Alcohol Dependence in Rats Is Associated with Global Changes in Gene Expression in the Central Amygdala
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Chronic Intermittent Ethanol (CIE) Exposure
2.2. Gene Expression Using RNA-Seq
2.3. Bioinformatics Analysis
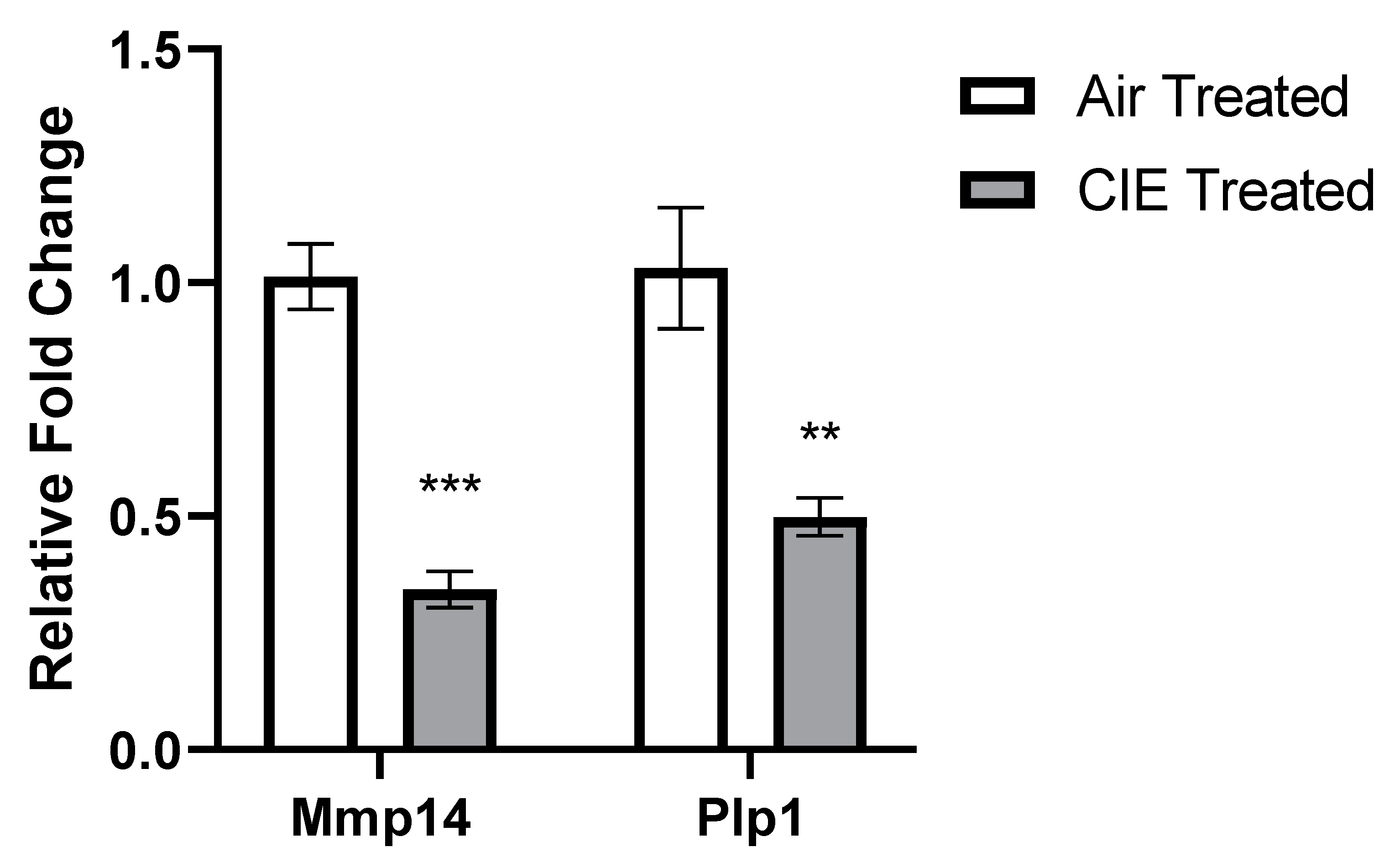
2.4. DEG Validation with qRT-PCR
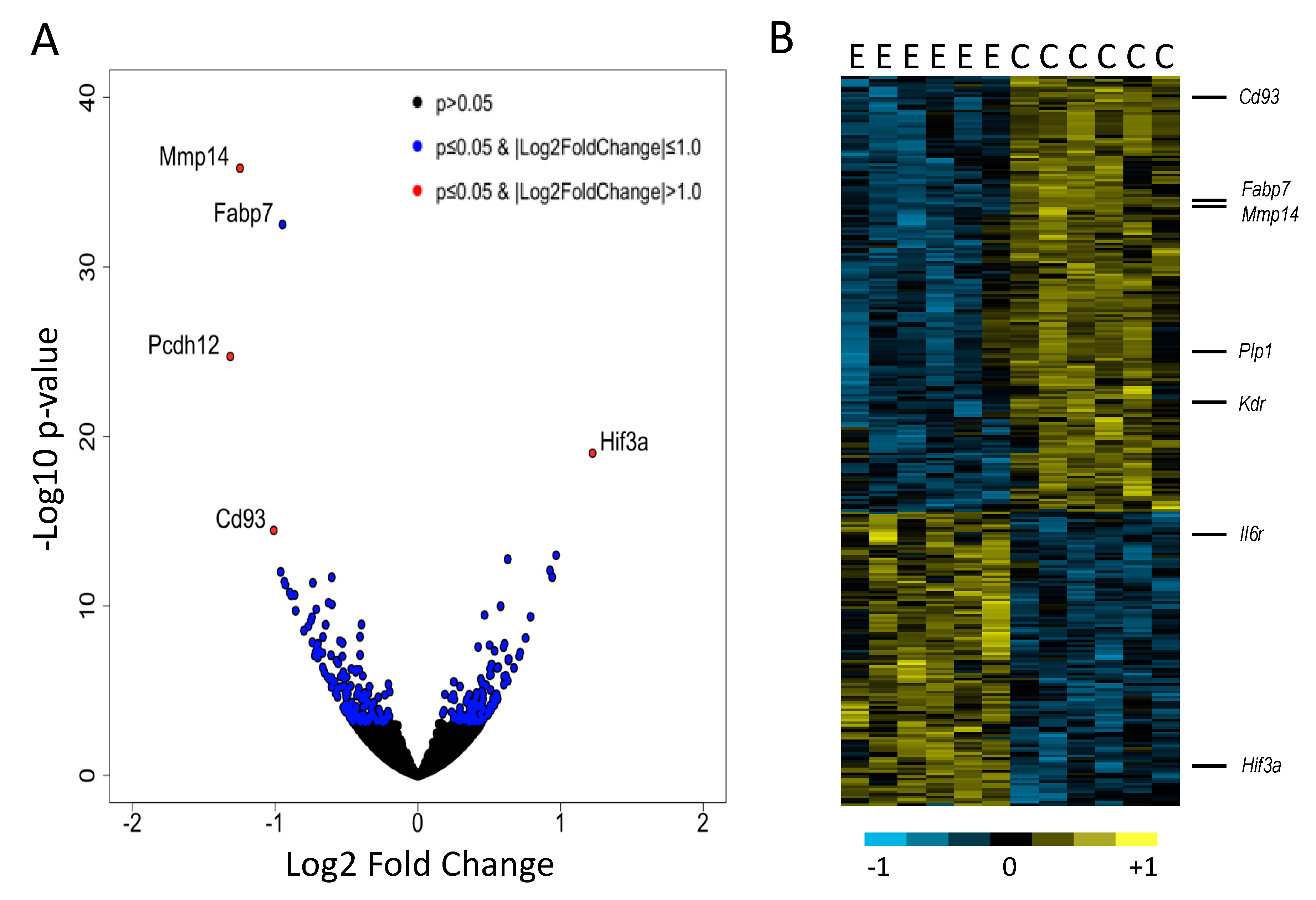
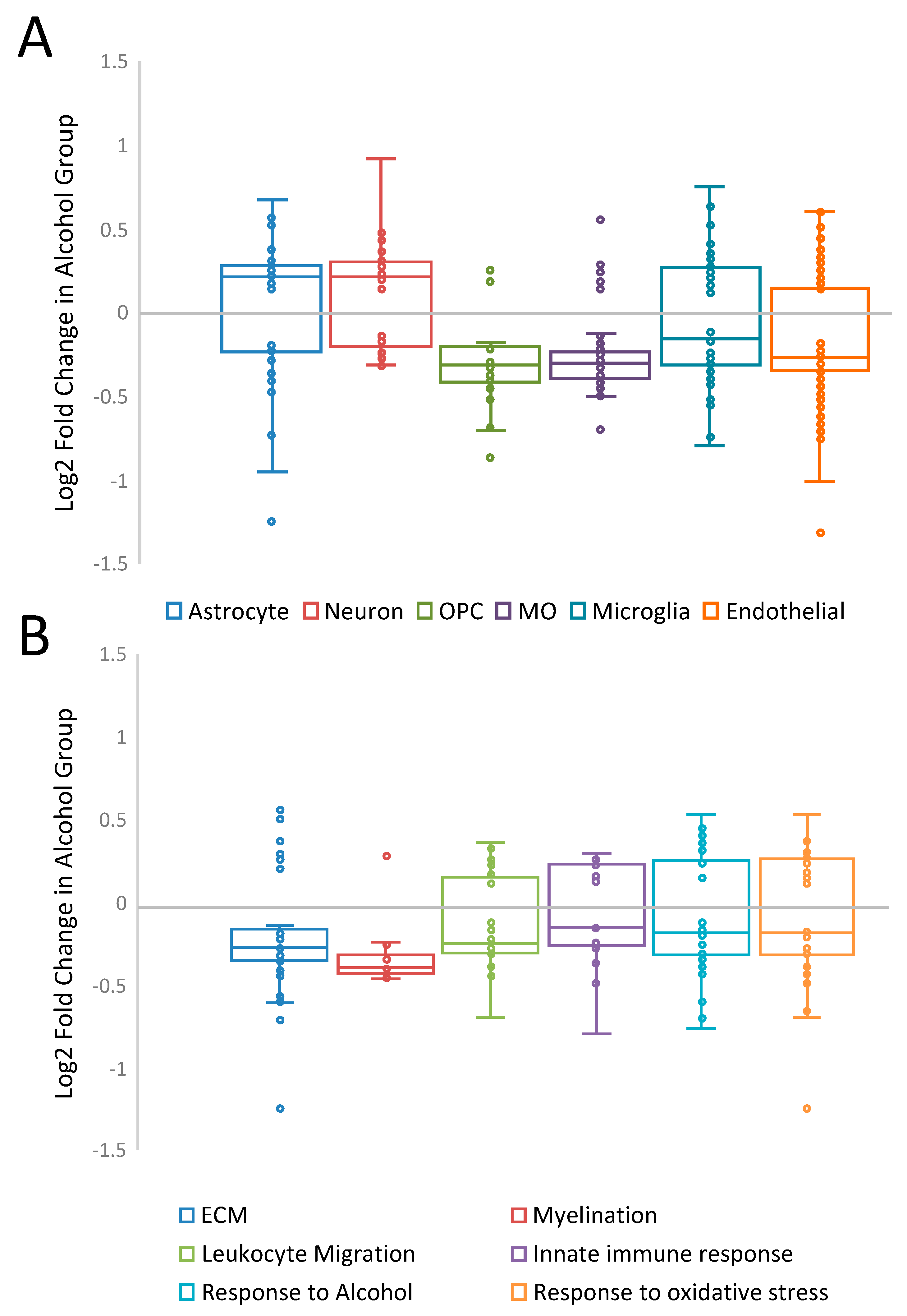
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Association: Arlington, VA, USA, 2015. [Google Scholar]
- O’Dell, L.E.; Roberts, A.J.; Smith, R.T.; Koob, G.F. Enhanced Alcohol Self-Administration after Intermittent versus Continuous Alcohol Vapor Exposure. Alcohol. Clin. Exp. Res. 2004, 28, 1676–1682. [Google Scholar] [CrossRef]
- Gilpin, N.W.; Stewart, R.B.; Badia-Elder, N.E. Neuropeptide Y Suppresses Ethanol Drinking in Ethanol-Abstinent, but Not Non-Ethanol-Abstinent, Wistar Rats. Alcohol 2008, 42, 541–551. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Roberto, M.; Cruz, M.T.; Gilpin, N.W.; Sabino, V.; Schweitzer, P.; Bajo, M.; Cottone, P.; Madamba, S.G.; Stouffer, D.G.; Zorrilla, E.P.; et al. Corticotropin Releasing Factor-Induced Amygdala Gamma-Aminobutyric Acid Release Plays a Key Role in Alcohol Dependence. Biol. Psychiatry 2010, 67, 831–839. [Google Scholar] [CrossRef]
- Varodayan, F.P.; de Guglielmo, G.; Logrip, M.L.; George, O.; Roberto, M. Alcohol Dependence Disrupts Amygdalar L-Type Voltage-Gated Calcium Channel Mechanisms. J. Neurosci. Off. J. Soc. Neurosci. 2017, 37, 4593–4603. [Google Scholar] [CrossRef] [PubMed]
- Herman, M.A.; Contet, C.; Roberto, M. A Functional Switch in Tonic GABA Currents Alters the Output of Central Amygdala Corticotropin Releasing Factor Receptor-1 Neurons Following Chronic Ethanol Exposure. J. Neurosci. 2016, 36, 10729–10741. [Google Scholar] [CrossRef] [PubMed]
- Becker, H.C.; Lopez, M.F. Increased Ethanol Drinking after Repeated Chronic Ethanol Exposure and Withdrawal Experience in C57BL/6 Mice. Alcohol. Clin. Exp. Res. 2004, 28, 1829–1838. [Google Scholar] [CrossRef]
- Ferguson, L.B.; Zhang, L.; Kircher, D.; Wang, S.; Mayfield, R.D.; Crabbe, J.C.; Morrisett, R.A.; Harris, R.A.; Ponomarev, I. Dissecting Brain Networks Underlying Alcohol Binge Drinking Using a Systems Genomics Approach. Mol. Neurobiol. 2019, 56, 2791–2810. [Google Scholar] [CrossRef]
- Pomrenze, M.B.; Giovanetti, S.M.; Maiya, R.; Gordon, A.G.; Kreeger, L.J.; Messing, R.O. Dissecting the Roles of GABA and Neuropeptides from Rat Central Amygdala CRF Neurons in Anxiety and Fear Learning. Cell Rep. 2019, 29, 13–21.e4. [Google Scholar] [CrossRef]
