth inhibition and limits functional recovery after spinal cord injury. J. Neurotrauma 2009, 26, 955–964. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Martos, C.M.; Gonzalez-Fernandez, C.; Gonzalez, P.; Maqueda, A.; Arenas, E.; Rodriguez, F.J. Differential expression of Wnts after spinal cord contusion injury in adult rats. PLoS ONE 2011, 6, e27000. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, P.; Fernandez-Martos, C.M.; Gonzalez-Fernandez, C.; Arenas, E.; Rodriguez, F.J. Spatio-temporal expression pattern of frizzled receptors after contusive spinal cord injury in adult rats. PLoS ONE 2012, 7, e50793. [Google Scholar] [CrossRef]
- Gonzalez, P.; Fernandez-Martos, C.M.; Arenas, E.; Rodriguez, F.J. The Ryk Receptor Is Expressed in Glial and Fibronectin-Expressing Cells after Spinal Cord Injury. J. Neurotrauma 2013, 30, 806–817. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Fernandez, C.; Fernandez-Martos, C.M.; Shields, S.; Arenas, E.; Rodriguez, F.J. Wnts are expressed in the spinal cord of adult mice and are differentially induced after injury. J. Neurotrauma 2013, 31, 565–581. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, P.; Gonzalez-Fernandez, C.; Campos-Martin, Y.; Mollejo, M.; Carballosa-Gautam, M.; Marcillo, A.; Norenberg, M.; Rodriguez, F.J. Frizzled 1 and Wnt1 as new potential therapeutic targets in the traumatically injured spinal cord. Cell Mol. Life Sci. 2020, 77, 4631–4662. [Google Scholar] [CrossRef]
- González, P.; González-Fernández, C.; Campos-Martín, Y.; Mollejo, M.; Carballosa-Gautam, M.; Marcillo, A.; Norenberg, M.; García-Ovejero, D.; Rodríguez, F.J. Spatio-temporal and Cellular Expression Patterns of PTK7 in the Healthy and Traumatically Injured Rat and Human Spinal Cord. Cell. Mol. Neurobiol. 2020, 40, 1087–1103. [Google Scholar] [CrossRef] [PubMed]
- Halleskog, C.; Dijksterhuis, J.P.; Kilander, M.B.; Becerril-Ortega, J.; Villaescusa, J.C.; Lindgren, E.; Arenas, E.; Schulte, G. Heterotrimeric G protein-dependent WNT-5A signaling to ERK1/2 mediates distinct aspects of microglia proinflammatory transformation. J. Neuroinflammation 2012, 9, 111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halleskog, C.; Mulder, J.; Dahlstrom, J.; Mackie, K.; Hortobagyi, T.; Tanila, H.; Kumar Puli, L.; Farber, K.; Harkany, T.; Schulte, G. WNT signaling in activated microglia is proinflammatory. Glia 2011, 59, 119–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, D.; Zhang, X.; Chen, J.; Liu, X.; Xue, J.; Zhang, L.; Lan, X. Wnt canonical pathway activator TWS119 drives microglial anti-inflammatory activation and facilitates neurological recovery following experimental stroke. J. Neuroinflammation 2019, 16, 256. [Google Scholar] [CrossRef]
