Functional Analysis of Zebrafish socs4a: Impacts on the Notochord and Sensory Function
Abstract
:1. Introduction
2. Materials and Methods
2.1. Zebrafish Methods
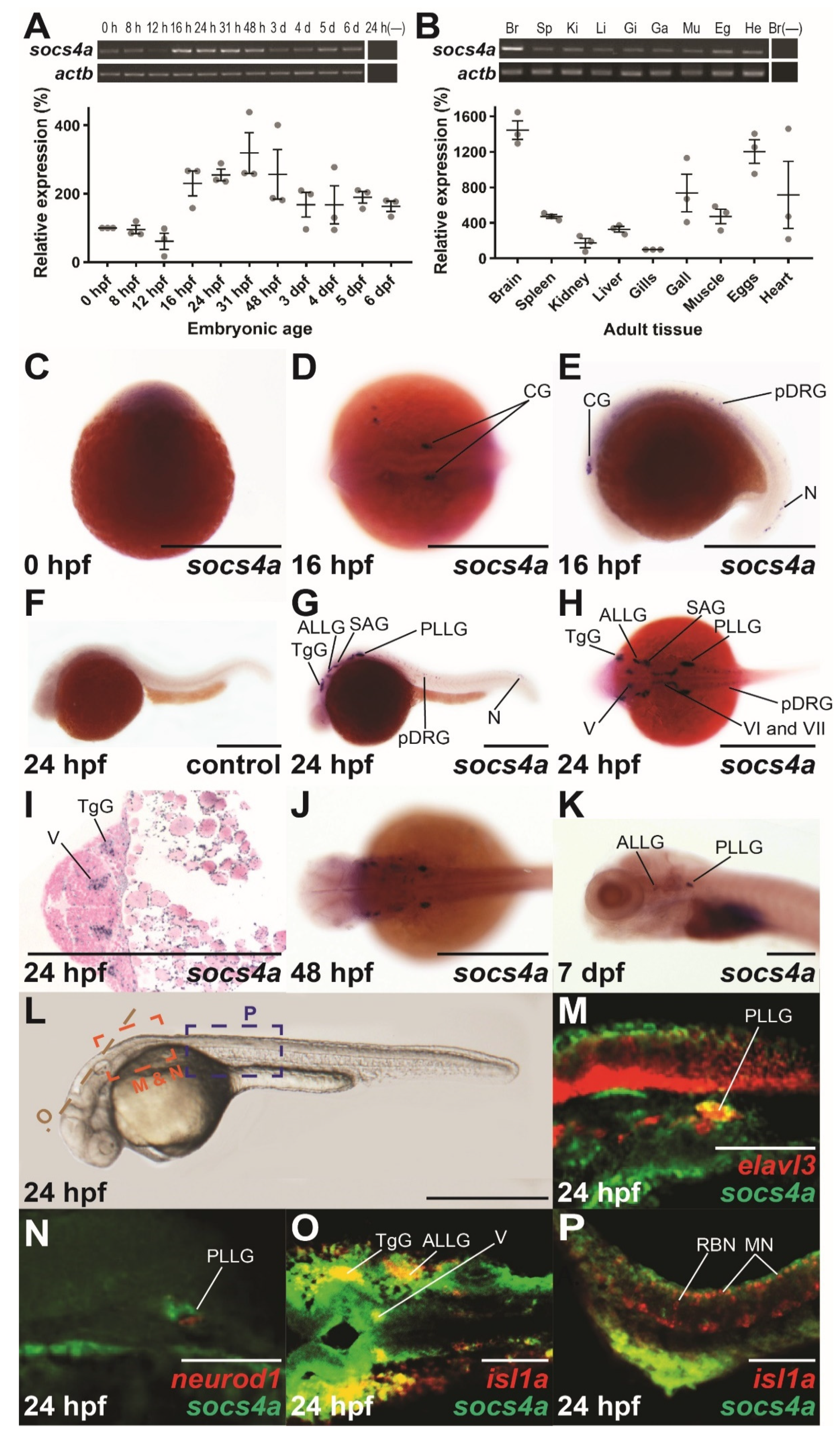
2.2. Reverse Transcription–Polymerase Chain Reaction (RT-PCR)
2.3. In Vitro Transcription and Translation
2.4. In Situ Hybridization
2.5. Imaging
3. Results
3.1. Expression Analysis of Zebrafish socs4a during Embryogenesis
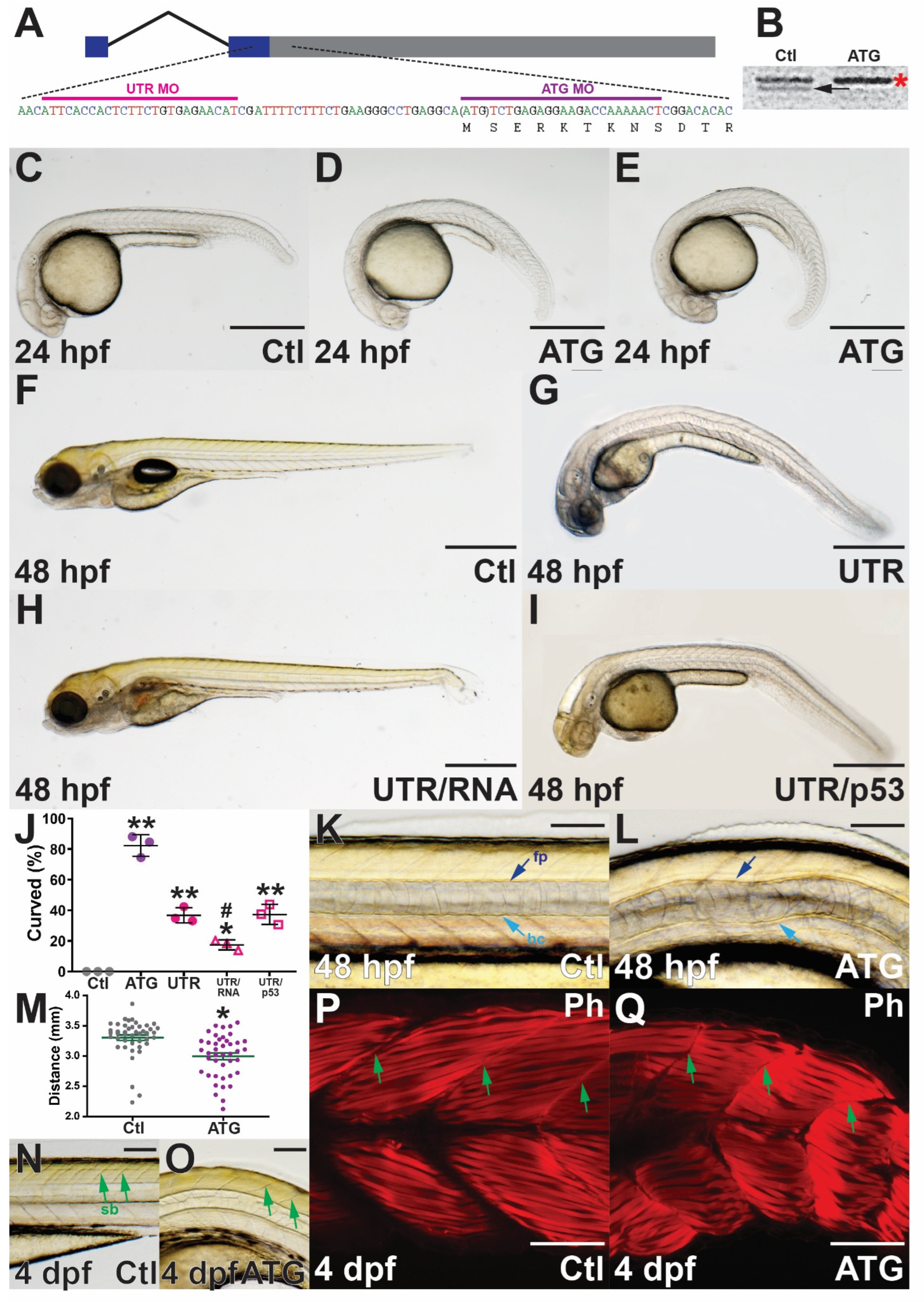
3.2. Targeted Knockdown of Zebrafish socs4a
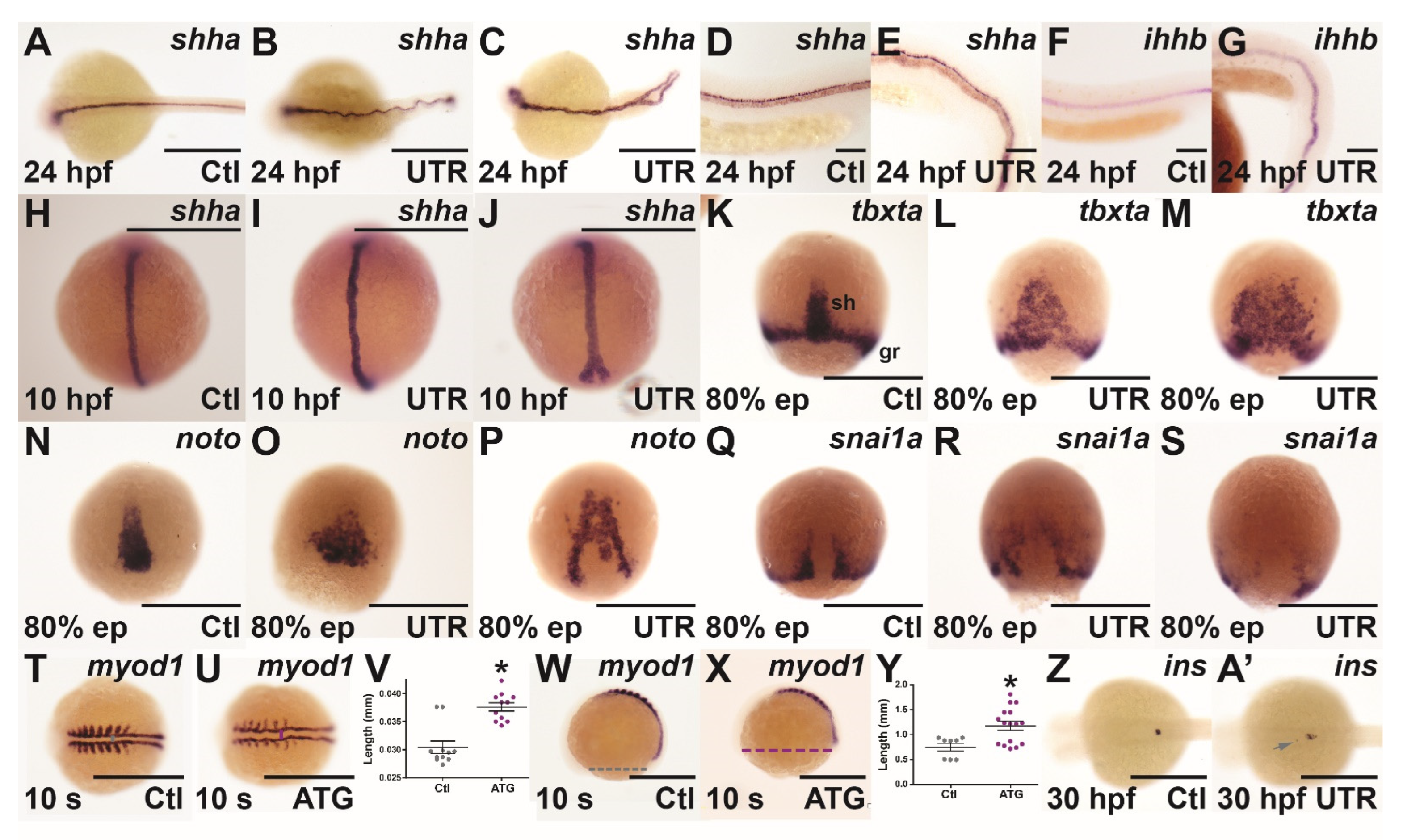
3.3. Molecular Analysis of socs4a Knockdown
3.4. Effect of socs4a Knockdown on the Sensory System
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yoshimura, A.; Nishinakamura, H.; Matsumura, Y.; Hanada, T. Negative regulation of cytokine signaling and immune responses by SOCS proteins. Arthritis Res. Ther. 2005, 7, 100–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexander, W.S.; Starr, R.; Metcalf, N.; Nicholson, S.E.; Farley, A.; Elefanty, A.G.; Brysha, M.; Kile, B.T.; Richardson, R.; Baca, M.; et al. Suppressors of cytokine signaling (SOCS): Negative regulators of signal transduction. J. Leukoc. Biol. 1999, 66, 588–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sobah, M.L.; Liongue, C.; Ward, A.C. SOCS proteins in immunity, inflammatory diseases and immune-related cancer. Front. Med. 2021, 8, 727987. [Google Scholar] [CrossRef] [PubMed]
- Trengove, M.C.; Ward, A.C. SOCS proteins in development and disease. Am. J. Clin. Exp. Immunol. 2013, 2, 1–29. [Google Scholar]
- Delgado-Ortega, M.; Melo, S.; Meurens, F. Expression of SOCS1-7 and CIS mRNA in porcine tissues. Vet. Immunol. Immunopathol. 2011, 144, 493–498. [Google Scholar] [CrossRef]
- Kedzierski, L.; Linossi, E.M.; Kolesnik, T.B.; Day, E.B.; Bird, N.L.; Kile, B.T.; Belz, G.T.; Metcalf, D.; Nicola, N.A.; Kedzierska, K.; et al. Suppressor of cytokine signaling 4 (SOCS4) protects against severe cytokine storm and enhances viral clearance during influenza infection. PLoS Pathog. 2014, 10, e1004134. [Google Scholar] [CrossRef]