- Gilpin, N.W.; Herman, M.A.; Roberto, M. The Central Amygdala as an Integrative Hub for Anxiety and Alcohol Use Disorders. Biol. Psychiatry 2015, 77, 859–869. [Google Scholar] [CrossRef] [PubMed]
- Bajo, M.; Madamba, S.G.; Roberto, M.; Blednov, Y.A.; Sagi, V.N.; Roberts, E.; Rice, K.C.; Harris, R.A.; Siggins, G.R. Innate Immune Factors Modulate Ethanol Interaction with GABAergic Transmission in Mouse Central Amygdala. Brain Behav. Immun. 2014, 40, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Kissler, J.L.; Sirohi, S.; Reis, D.J.; Jansen, H.T.; Quock, R.M.; Smith, D.G.; Walker, B.M. The One-Two Punch of Alcoholism: Role of Central Amygdala Dynorphins/Kappa-Opioid Receptors. Biol. Psychiatry 2014, 75, 774–782. [Google Scholar] [CrossRef]
- Roberto, M.; Kirson, D.; Khom, S. The Role of the Central Amygdala in Alcohol Dependence. Cold Spring Harb. Perspect. Med. 2020, 11, a039339. [Google Scholar] [CrossRef]
- Logrip, M.L.; Oleata, C.; Roberto, M. Sex Differences in Responses of the Basolateral-Central Amygdala Circuit to Alcohol, Corticosterone and Their Interaction. Neuropharmacology 2017, 114, 123–134. [Google Scholar] [CrossRef]
- Roberto, G.; Gadens Zamboni, C.; Peretti, C.; Correia, D.; Veloso, A.; Rueda, L.; Camarini, R.; Brunialti-Godard, A.L.; Boerngen-Lacerda, R. GABA B Receptor Agonist Only Reduces Ethanol Drinking in Light-Drinking Mice. Pharmacol. Biochem. Behav. 2012, 102, 223–240. [Google Scholar] [CrossRef]
- Avegno, E.M.; Lobell, T.D.; Itoga, C.A.; Baynes, B.B.; Whitaker, A.M.; Weera, M.M.; Edwards, S.; Middleton, J.W.; Gilpin, N.W. Central Amygdala Circuits Mediate Hyperalgesia in Alcohol-Dependent Rats. J. Neurosci. 2018, 38, 7761–7773. [Google Scholar] [CrossRef] [PubMed]
- Agoglia, A.E.; Zhu, M.; Quadir, S.G.; Bluitt, M.N.; Douglass, E.; Hanback, T.; Tella, J.; Ying, R.; Hodge, C.W.; Herman, M.A. Sex-Specific Plasticity in CRF Regulation of Inhibitory Control in Central Amygdala CRF1 Neurons after Chronic Voluntary Alcohol Drinking. Addict. Biol. 2021, e13067. [Google Scholar] [CrossRef]
- Aroni, S.; Marino, R.A.M.; Girven, K.S.; Irving, J.M.; Cheer, J.F.; Sparta, D.R. Repeated Binge Ethanol Drinking Enhances Electrical Activity of Central Amygdala Corticotropin Releasing Factor Neurons in Vivo. Neuropharmacology 2021, 189, 108527. [Google Scholar] [CrossRef]
- Koob, G.F.; Volkow, N.D. Neurobiology of Addiction: A Neurocircuitry Analysis. Lancet Psychiatry 2016, 3, 760–773. [Google Scholar] [CrossRef]