- Van Steenwinckel, J.; Schang, A.L.; Krishnan, M.L.; Degos, V.; Delahaye-Duriez, A.; Bokobza, C.; Csaba, Z.; Verdonk, F.; Montane, A.; Sigaut, S.; et al. Decreased microglial Wnt/beta-catenin signalling drives microglial pro-inflammatory activation in the developing brain. Brain 2019, 142, 3806–3833. [Google Scholar] [CrossRef]
- Zhang, D.; Lu, Z.; Man, J.; Cui, K.; Fu, X.; Yu, L.; Gao, Y.; Liao, L.; Xiao, Q.; Guo, R.; et al. Wnt-3a alleviates neuroinflammation after ischemic stroke by modulating the responses of microglia/macrophages and astrocytes. Int. Immunopharmacol. 2019, 75, 105760. [Google Scholar] [CrossRef]
- Han, D.; Zhou, Y. YY1-induced upregulation of lncRNA NEAT1 contributes to OGD/R injury-induced inflammatory response in cerebral microglial cells via Wnt/beta-catenin signaling pathway. In Vitro Cell. Dev. Biol. Anim. 2019, 55, 501–511. [Google Scholar] [CrossRef]
- Yao, Y.Y.; Bian, L.G.; Yang, P.; Sui, Y.; Li, R.; Chen, Y.L.; Sun, L.; Ai, Q.L.; Zhong, L.M.; Lu, D. Gastrodin attenuates proliferation and inflammatory responses in activated microglia through Wnt/beta-catenin signaling pathway. Brain Res. 2019, 1717, 190–203. [Google Scholar] [CrossRef]
- Cao, W.; Dong, Y.; Zhao, W.; Lu, X.; Sun, L. Mulberrin attenuates 1-methyl-4-phenyl-1,2,3,6- tetrahydropyridine (MPTP)-induced Parkinson’s disease by promoting Wnt/beta-catenin signaling pathway. J. Chem. Neuroanat. 2019, 98, 63–70. [Google Scholar] [CrossRef]
- Matias, D.; Dubois, L.G.; Pontes, B.; Rosario, L.; Ferrer, V.P.; Balca-Silva, J.; Fonseca, A.C.C.; Macharia, L.W.; Romao, L.; TCLS, E.S.; et al. GBM-Derived Wnt3a Induces M2-Like Phenotype in Microglial Cells Through Wnt/beta-Catenin Signaling. Mol. Neurobiol. 2019, 56, 1517–1530. [Google Scholar] [CrossRef]
- Halleskog, C.; Schulte, G. WNT-3A and WNT-5A counteract lipopolysaccharide-induced pro-inflammatory changes in mouse primary microglia. J. Neurochem. 2013, 125, 803–808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halleskog, C.; Schulte, G. Pertussis toxin-sensitive heterotrimeric G(alphai/o) proteins mediate WNT/beta-catenin and WNT/ERK1/2 signaling in mouse primary microglia stimulated with purified WNT-3A. Cell Signal 2013, 25, 822–828. [Google Scholar] [CrossRef]
- Mulder, J.; Zilberter, M.; Pasquare, S.J.; Alpar, A.; Schulte, G.; Ferreira, S.G.; Kofalvi, A.; Martin-Moreno, A.M.; Keimpema, E.; Tanila, H.; et al. Molecular reorganization of endocannabinoid signalling in Alzheimer’s disease. Brain 2011, 134, 1041–1060. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bao, D.J.; Xu, B.; Cheng, C.D.; Dong, Y.F.; Wei, X.P.; Niu, C.S. Neuroprotection mediated by the Wnt/Frizzled signaling pathway in early brain injury induced by subarachnoid hemorrhage. Neural Regen. Res. 2019, 14, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Chong, Z.Z.; Li, F.; Maiese, K. Cellular demise and inflammatory microglial activation during beta-amyloid toxicity are governed by Wnt1 and canonical signaling pathways. Cell Signal 2007, 19, 1150–1162. [Google Scholar] [CrossRef] [Green Version]