- Kario, E.; Marmor, M.D.; Adamsky, K.; Citri, A.; Amit, I.; Amariglio, N.; Rechavi, G.; Yarden, Y. Suppressors of cytokine signaling 4 and 5 regulate epidermal growth factor receptor signaling. J. Biol. Chem. 2005, 280, 7038–7048. [Google Scholar] [CrossRef] [Green Version]
- Hu, G.; Zhou, R.; Liu, J.; Gong, A.Y.; Chen, X.M. MicroRNA-98 and let-7 regulate expression of suppressor of cytokine signaling 4 in biliary epithelial cells in response to Cryptosporidium parvum infection. J. Infect. Dis. 2010, 202, 125–135. [Google Scholar] [CrossRef] [Green Version]
- Sutherland, J.; Keightley, R.; Nixon, B.; Roman, S.; Robker, R.; Russell, D.; McLaughlin, E. Suppressor of cytokine signaling 4 (SOCS4): Moderator of ovarian primordial follicle activation. J. Cell. Physiol. 2011, 227, 1188–1198. [Google Scholar] [CrossRef]
- Liongue, C.; O’Sullivan, L.A.; Trengove, M.C.; Ward, A.C. Evolution of JAK-STAT pathway components: Mechanisms and role in immune system development. PLoS ONE 2012, 7, e32777. [Google Scholar] [CrossRef]
- O’Sullivan, L.A.; Noor, S.M.; Trengove, M.C.; Lewis, R.S.; Liongue, C.; Sprigg, N.S.; Nicholson, S.E.; Ward, A.C. Suppressor of cytokine signaling 1 regulates embryonic myelopoiesis independently of its effects on T cell development. J. Immunol. 2011, 186, 4751–4761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robu, M.E.; Larson, J.D.; Nasevicius, A.; Beiraghi, S.; Brenner, C.; Farber, S.A.; Ekker, S.C. p53 activation by knockdown technologies. PLoS Genet. 2007, 3, e78. [Google Scholar] [CrossRef] [PubMed]
- Goody, M.; Henry, C. Phalloidin staining and immunohistochemistry of zebrafish embryos. Bio-Protocol 2013, 3, e786. [Google Scholar] [CrossRef]