- Herman, M.A.; Roberto, M. Cell-Type-Specific Tonic GABA Signaling in the Rat Central Amygdala Is Selectively Altered by Acute and Chronic Ethanol. Addict. Biol. 2016, 21, 72–86. [Google Scholar] [CrossRef]
- Avegno, E.M.; Middleton, J.W.; Gilpin, N.W. Synaptic GABAergic Transmission in the Central Amygdala (CeA) of Rats Depends on Slice Preparation and Recording Conditions. Physiol. Rep. 2019, 7, e14245. [Google Scholar] [CrossRef]
- Roberto, M.; Madamba, S.G.; Stouffer, D.G.; Parsons, L.H.; Siggins, G.R. Increased GABA Release in the Central Amygdala of Ethanol-Dependent Rats. J. Neurosci. 2004, 24, 10159–10166. [Google Scholar] [CrossRef] [PubMed]
- Roberto, M.; Schweitzer, P.; Madamba, S.G.; Stouffer, D.G.; Parsons, L.H.; Siggins, G.R. Acute and Chronic Ethanol Alter Glutamatergic Transmission in Rat Central Amygdala: An In Vitro and In Vivo Analysis. J. Neurosci. 2004, 24, 1594–1603. [Google Scholar] [CrossRef]
- Varodayan, F.P.; Logrip, M.L.; Roberto, M. P/Q-Type Voltage-Gated Calcium Channels Mediate the Ethanol and CRF Sensitivity of Central Amygdala GABAergic Synapses. Neuropharmacology 2017, 125, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Gilpin, N.W.; Roberto, M. Neuropeptide Modulation of Central Amygdala Neuroplasticity Is a Key Mediator of Alcohol Dependence. Neurosci. Biobehav. Rev. 2012, 36, 873–888. [Google Scholar] [CrossRef]
- Roberto, M.; Gilpin, N.W.; Siggins, G.R. The Central Amygdala and Alcohol: Role of γ-Aminobutyric Acid, Glutamate, and Neuropeptides. Cold Spring Harb. Perspect. Med. 2012, 2, a012195. [Google Scholar] [CrossRef] [PubMed]
- de Guglielmo, G.; Crawford, E.; Kim, S.; Vendruscolo, L.F.; Hope, B.T.; Brennan, M.; Cole, M.; Koob, G.F.; George, O. Recruitment of a Neuronal Ensemble in the Central Nucleus of the Amygdala Is Required for Alcohol Dependence. J. Neurosci. 2016, 36, 9446–9453. [Google Scholar] [CrossRef]
- Patel, R.R.; Wolfe, S.A.; Bajo, M.; Abeynaike, S.; Pahng, A.; Borgonetti, V.; D’Ambrosio, S.; Nikzad, R.; Edwards, S.; Paust, S.; et al. IL-10 Normalizes Aberrant Amygdala GABA Transmission and Reverses Anxiety-like Behavior and Dependence-Induced Escalation of Alcohol Intake. Prog. Neurobiol. 2021, 199, 101952. [Google Scholar] [CrossRef]
- Adke, A.P.; Khan, A.; Ahn, H.-S.; Becker, J.J.; Wilson, T.D.; Valdivia, S.; Sugimura, Y.K.; Gonzalez, S.M.; Carrasquillo, Y. Cell-Type Specificity of Neuronal Excitability and Morphology in the Central Amygdala. eNeuro 2021, 8, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Amano, T.; Amir, A.; Goswami, S.; Paré, D. Morphology, PKCδ Expression, and Synaptic Responsiveness of Different Types of Rat Central Lateral Amygdala Neurons. J. Neurophysiol. 2012, 108, 3196–3205. [Google Scholar] [CrossRef]