- L’Episcopo, F.; Tirolo, C.; Peruzzotti-Jametti, L.; Serapide, M.F.; Testa, N.; Caniglia, S.; Balzarotti, B.; Pluchino, S.; Marchetti, B. Neural Stem Cell Grafts Promote Astroglia-Driven Neurorestoration in the Aged Parkinsonian Brain via Wnt/beta-Catenin Signaling. Stem Cells 2018, 36, 1179–1197. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, J.P.; Coulter, M.; Miotke, J.; Meyer, R.L.; Takemaru, K.; Levine, J.M. Abrogation of beta-catenin signaling in oligodendrocyte precursor cells reduces glial scarring and promotes axon regeneration after CNS injury. J. Neurosci. 2014, 34, 10285–10297. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; Jia, L.; Liu, C.C.; Rong, Z.; Zhong, L.; Yang, L.; Chen, X.F.; Fryer, J.D.; Wang, X.; Zhang, Y.W.; et al. TREM2 Promotes Microglial Survival by Activating Wnt/beta-Catenin Pathway. J. Neurosci. 2017, 37, 1772–1784. [Google Scholar] [CrossRef] [Green Version]
- Shang, Y.C.; Chong, Z.Z.; Wang, S.; Maiese, K. Erythropoietin and Wnt1 govern pathways of mTOR, Apaf-1, and XIAP in inflammatory microglia. Curr. Neurovascular Res. 2011, 8, 270–285. [Google Scholar] [CrossRef]
- Shang, Y.C.; Chong, Z.Z.; Hou, J.; Maiese, K. Wnt1, FoxO3a, and NF-kappaB oversee microglial integrity and activation during oxidant stress. Cell Signal 2010, 22, 1317–1329. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Gibb, S.L.; Zhao, J.; Moore, A.N.; Hylin, M.J.; Menge, T.; Xue, H.; Baimukanova, G.; Potter, D.; Johnson, E.M.; et al. Wnt3a, a Protein Secreted by Mesenchymal Stem Cells Is Neuroprotective and Promotes Neurocognitive Recovery Following Traumatic Brain Injury. Stem Cells 2016, 34, 1263–1272. [Google Scholar] [CrossRef] [Green Version]
- Dohare, P.; Cheng, B.; Ahmed, E.; Yadala, V.; Singla, P.; Thomas, S.; Kayton, R.; Ungvari, Z.; Ballabh, P. Glycogen synthase kinase-3beta inhibition enhances myelination in preterm newborns with intraventricular hemorrhage, but not recombinant Wnt3A. Neurobiol. Dis. 2018, 118, 22–39. [Google Scholar] [CrossRef] [PubMed]
- Kilander, M.B.; Halleskog, C.; Schulte, G. Recombinant WNTs differentially activate beta-catenin-dependent and -independent signalling in mouse microglia-like cells. Acta Physiol. 2011, 203, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, P.; Gonzalez-Fernandez, C.; Javier Rodriguez, F. Effects of Wnt5a overexpression in spinal cord injury. J. Cell. Mol. Med. 2021, 25, 5150–5163. [Google Scholar] [CrossRef] [PubMed]