- Fraher, D.; Ellis, M.K.; Morrison, S.; McGee, S.L.; Ward, A.; Walder, K.; Gibert, Y. Lipid abundance in zebrafish embryos is regulated by complementary actions of the endocannabinoid system and retinoic acid pathway. Endocrinology 2015, 156, 3596–3609. [Google Scholar] [CrossRef] [Green Version]
- Granato, M.; Van Eeden, F.J.; Schach, U.; Trowe, T.; Brand, M.; Furutani-Seiki, M.; Haffter, P.; Hammerschmidt, M.; Heisenberg, C.P.; Jiang, Y.J.; et al. Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development 1996, 123, 399–413. [Google Scholar] [CrossRef]
- Ma, A.C.H.; Ward, A.C.; Liang, R.; Leung, A.Y.H. The role of jak2a in zebrafish hematopoiesis. Blood 2007, 110, 1824–1830. [Google Scholar] [CrossRef] [Green Version]
- Lewis, R.S.; Noor, S.M.; Fraser, F.W.; Sertori, R.; Liongue, C.; Ward, A.C. Regulation of embryonic hematopoiesis by a cytokine-inducible SH2 domain homolog in zebrafish. J. Immunol. 2014, 192, 5739–5748. [Google Scholar] [CrossRef] [Green Version]
- Yoong, S.; O’Connell, B.; Soanes, A.; Crowhurst, M.O.; Lieschke, G.J.; Ward, A.C. Characterization of the zebrafish matrix metalloproteinase 9 gene and its developmental expression pattern. Patterns 2007, 7, 39–46. [Google Scholar] [CrossRef]
- Lauter, G.; Söll, I.; Hauptmann, G. Sensitive whole-mount fluorescent in situ hybridization in zebrafish using enhanced tyramide signal amplification. In Brain Development; Humana Press: Totowa, NJ, USA, 2014; pp. 175–185. [Google Scholar]
- Kim, C.H.; Ueshima, E.; Muraoka, O.; Tanaka, H.; Yeo, S.Y.; Huh, T.L.; Miki, N. Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci. Lett. 1996, 216, 109–112. [Google Scholar] [CrossRef]
- Mueller, T.; Wullimann, M.F. Expression domains of neuroD (nrd) in the early postembryonic zebrafish brain. Brain Res. Bull. 2002, 57, 377–379. [Google Scholar] [CrossRef]