- McBride, W.J.; Kimpel, M.W.; Schultz, J.A.; McClintick, J.N.; Edenberg, H.J.; Bell, R.L. Changes in Gene Expression in Regions of the Extended Amygdala of Alcohol-Preferring Rats after Binge-like Alcohol Drinking. Alcohol 2010, 44, 171–183. [Google Scholar] [CrossRef]
- McBride, W.J.; Kimpel, M.W.; McClintick, J.N.; Ding, Z.M.; Edenberg, H.J.; Liang, T.; Rodd, Z.A.; Bell, R.L. Changes in Gene Expression within the Extended Amygdala Following Binge-like Alcohol Drinking by Adolescent Alcohol-Preferring (P) Rats. Pharmacol. Biochem. Behav. 2014, 117, 52–60. [Google Scholar] [CrossRef][Green Version]
- Ferguson, L.B.; Ozburn, A.R.; Ponomarev, I.; Metten, P.; Reilly, M.; Crabbe, J.C.; Harris, R.A.; Mayfield, R.D. Genome-Wide Expression Profiles Drive Discovery of Novel Compounds That Reduce Binge Drinking in Mice. Neuropsychopharmacology 2018, 43, 1257–1266. [Google Scholar] [CrossRef] [PubMed]
- Farris, S.P.; Arasappan, D.; Hunicke-Smith, S.; Harris, R.A.; Mayfield, R.D. Transcriptome Organization for Chronic Alcohol Abuse in Human Brain. Mol. Psychiatry 2015, 20, 1438–1447. [Google Scholar] [CrossRef]
- Mulligan, M.K.; Ponomarev, I.; Hitzemann, R.J.; Belknap, J.K.; Tabakoff, B.; Harris, R.A.; Crabbe, J.C.; Blednov, Y.A.; Grahame, N.J.; Phillips, T.J.; et al. Toward Understanding the Genetics of Alcohol Drinking through Transcriptome Meta-Analysis. Proc. Natl. Acad. Sci. USA 2006, 103, 6368–6373. [Google Scholar] [CrossRef]
- Mulligan, M.K.; Rhodes, J.S.; Crabbe, J.C.; Mayfield, R.D.; Adron Harris, R.; Ponomarev, I. Molecular Profiles of Drinking Alcohol to Intoxication in C57BL/6J Mice. Alcohol. Clin. Exp. Res. 2011, 35, 659–670. [Google Scholar] [CrossRef]
- Agrawal, R.G.; Owen, J.A.; Levin, P.S.; Hewetson, A.; Berman, A.E.; Franklin, S.R.; Hogue, R.J.; Chen, Y.; Walz, C.; Colvard, B.D.; et al. Bioinformatics Analyses Reveal Age-Specific Neuroimmune Modulation as a Target for Treatment of High Ethanol Drinking. Alcohol. Clin. Exp. Res. 2014, 38, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Iancu, O.D.; Colville, A.; Walter, N.A.R.; Darakjian, P.; Oberbeck, D.L.; Daunais, J.B.; Zheng, C.L.; Searles, R.P.; McWeeney, S.K.; Grant, K.A.; et al. On the Relationships in Rhesus Macaques between Chronic Ethanol Consumption and the Brain Transcriptome. Addict. Biol. 2018, 23, 196–205. [Google Scholar] [CrossRef]