- Bohlen, C.J.; Bennett, F.C.; Bennett, M.L. Isolation and Culture of Microglia. Curr. Protoc. Immunol. 2019, 125, e70. [Google Scholar] [CrossRef] [PubMed]
- Lively, S.; Schlichter, L.C. Microglia Responses to Pro-inflammatory Stimuli (LPS, IFNgamma+TNFalpha) and Reprogramming by Resolving Cytokines (IL-4, IL-10). Front. Cell Neurosci. 2018, 12, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, D.; Lively, S.; Schlichter, L.C. Responses of rat and mouse primary microglia to pro- and anti-inflammatory stimuli: Molecular profiles, K(+) channels and migration. J. Neuroinflammation 2017, 14, 166. [Google Scholar] [CrossRef] [Green Version]
- David, S.; Kroner, A. Repertoire of microglial and macrophage responses after spinal cord injury. Nat. Rev. Neurosci. 2011, 12, 388–399. [Google Scholar] [CrossRef]
- Gonzalez, P.; Rodriguez, F.J. Analysis of the expression of the Wnt family of proteins and its modulatory role on cytokine expression in non activated and activated astroglial cells. Neurosci. Res. 2017, 114, 16–29. [Google Scholar] [CrossRef]
- Hernandez, J.; Francos-Quijorna, I.; Redondo-Castro, E.; Lopez-Vales, R.; Navarro, X. Microglia Stimulation by Protein Extract of Injured Rat Spinal Cord. A Novel In vitro Model for Studying Activated Microglia. Front. Mol. Neurosci. 2021, 14, 582497. [Google Scholar] [CrossRef]
- Bowes, A.L.; Yip, P.K. Modulating inflammatory cell responses to spinal cord injury: All in good time. J. Neurotrauma 2014, 31, 1753–1766. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Fernandez, C.; Gonzalez, P.; Andres-Benito, P.; Ferrer, I.; Rodriguez, F.J. Wnt Signaling Alterations in the Human Spinal Cord of Amyotrophic Lateral Sclerosis Cases: Spotlight on Fz2 and Wnt5a. Mol. Neurobiol. 2019, 56, 6777–6791. [Google Scholar] [CrossRef] [PubMed]
- Sharif-Alhoseini, M.; Khormali, M.; Rezaei, M.; Safdarian, M.; Hajighadery, A.; Khalatbari, M.M.; Safdarian, M.; Meknatkhah, S.; Rezvan, M.; Chalangari, M.; et al. Animal models of spinal cord injury: A systematic review. Spinal Cord 2017, 55, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Rowland, J.W.; Hawryluk, G.W.; Kwon, B.; Fehlings, M.G. Current status of acute spinal cord injury pathophysiology and emerging therapies: Promise on the horizon. Neurosurg. Focus 2008, 25, E2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beck, K.D.; Nguyen, H.X.; Galvan, M.D.; Salazar, D.L.; Woodruff, T.M.; Anderson, A.J. Quantitative analysis of cellular inflammation after traumatic spinal cord injury: Evidence for a multiphasic inflammatory response in the acute to chronic environment. Brain 2010, 133, 433–447. [Google Scholar] [CrossRef]