- Inoue, A.; Takahashi, M.; Hatta, K.; Hotta, Y.; Okamoto, H. Developmental regulation of Islet-1 mRNA expression during neuronal differentiation in embryonic zebrafish. Dev. Dyn. 1994, 199, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nasevicius, A.; Ekker, S.C. Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 2000, 26, 216–220. [Google Scholar] [CrossRef] [PubMed]
- Stemple, D.L. Structure and function of the notochord: An essential organ for chordate development. Development 2005, 132, 2503–2512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stemple, D.L.; Solnica-Krezel, L.; Zwartkruis, F.; Neuhauss, S.C.; Schier, A.F.; Malicki, J.; Driever, W. Mutations affecting development of the notochord in zebrafish. Development 1996, 123, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Chung, A.-Y.; Kim, S.; Kim, E.; Kim, H.; Jeong, I.; Cha, Y.R.; Bae, Y.-K.; Park, S.W.; Lee, J.; Park, H.-C. Indian hedgehog b function is required for the specification of oligodendrocyte progenitor cells in the zebrafish CNS. J. Neurosci. 2013, 33, 1728–1733. [Google Scholar] [CrossRef]
- Schulte-Merker, S.; Van Eeden, F.J.; Halpern, M.E.; Kimmel, C.B.; Nüsslein-Volhard, C. No tail (ntl) is the zebrafish homologue of the mouse T (Brachyury) gene. Development 1994, 120, 1009–1015. [Google Scholar] [CrossRef]
- Hammerschmidt, M.; Nüsslein-Volhard, C. The expression of a zebrafish gene homologous to Drosophila snail suggests a conserved function in invertebrate and vertebrate gastrulation. Development 1993, 119, 1107–1118. [Google Scholar] [CrossRef]
- Weinberg, E.; Allende, M.; Kelly, C.; Abdelhamid, A.; Murakami, T.; Andermann, P.; Doerre, O.; Grunwald, D.; Riggleman, B. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development 1996, 122, 271–280. [Google Scholar] [CrossRef]
- Cleaver, O.; Krieg, P.A. Notochord patterning of the endoderm. Dev. Biol. 2001, 234, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Papasani, M.R.; Robison, B.D.; Hardy, R.W.; Hill, R.A. Early developmental expression of two insulins in zebrafish (Danio rerio). Physiol. Genom. 2006, 27, 79–85. [Google Scholar] [CrossRef]