- Kozell, L.B.; Lockwood, D.; Darakjian, P.; Edmunds, S.; Shepherdson, K.; Buck, K.J.; Hitzemann, R. RNA-Seq Analysis of Genetic and Transcriptome Network Effects of Dual-Trait Selection for Ethanol Preference and Withdrawal Using SOT and NOT Genetic Models. Alcohol. Clin. Exp. Res. 2020, 44, 820–830. [Google Scholar] [CrossRef]
- Colville, A.M.; Iancu, O.D.; Lockwood, D.R.; Darakjian, P.; McWeeney, S.K.; Searles, R.; Zheng, C.; Hitzemann, R. Regional Differences and Similarities in the Brain Transcriptome for Mice Selected for Ethanol Preference From HS-CC Founders. Front. Genet. 2018, 9, 300. [Google Scholar] [CrossRef] [PubMed]
- Ponomarev, I.; Maiya, R.; Harnett, M.T.; Schafer, G.L.; Ryabinin, A.E.; Blednov, Y.A.; Morikawa, H.; Boehm, S.L.; Homanics, G.E.; Berman, A.; et al. Transcriptional Signatures of Cellular Plasticity in Mice Lacking the A1 Subunit of GABAA Receptors. J. Neurosci. 2006, 26, 5673–5683. [Google Scholar] [CrossRef]
- Mulligan, M.K.; Mozhui, K.; Pandey, A.K.; Smith, M.L.; Gong, S.; Ingels, J.; Miles, M.F.; Lopez, M.F.; Lu, L.; Williams, R.W. Genetic Divergence in the Transcriptional Engram of Chronic Alcohol Abuse: A Laser-Capture RNA-Seq Study of the Mouse Mesocorticolimbic System. Alcohol 2017, 58, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Iancu, O.D.; Colville, A.M.; Wilmot, B.; Searles, R.; Darakjian, P.; Zheng, C.; McWeeney, S.; Kawane, S.; Crabbe, J.C.; Metten, P.; et al. Gender-Specific Effects of Selection for Drinking in the Dark on the Network Roles of Coding and Noncoding RNAs. Alcohol. Clin. Exp. Res. 2018, 42, 1454–1465. [Google Scholar] [CrossRef]
- Bogenpohl, J.W.; Smith, M.L.; Farris, S.P.; Dumur, C.I.; Lopez, M.F.; Becker, H.C.; Grant, K.A.; Miles, M.F. Cross-Species Co-Analysis of Prefrontal Cortex Chronic Ethanol Transcriptome Responses in Mice and Monkeys. Front. Mol. Neurosci. 2019, 12, 197. [Google Scholar] [CrossRef]
- Wolstenholme, J.T.; Warner, J.A.; Capparuccini, M.I.; Archer, K.J.; Shelton, K.L.; Miles, M.F. Genomic Analysis of Individual Differences in Ethanol Drinking: Evidence for Non-Genetic Factors in C57Bl/6 Mice. PLoS ONE 2011, 6, e21100. [Google Scholar] [CrossRef] [PubMed]
- Colville, A.M.; Iancu, O.D.; Oberbeck, D.L.; Darakjian, P.; Zheng, C.L.; Walter, N.A.R.; Harrington, C.A.; Searles, R.P.; McWeeney, S.; Hitzemann, R.J. Effects of Selection for Ethanol Preference on Gene Expression in the Nucleus Accumbens of HS-CC Mice. Genes Brain Behav. 2017, 16, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Marballi, K.; Genabai, N.K.; Blednov, Y.A.; Harris, R.A.; Ponomarev, I. Alcohol Consumption Induces Global Gene Expression Changes in VTA Dopaminergic Neurons. Genes Brain Behav. 