- Kramps, J.A.; van der Valk, P.; van der Sandt, M.M.; Lindeman, J.; Meijer, C.J. Elastase as a marker for neutrophilic myeloid cells. J. Histochem. Cytochem. 1984, 32, 389–394. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Orihuela, R.; McPherson, C.A.; Harry, G.J. Microglial M1/M2 polarization and metabolic states. Br. J. Pharmacol. 2016, 173, 649–665. [Google Scholar] [CrossRef]
- David, S.; Greenhalgh, A.D.; Kroner, A. Macrophage and microglial plasticity in the injured spinal cord. Neuroscience 2015, 307, 311–318. [Google Scholar] [CrossRef]
- Kjell, J.; Olson, L. Rat models of spinal cord injury: From pathology to potential therapies. Dis. Model. Mech. 2016, 9, 1125–1137. [Google Scholar] [CrossRef] [Green Version]
- Jurga, A.M.; Paleczna, M.; Kuter, K.Z. Overview of General and Discriminating Markers of Differential Microglia Phenotypes. Front. Cell. Neurosci. 2020, 14, 198. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.L.; Bennett, F.C.; Liddelow, S.A.; Ajami, B.; Zamanian, J.L.; Fernhoff, N.B.; Mulinyawe, S.B.; Bohlen, C.J.; Adil, A.; Tucker, A.; et al. New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. USA 2016, 113, E1738–E1746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, C.; Kros, J.M.; van der Weiden, M.; Zheng, P.; Cheng, C.; Mustafa, D.A. Expression site of P2RY12 in residential microglial cells in astrocytomas correlates with M1 and M2 marker expression and tumor grade. Acta Neuropathol. Commun. 2017, 5, 4. [Google Scholar] [CrossRef] [Green Version]
- Toledano Furman, N.E.; Prabhakara, K.S.; Bedi, S.; Cox, C.S., Jr.; Olson, S.D. OMIP-041: Optimized multicolor immunofluorescence panel rat microglial staining protocol. Cytometry A 2018, 93, 182–185. [Google Scholar] [CrossRef] [Green Version]
- Benmamar-Badel, A.; Owens, T.; Wlodarczyk, A. Protective Microglial Subset in Development, Aging, and Disease: Lessons From Transcriptomic Studies. Front. Immunol. 2020, 11, 430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noristani, H.N.; Gerber, Y.N.; Sabourin, J.C.; Le Corre, M.; Lonjon, N.; Mestre-Frances, N.; Hirbec, H.E.; Perrin, F.E. RNA-Seq Analysis of Microglia Reveals Time-Dependent Activation of Specific Genetic Programs following Spinal Cord Injury. Front. Mol. Neurosci. 2017, 10, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wahane, S.; Zhou, X.; Zhou, X.; Guo, L.; Friedl, M.S.; Kluge, M.; Ramakrishnan, A.; Shen, L.; Friedel, C.C.; Zhang, B.; et al. Diversified transcriptional responses of myeloid and glial cells in spinal cord injury shaped by HDAC3 activity. Sci. Adv. 2021, 7, eabd8811. [Google Scholar] [CrossRef]
- Zhu, Y.; Lyapichev, K.; Lee, D.H.; Motti, D.; Ferraro, N.M.; Zhang, Y.; Yahn, S.; Soderblom, C.; Zha, J.; Bethea, J.R.; et al. Macrophage Transcriptional Profile Identifies Lipid Catabolic Pathways That Can Be Therapeutically Targeted after Spinal Cord Injury. J. Neurosci. 2017, 37, 2362–2376. [Google Scholar] [CrossRef]