- Dijkgraaf, S. The functioning and significance of the lateral-line organs. Biol. Rev. 1963, 38, 51–105. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.; Strähle, U.; Scholpp, S. Neurogenesis in zebrafish—From embryo to adult. Neural Dev. 2013, 8, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isogai, S.; Horiguchi, M.; Weinstein, B.M. The vascular anatomy of the developing zebrafish: An atlas of embryonic and early larval development. Dev. Biol. 2001, 230, 278–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, S.; Shekhar, K.; Regev, A.; Schier, A.F. Comprehensive identification and spatial mapping of habenular neuronal types using single-cell RNA-Seq. Curr. Biol. 2018, 28, 1052–1065.e7. [Google Scholar] [CrossRef] [Green Version]
- Hebrok, M. Hedgehog signaling in pancreas development. Mech. Dev. 2002, 120, 45–57. [Google Scholar] [CrossRef]
- Boczonadi, V.; Müller, J.S.; Pyle, A.; Munkley, J.; Dor, T.; Quartararo, J.; Ferrero, I.; Karcagi, V.; Giunta, M.; Polvikoski, T.; et al. EXOSC8 mutations alter mRNA metabolism and cause hypomyelination with spinal muscular atrophy and cerebellar hypoplasia. Nat. Commun. 2014, 5, 4287. [Google Scholar] [CrossRef] [Green Version]
- Thomas, E.; Cruz, I.A.; Hailey, D.W.; Raible, D.W. There and back again: Development and regeneration of the zebrafish lateral line system. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 160. [Google Scholar] [CrossRef] [Green Version]
- Ohno, S. Gene duplication and the uniqueness of vertebrate genomes circa 1970–1999. Semin. Cell Dev. Biol. 1999, 10, 517–522. [Google Scholar] [CrossRef]
- Yamashita, S.; Miyagi, C.; Carmany-Rampey, A.; Shimizu, T.; Fujii, R.; Schier, A.F.; Hirano, T. Stat3 controls cell movements during zebrafish gastrulation. Dev. Cell 2002, 2, 363–375. [Google Scholar] [CrossRef] [Green Version]