2016, 15, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Bell, R.L.; Kimpel, M.W.; McClintick, J.N.; Strother, W.N.; Carr, L.G.; Liang, T.; Rodd, Z.A.; Mayfield, R.D.; Edenberg, H.J.; McBride, W.J. Gene Expression Changes in the Nucleus Accumbens of Alcohol-Preferring Rats Following Chronic Ethanol Consumption. Pharmacol. Biochem. Behav. 2009, 94, 131–147. [Google Scholar] [CrossRef]
- Harris, R.A.; Bajo, M.; Bell, R.L.; Blednov, Y.A.; Varodayan, F.P.; Truitt, J.M.; de Guglielmo, G.; Lasek, A.W.; Logrip, M.L.; Vendruscolo, L.F.; et al. Genetic and Pharmacologic Manipulation of TLR4 Has Minimal Impact on Ethanol Consumption in Rodents. J. Neurosci. 2017, 37, 1139–1155. [Google Scholar] [CrossRef] [PubMed]
- Farris, S.P.; Pietrzykowski, A.Z.; Miles, M.F.; O’Brien, M.A.; Sanna, P.P.; Zakhari, S.; Mayfield, R.D.; Harris, R.A. Applying the New Genomics to Alcohol Dependence. Alcohol 2015, 49, 825–836. [Google Scholar] [CrossRef]
- Ponomarev, I.; Wang, S.; Zhang, L.; Adron Harris, R.; Dayne Mayfield, R. Gene Coexpression Networks in Human Brain Identify Epigenetic Modifications in Alcohol Dependence. J. Neurosci. 2012, 32, 1884–1897. [Google Scholar] [CrossRef]
- Tapocik, J.D.; Solomon, M.; Flanigan, M.; Meinhardt, M.; Barbier, E.; Schank, J.; Schwandt, M.; Sommer, W.H.; Heilig, M. Coordinated Dysregulation of MRNAs and MicroRNAs in the Rat Medial Prefrontal Cortex Following a History of Alcohol Dependence. Pharm. J. 2012, 13, 286–296. [Google Scholar] [CrossRef] [PubMed]
- Repunte-Canonigo, V.; Shin, W.; Vendruscolo, L.F.; Lefebvre, C.; van der Stap, L.; Kawamura, T.; Schlosburg, J.E.; Alvarez, M.; Koob, G.F.; Califano, A.; et al. Identifying Candidate Drivers of Alcohol Dependence-Induced Excessive Drinking by Assembly and Interrogation of Brain-Specific Regulatory Networks. Genome Biol. 2015, 16, 1–13. [Google Scholar] [CrossRef]
- Walter, N.; Cervera-Juanes, R.; Zheng, C.; Darakjian, P.; Lockwood, D.; Cuzon-Carlson, V.; Ray, K.; Fei, S.; Conrad, D.; Searles, R.; et al. Effect of Chronic Ethanol Consumption in Rhesus Macaques on the Nucleus Accumbens Core Transcriptome. Addict. Biol. 2021, 26, e13021. [Google Scholar] [CrossRef]
- Hitzemann, R.; Bergeson, S.E.; Berman, A.E.; Bubier, J.A.; Chesler, E.J.; Finn, D.A.; Hein, L.; Hoffman, P.; Holmes, A.; Kisby, B.R.; et al. Sex Differences in the Brain Transcriptome Related to Alcohol Effects and Alcohol Use Disorder. Biol. Psychiatry 2021. [Google Scholar] [CrossRef] [PubMed]