- Milich, L.M.; Choi, J.S.; Ryan, C.; Cerqueira, S.R.; Benavides, S.; Yahn, S.L.; Tsoulfas, P.; Lee, J.K. Single-cell analysis of the cellular heterogeneity and interactions in the injured mouse spinal cord. J. Exp. Med. 2021, 218, e20210040. [Google Scholar] [CrossRef]
- Peng, Y.; Chu, S.; Yang, Y.; Zhang, Z.; Pang, Z.; Chen, N. Neuroinflammatory In Vitro Cell Culture Models and the Potential Applications for Neurological Disorders. Front. Pharmacol. 2021, 12, 671734. [Google Scholar] [CrossRef]
- Ouali Alami, N.; Schurr, C.; Olde Heuvel, F.; Tang, L.; Li, Q.; Tasdogan, A.; Kimbara, A.; Nettekoven, M.; Ottaviani, G.; Raposo, C.; et al. NF-kappaB activation in astrocytes drives a stage-specific beneficial neuroimmunological response in ALS. EMBO J. 2018, 37, e98697. [Google Scholar] [CrossRef] [PubMed]
- Mecha, M.; Yanguas-Casas, N.; Feliu, A.; Mestre, L.; Carrillo-Salinas, F.J.; Riecken, K.; Gomez-Nicola, D.; Guaza, C. Involvement of Wnt7a in the role of M2c microglia in neural stem cell oligodendrogenesis. J. Neuroinflammation 2020, 17, 88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butovsky, O.; Jedrychowski, M.P.; Moore, C.S.; Cialic, R.; Lanser, A.J.; Gabriely, G.; Koeglsperger, T.; Dake, B.; Wu, P.M.; Doykan, C.E.; et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat. Neurosci. 2014, 17, 131–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellwig, S.; Heinrich, A.; Biber, K. The brain’s best friend: Microglial neurotoxicity revisited. Front. Cell. Neurosci. 2013, 7, 71. [Google Scholar] [CrossRef] [Green Version]
- Haimon, Z.; Volaski, A.; Orthgiess, J.; Boura-Halfon, S.; Varol, D.; Shemer, A.; Yona, S.; Zuckerman, B.; David, E.; Chappell-Maor, L.; et al. Re-evaluating microglia expression profiles using RiboTag and cell isolation strategies. Nat. Immunol. 2018, 19, 636–644. [Google Scholar] [CrossRef]
- Bohlen, C.J.; Bennett, F.C.; Tucker, A.F.; Collins, H.Y.; Mulinyawe, S.B.; Barres, B.A. Diverse Requirements for Microglial Survival, Specification, and Function Revealed by Defined-Medium Cultures. Neuron 2017, 94, 759–773 e758. [Google Scholar] [CrossRef] [Green Version]
- Devanney, N.A.; Stewart, A.N.; Gensel, J.C. Microglia and macrophage metabolism in CNS injury and disease: The role of immunometabolism in neurodegeneration and neurotrauma. Exp. Neurol. 2020, 329, 113310. [Google Scholar] [CrossRef]
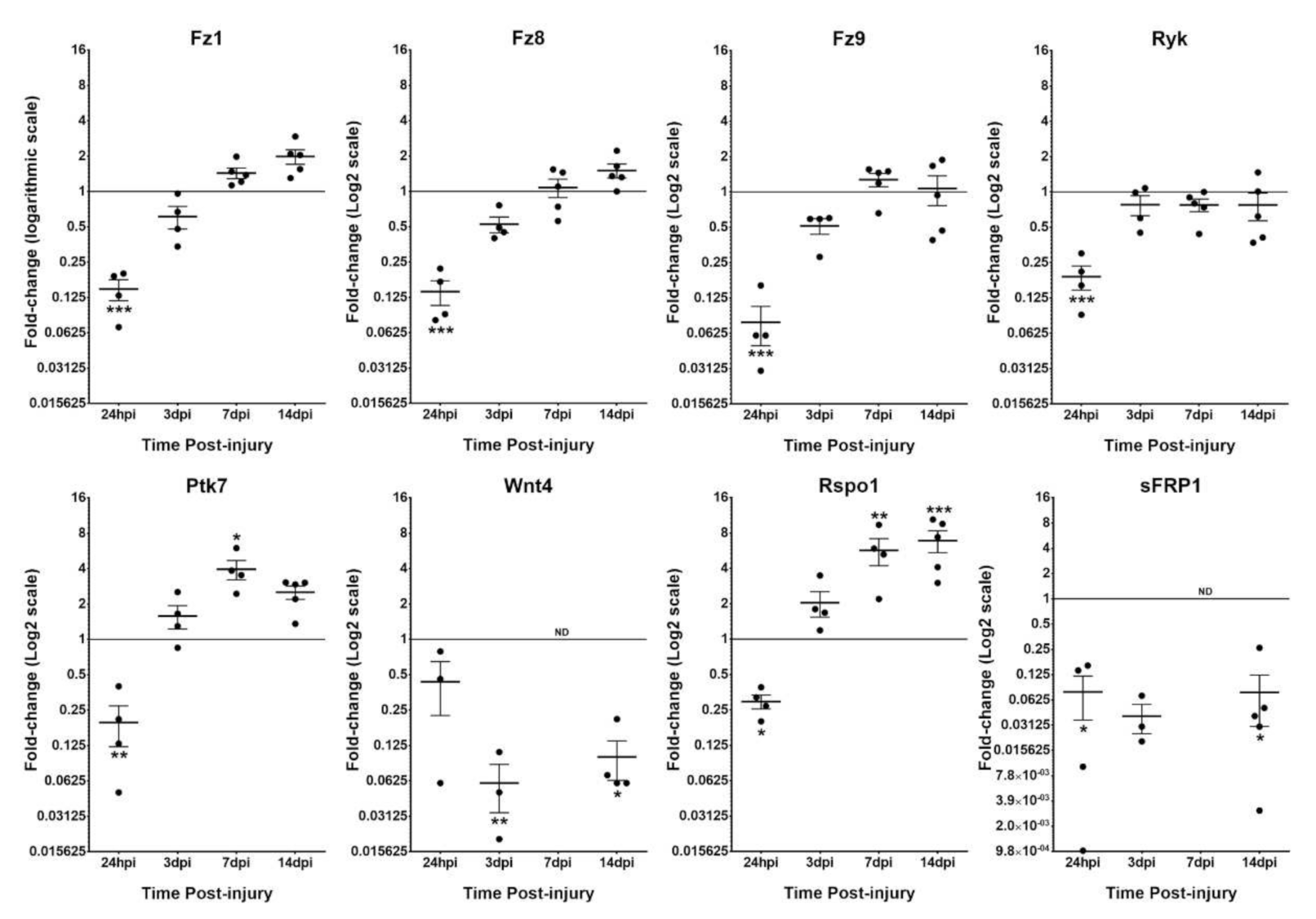
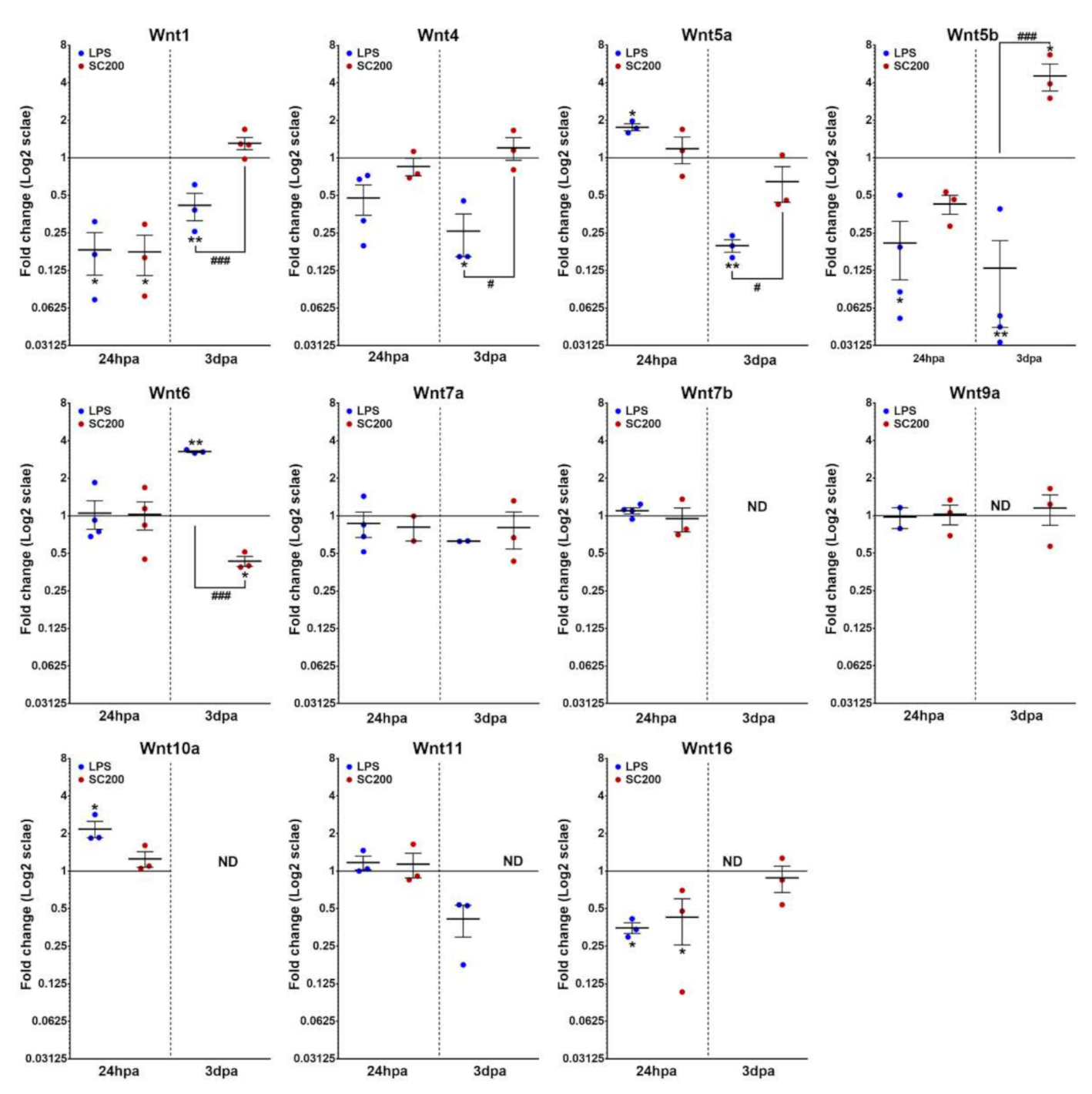
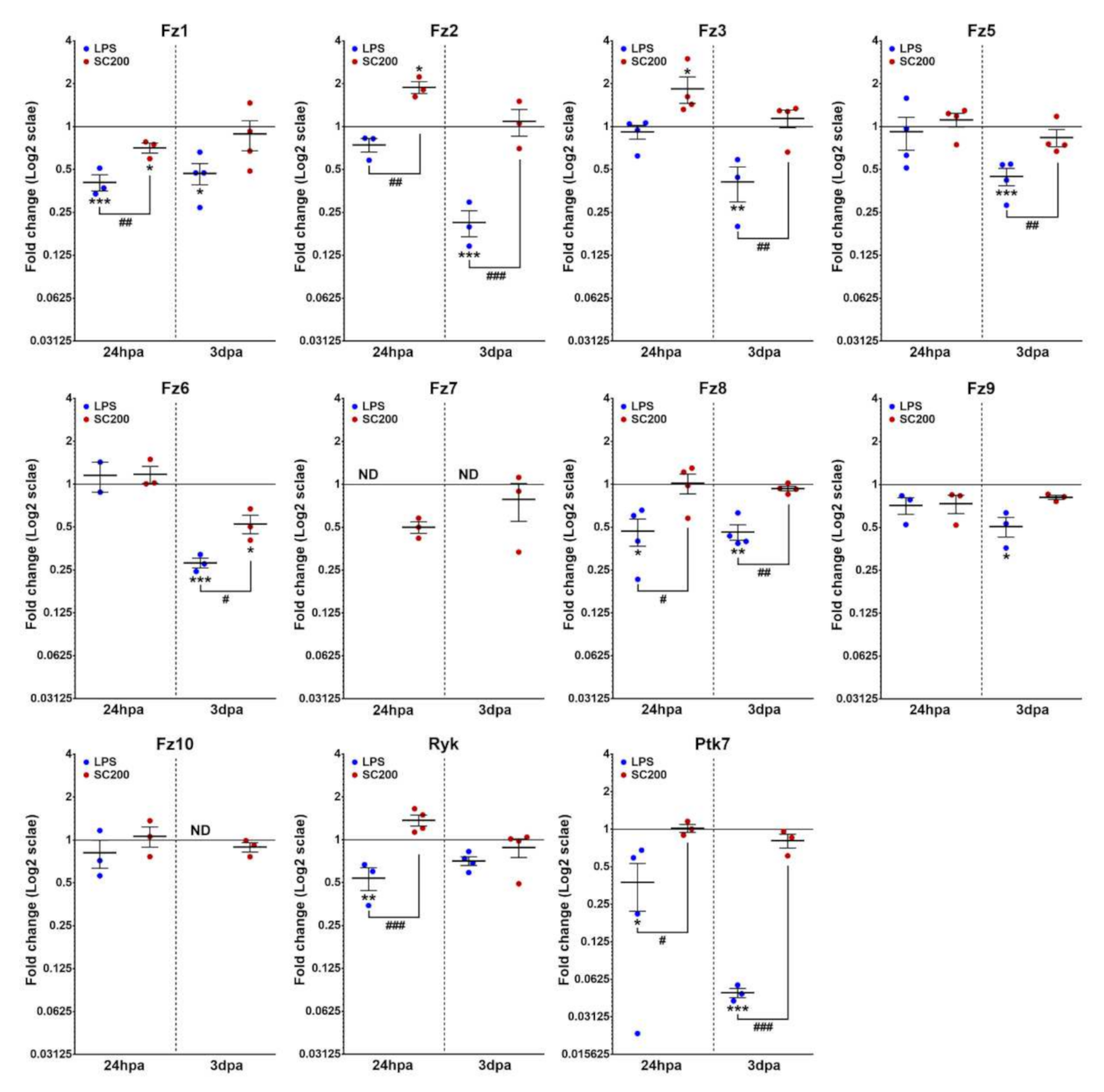
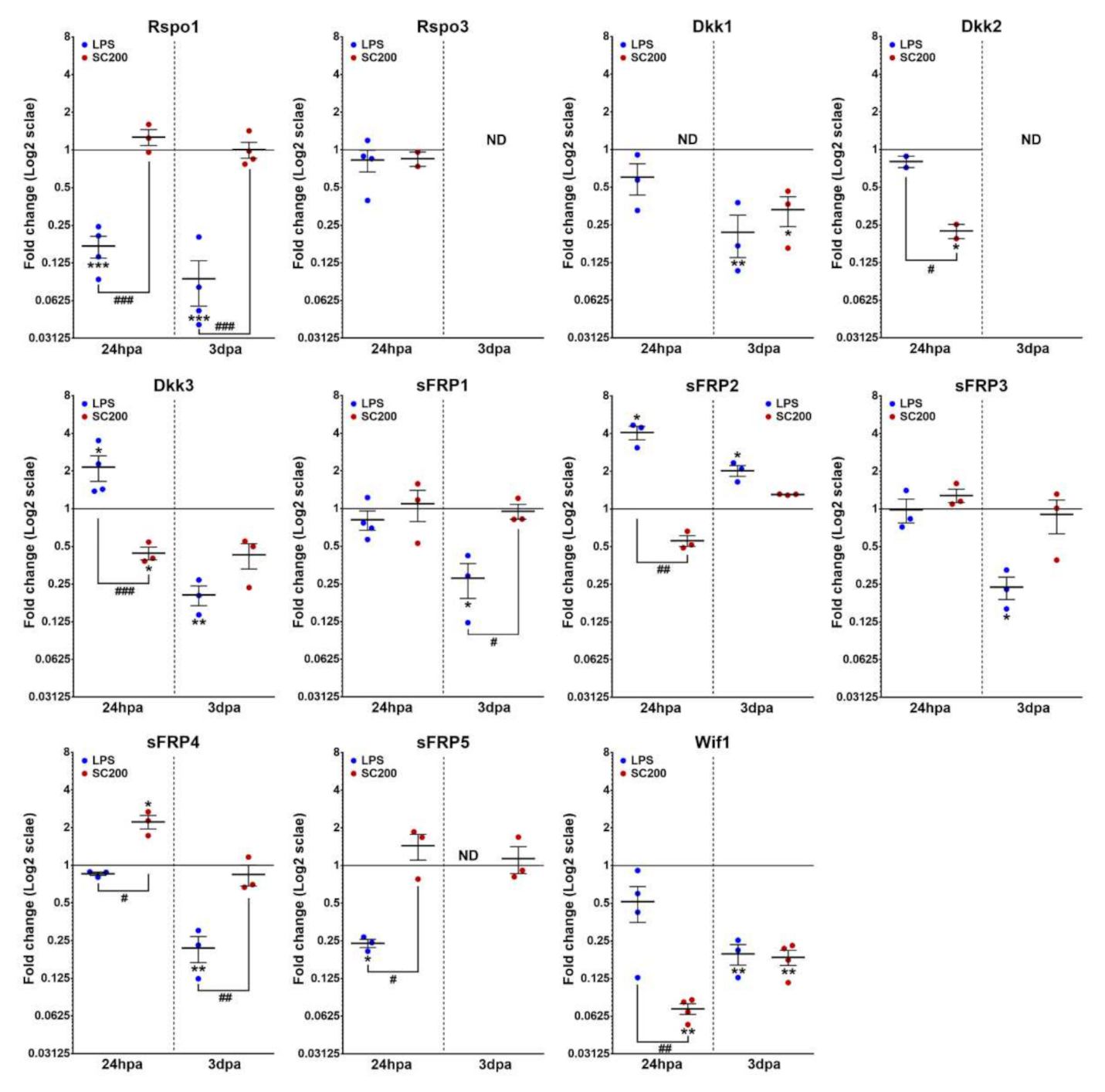
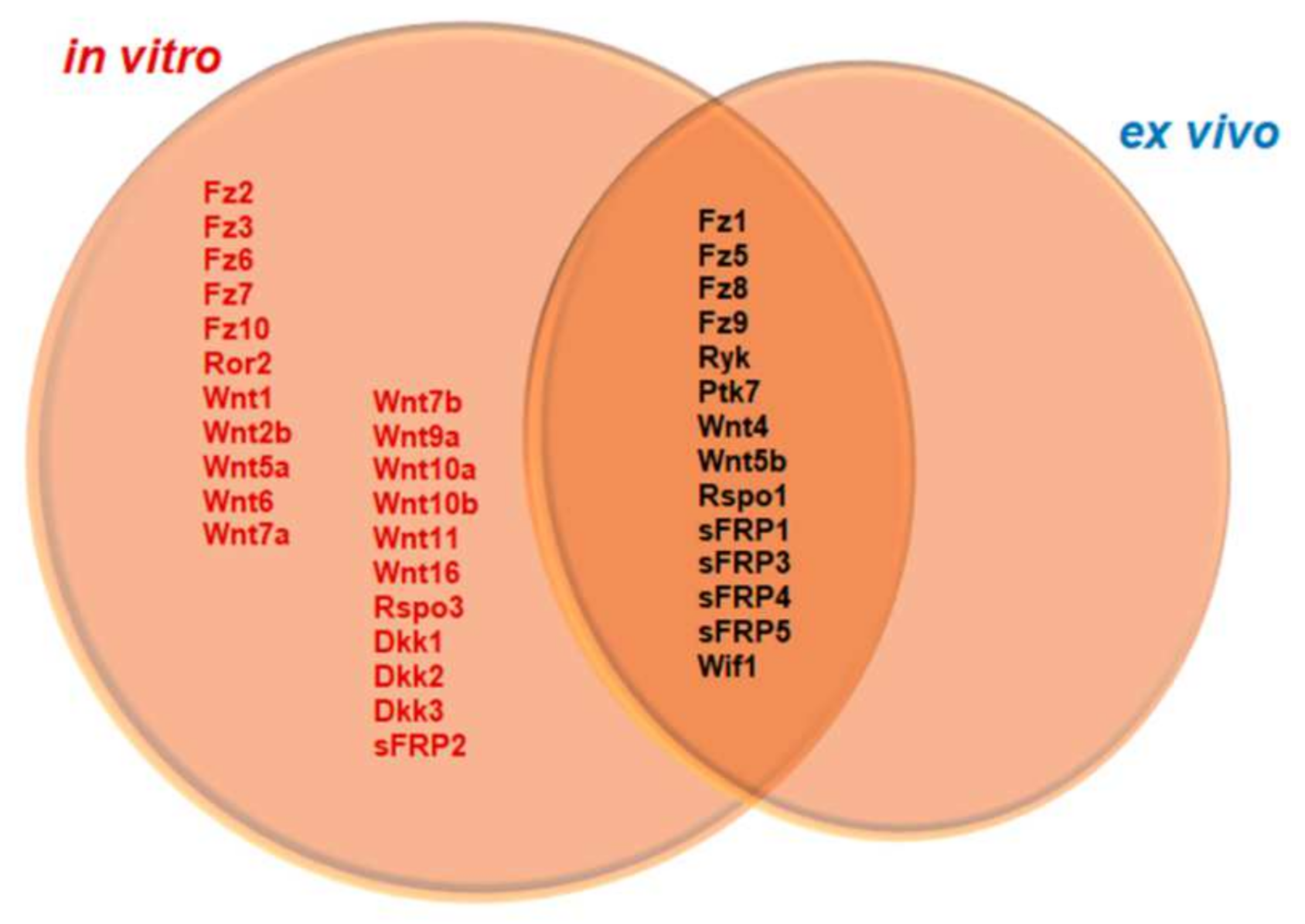
| Gene | NL | 24 hpi | 3 dpi | 7 dpi | 14 dpi |
|---|---|---|---|---|---|
| Fz1 | −3.77 ± 0.51 | −6.63 ± 0.34 | −4.58 ± 0.32 | −3.28 ± 0.14 | −2.84 ± 0.20 |