- Oates, A.C.; Wollberg, P.; Pratt, S.J.; Paw, B.; Johnson, S.L.; Ho, R.K.; Postlethwait, J.H.; Zon, L.I.; Wilks, A.F. Zebrafish stat3 is expressed in restricted tissues during embryogenesis and stat1 rescues cytokine signaling in a STAT1-deficient human cell line. Dev. Dyn. 1999, 215, 352–370. [Google Scholar] [CrossRef]
- Liu, Y.; Sepich, D.S.; Solnica-Krezel, L. Stat3/Cdc25a-dependent cell proliferation promotes embryonic axis extension during zebrafish gastrulation. PLoS Genet. 2017, 13, e1006564. [Google Scholar] [CrossRef] [PubMed]
- Bullock, A.N.; Rodriguez, M.C.; Debreczeni, J.; Songyang, Z.; Knapp, S. Structure of the SOCS4-elonginB/C complex reveals a distinct SOCS box interface and the molecular basis for SOCS-dependent EGFR degradation. Structure 2007, 15, 1493–1504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheitz, C.J.F.; Lee, T.S.; McDermitt, D.J.; Tumbar, T. Defining a tissue stem cell-driven Runx1/Stat3 signalling axis in epithelial cancer. EMBO J. 2012, 31, 4124–4139. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trengove, M.; Wyett, R.; Liongue, C.; Ward, A.C. Functional Analysis of Zebrafish socs4a: Impacts on the Notochord and Sensory Function. Brain Sci. 2022, 12, 241. https://doi.org/10.3390/brainsci12020241
Trengove M, Wyett R, Liongue C, Ward AC. Functional Analysis of Zebrafish socs4a: Impacts on the Notochord and Sensory Function. Brain Sciences. 2022; 12(2):241. https://doi.org/10.3390/brainsci12020241
Chicago/Turabian StyleTrengove, Monique, Ruby Wyett, Clifford Liongue, and Alister C. Ward. 2022. "Functional Analysis of Zebrafish socs4a: Impacts on the Notochord and Sensory Function" Brain Sciences 12, no. 2: 241. https://doi.org/10.3390/brainsci12020241
APA StyleTrengove, M., Wyett, R., Liongue, C., & Ward, A. C. (2022). Functional Analysis of Zebrafish socs4a: Impacts on the Notochord and Sensory Function. Brain Sciences, 12(2), 241. https://doi.org/10.3390/brainsci12020241