- Paxinos, G.; Watson, C. The Rat Brain in Stereotaxic Coordinates; Elsevier: Amsterdam, The Netherlands, 2007; ISBN 9780080475158. [Google Scholar]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, K.; Sloan, S.A.; Bennett, M.L.; Scholze, A.R.; O’Keeffe, S.; Phatnani, H.P.; Guarnieri, P.; Caneda, C.; Ruderisch, N.; et al. An RNA-Sequencing Transcriptome and Splicing Database of Glia, Neurons, and Vascular Cells of the Cerebral Cortex. J. Neurosci. 2014, 34, 11929–11947. [Google Scholar] [CrossRef]
- Li, Q.; Cheng, Z.; Zhou, L.; Darmanis, S.; Neff, N.F.; Okamoto, J.; Gulati, G.; Bennett, M.L.; Sun, L.O.; Clarke, L.E.; et al. Developmental Heterogeneity of Microglia and Brain Myeloid Cells Revealed by Deep Single-Cell RNA Sequencing. Neuron 2019, 101, 207–223.e10. [Google Scholar] [CrossRef]
- Roberts, A.J.; Khom, S.; Bajo, M.; Vlkolinsky, R.; Polis, I.; Cates-Gatto, C.; Roberto, M.; Gruol, D.L. Increased IL-6 Expression in Astrocytes Is Associated with Emotionality, Alterations in Central Amygdala GABAergic Transmission, and Excitability during Alcohol Withdrawal. Brain Behav. Immun. 2019, 82, 188–202. [Google Scholar] [CrossRef]
- Roberto, M.; Patel, R.R.; Bajo, M. Ethanol and Cytokines in the Central Nervous System. Handb. Exp. Pharmacol. 2017, 248, 397–431. [Google Scholar] [CrossRef]
- Barney, T.M.; Vore, A.S.; Gano, A.; Mondello, J.E.; Deak, T. The Influence of Central Interleukin-6 on Behavioral Changes Associated with Acute Alcohol Intoxication in Adult Male Rats. Alcohol 2019, 79, 37–45. [Google Scholar] [CrossRef]
- Ferguson, L.B.; Patil, S.; Moskowitz, B.A.; Ponomarev, I.; Harris, R.A.; Mayfield, R.D.; Messing, R.O. A Pathway-Based Genomic Approach to Identify Medications: Application to Alcohol Use Disorder. Brain Sci. 2019, 9, 381. [Google Scholar] [CrossRef] [PubMed]
- Go, B.S.; Sirohi, S.; Walker, B.M. The Role of Matrix Metalloproteinase-9 in Negative Reinforcement Learning and Plasticity in Alcohol Dependence. Addict. Biol. 2019, 25, e12715. [Google Scholar] [CrossRef] [PubMed]
- Nie, Z.; Schweitzer, P.; Roberts, A.J.; Madamba, S.G.; Moore, S.D.; Siggins, G.R. Ethanol Augments GABAergic Transmission in the Central Amygdala via CRF1 Receptors. Science 2004, 303, 1512–1514. [Google Scholar] [CrossRef]
- Pleil, K.E.; Helms, C.M.; Sobus, J.R.; Daunais, J.B.; Grant, K.A.; Kash, T.L. Effects of Chronic Alcohol Consumption on Neuronal Function in the Non-Human Primate BNST. Addict. Biol. 2016, 21, 1151–1167. [Google Scholar] [CrossRef]