| Fz5 | ? | −9.31 ± 0.57 | −6.05 ± 0.35 | −5.71 ± 0.90 | −6.63 ± 0.94 |
| Fz8 | 1.20 ± 0.22 | −1.77 ± 0.36 | 0.22 ± 0.20 | 1.21 ± 0.28 | 1.74 ± 0.19 |
| Fz9 | −3.06 ± 0.12 | −6.95 ± 0.46 | −4.08 ± 0.27 | −2.78 ± 0.23 | −3.24 ± 0.46 |
| Ryk | 1.81 ± 0.16 | −0.74 ± 0.38 | 1.37 ± 0.30 | 1.39 ± 0.21 | 1.24 ± 0.38 |
| Ptk7 | −5.72 ± 0.61 | −8.40 ± 0.60 | −5.17 ± 0.33 | −3.81 ± 0.27 | −4.45 ± 0.22 |
| Wnt4 | −3.90 ± 0.24 | −5.74 ± 1.14 | −8.21 ± 0.66 | ND | −7.49 ± 0.45 |
| Wnt5b | ND | ND | −7.49 ± 1.02 | ND | ND |
| Rspo1 | −1.89 ± 0.41 | −3.69 ± 0.20 | −0.98 ± 0.32 | −0.10 ± 0.63 | 0.74 ± 0.35 |
| sFRP1 | −0.21 ± 0.46 | −5.55 ± 1.64 | −5.14 ± 0.61 | ND | −5.03 ± 1.03 |
| sFRP3 | ? | ? | −6.89 ± 0.31 | ND | ND |
| sFRP4 | −5.53 ± 0.69 | ND | ND | ND | ND |
| sFRP5 | ND | ND | ND | ND | −6.99 ± 0.58 |
| Wif1 | ND | −8.40 ± 0.95 | −8.35 ± 0.50 | ND | ND |
| 24 hpa | 3 dpa | |||||
|---|---|---|---|---|---|---|
| Gene | NA | LPS | SCE | NA | LPS | SCE |
| Fz1 | 5.30 ± 0.05 | −6.66 ± 0.13 | −5.85 ± 0.16 | −4.11 ± 0.07 | −5.27 ± 0.30 | −4.40 ± 0.29 |
| Fz2 | −13.50 ± 0.12 | −13.94 ± 0.19 | −12.59 ± 0.22 | −11.11 ± 0.28 | −13.40 ± 0.19 | −11.06 ± 0.12 |
| Fz3 | −11.24 ± 0.06 | −11.02 ± 0.19 | −10.07 ± 0.34 | −9.61 ± 0.22 | −11.27 ± 0.21 | −9.47 ± 0.19 |
| Fz5 | −11.56 ± 0.23 | −11.54 ± 0.21 | −11.16 ± 0.33 | −10.90 ± 0.15 | −12.11 ± 0.10 | −11.19 ± 0.07 |
| Fz6 | −14.33 ± 0.30 | −14.17 ± 0.35 | −14.12 ± 0.18 | −11.55 ± 0.14 | −13.40 ± 0.11 | −12.51 ± 0.21 |
| Fz7 | −15.73 ± 0.08 | ND | −16.74 ± 0.12 | −14.74 ± 0.07 | ND | −15.27 ± 0.40 |
| Fz8 | −5.39 ± 0.27 | −6.61 ± 0.34 | −5.43 ± 0.14 | −4.73 ± 0.12 | −5.88 ± 0.26 | −4.84 ± 0.10 |
| Fz9 | −11.38 ± 0.17 | −11.89 ± 0.32 | −11.86 ± 0.35 | −11.45 ± 0.26 | −12.46 ± 0.08 | −11.75 ± 0.28 |
| Fz10 | −16.01 ± 0.17 | −16.37 ± 0.29 | −15.96 ± 0.20 | −15.43 ± 0.03 | ND | −15.60 ± 0.15 |
| Ryk | −6.19 ± 0.05 | −7.13 ± 0.23 | −5.75 ± 0.16 | −4.09 ± 0.18 | −4.60 ± 0.08 | −4.34 ± 0.10 |
| Ror2 | ND | −16.66 ± 0.48 | −16.57 ± 0.62 | ND | ND | ND |
| Ptk7 | −11.67 ± 0.26 | −13.11 ± 0.32 | −11.65 ± 0.36 | −7.62 ± 0.30 | −11.99 ± 0.34 | −7.95 ± 0.49 |
| Wnt1 | −11.64 ± 0.24 | −14.23 ± 0.80 | −14.50 ± 0.47 | −14.06 ± 0.06 | −15.39 ± 0.23 | −13.70 ± 0.18 |
| Wnt2b | ND | −17.40 ± 0.20 | −17.31 ± 0.11 | ND | ND | ND |
| Wnt4 | −12.26 ± 0.27 | −13.52 ± 0.56 | −12.71 ± 0.18 | −13.58 ± 0.16 | −15.71 ± 0.56 | −13.37 ± 0.38 |
| Wnt5a | −15.12 ± 0.16 | −14.31 ± 0.09 | −14.97 ± 0.22 | −12.90 ± 0.12 | −15.26 ± 0.14 | −13.67 ± 0.52 |
| Wnt5b | −13.28 ± 0.18 | −15.82 ± 0.66 | −14.26 ± 0.14 | −11.88 ± 0.21 | −15.62 ± 0.69 | −9.63 ± 0.11 |
| Wnt6 | −8.67 ± 0.33 | −8.72 ± 0.49 | −8.78 ± 0.35 | −11.06 ± 0.14 | −9.35 ± 0.12 | −12.28 ± 0.26 |
| Wnt7a | −15.92 ± 0.50 | −15.27 ± 1.03 | −16.02 ± 0.08 | −14.58 ± 0.29 | −15.77 ± 0.23 | −13.66 ± 0.96 |
| Wnt7b | −17.02 ± 0.17 | −16.16 ± 0.66 | −16.30 ± 0.75 | ND | ND | ND |
| Wnt9a | −16.61 ± 0.25 | −16.85 ± 0.10 | −16.54 ± 0.47 | −15.27 ± 0.25 | ND | −15.36 ± 0.45 |
| Wnt10a | −15.99 ± 0.12 | −14.90 ± 0.26 | −15.69 ± 0.24 | ND | ND | ND |
| Wnt10b | ND | −13.31 ± 1.34 | ND | −11.39 ± 0.18 | −13.12 ± 0.33 | −13.48 ± 0.33 |
| Wnt11 | −16.72 ± 0.18 | −16.58 ± 0.26 | −16.66 ± 0.35 | −15.49 ± 0.11 | −16.37 ± 0.10 | ND |
| Wnt16 | −15.52 ± 0.06 | −17.05 ± 0.11 | −16.76 ± 0.37 | −15.24 ± 0.13 | ND | −15.59 ± 0.30 |
| Rspo1 | −6.56 ± 0.25 | −9.20 ± 0.15 | −6.16 ± 0.44 | −3.52 ± 0.15 | −7.52 ± 0.25 | −3.84 ± 0.40 |
| Rspo3 | −16.74 ± 0.04 | −16.60 ± 0.66 | −17.02 ± 0.23 | −15.45 ± 0.11 | ND | ND |
| Dkk1 | −15.82 ± 0.14 | −16.67 ± 0.35 | ND | −13.32 ± 0.26 | −15.71 ± 0.30 | −15.04 ± 0.32 |
| Dkk2 | −14.11 ± 0.22 | −14.43 ± 0.15 | −16.28 ± 0.18 | ND | ND | ND |
| Dkk3 | −15.79 ± 0.13 | −14.80 ± 0.28 | −16.90 ± 0.27 | −12.91 ± 0.22 | −15.24 ± 0.45 | −14.23 ± 0.28 |
| sFRP1 | −13.74 ± 0.20 | −13.17 ± 0.94 | −12.60 ± 1.52 | −12.29 ± 0.16 | −14.31 ± 0.35 | −12.38 ± 0.35 |
| sFRP2 | −12.52 ± 0.23 | −10.49 ± 0.51 | −13.19 ± 0.32 | −10.58 ± 0.17 | −9.73 ± 0.17 | −10.07 ± 0.17 |
| sFRP3 | −14.37 ± 0.19 | −14.88 ± 0.45 | −14.21 ± 0.12 | −14.21 ± 0.12 | −16.34 ± 0.30 | −14.52 ± 0.65 |
| sFRP4 | −13.08 ± 0.19 | −13.31 ± 0.15 | −11.95 ± 0.37 | −10.79 ± 0.12 | −13.07 ± 0.29 | −11.08 ± 0.37 |
| sFRP5 | −13.77 ± 0.27 | −15.84 ± 0.27 | −13.34 ± 0.54 | −13.42 ± 0.01 | ND | −13.79 ± 0.17 |
| Wif1 | −9.93 ± 0.09 | −10.50 ± 0.32 | −13.06 ± 0.56 | −6.67 ± 0.11 | −8.82 ± 0.07 | −8.91 ± 0.37 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Fernández, C.; González, P.; González-Pérez, F.; Rodríguez, F.J. Characterization of Ex Vivo and In Vitro Wnt Transcriptome Induced by Spinal Cord Injury in Rat Microglial Cells. Brain Sci. 2022, 12, 708. https://doi.org/10.3390/brainsci12060708
González-Fernández C, González P, González-Pérez F, Rodríguez FJ. Characterization of Ex Vivo and In Vitro Wnt Transcriptome Induced by Spinal Cord Injury in Rat Microglial Cells. Brain Sciences. 2022; 12(6):708. https://doi.org/10.3390/brainsci12060708
Chicago/Turabian StyleGonzález-Fernández, Carlos, Pau González, Francisco González-Pérez, and Francisco Javier Rodríguez. 2022. "Characterization of Ex Vivo and In Vitro Wnt Transcriptome Induced by Spinal Cord Injury in Rat Microglial Cells" Brain Sciences 12, no. 6: 708. https://doi.org/10.3390/brainsci12060708
APA StyleGonzález-Fernández, C., González, P., González-Pérez, F., & Rodríguez, F. J. (2022). Characterization of Ex Vivo and In Vitro Wnt Transcriptome Induced by Spinal Cord Injury in Rat Microglial Cells. Brain Sciences, 12(6), 708. https://doi.org/10.3390/brainsci12060708