- Jimenez, V.A.; Herman, M.A.; Cuzon Carlson, V.C.; Walter, N.A.; Grant, K.A.; Roberto, M. Synaptic Adaptations in the Central Amygdala and Hypothalamic Paraventricular Nucleus Associated with Protracted Ethanol Abstinence in Male Rhesus Monkeys. Neuropsychopharmacology 2019, 44, 982–993. [Google Scholar] [CrossRef]
- Gorini, G.; Roberts, A.J.; Mayfield, R.D. Neurobiological Signatures of Alcohol Dependence Revealed by Protein Profiling. PLoS ONE 2013, 8, e82656. [Google Scholar] [CrossRef]
- Rice, J.; Gu, C. Function and Mechanism of Myelin Regulation in Alcohol Abuse and Alcoholism. BioEssays 2019, 41, e1800255. [Google Scholar] [CrossRef] [PubMed]
- Miguel-Hidalgo, J.J. Molecular Neuropathology of Astrocytes and Oligodendrocytes in Alcohol Use Disorders. Front. Mol. Neurosci. 2018, 11, 78. [Google Scholar] [CrossRef] [PubMed]
- Lasek, A.W. Effects of Ethanol on Brain Extracellular Matrix: Implications for Alcohol Use Disorder. Alcohol. Clin. Exp. Res. 2016, 40, 2030–2042. [Google Scholar] [CrossRef] [PubMed]
- Warden, A.S.; Azzam, M.; DaCosta, A.; Mason, S.; Blednov, Y.A.; Messing, R.O.; Mayfield, R.D.; Harris, R.A. Toll-like Receptor 3 Dynamics in Female C57BL/6J Mice: Regulation of Alcohol Intake. Brain Behav. Immun. 2019, 77, 66–76. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, G.M.; Warden, A.S.; Bridges, C.R.; Blednov, Y.A.; Harris, R.A. Chronic Ethanol Consumption: Role of TLR3/TRIF-Dependent Signaling. Addict. Biol. 2018, 23, 889–903. [Google Scholar] [CrossRef] [PubMed]
- Ponomarev, I.; Stelly, C.E.; Morikawa, H.; Blednov, Y.A.; Mayfield, R.D.; Harris, R.A. Mechanistic Insights into Epigenetic Modulation of Ethanol Consumption. Alcohol 2017, 60, 95–101. [Google Scholar] [CrossRef]
- Erickson, E.K.; Blednov, Y.A.; Harris, R.A.; Mayfield, R.D. Glial Gene Networks Associated with Alcohol Dependence. Sci. Rep. 2019, 9, 10949. [Google Scholar] [CrossRef] [PubMed]
- Warden, A.S.; Wolfe, S.A.; Khom, S.; Varodayan, F.P.; Patel, R.R.; Steinman, M.Q.; Bajo, M.; Montgomery, S.; Vlkolinsky, R.; Nadav, T.; et al. Microglia Control Escalation of Drinking in Alcohol Dependent Mice: Genomic and Synaptic Drivers. Biol. Psychiatry 2020, 88, 910–921. [Google Scholar] [CrossRef]
- Warden, A.S.; Triplett, T.A.; Lyu, A.; Grantham, E.K.; Azzam, M.M.; DaCosta, A.; Mason, S.; Blednov, Y.A.; Ehrlich, L.I.R.; Mayfield, R.D.; et al. Microglia Depletion and Alcohol: Transcriptome and Behavioral Profiles. Addict. Biol. 2020, 26, e12889. [Google Scholar] [CrossRef]
- Erickson, E.K.; DaCosta, A.J.; Mason, S.C.; Blednov, Y.A.; Mayfield, R.D.; Harris, R.A. Cortical Astrocytes Regulate Ethanol Consumption and Intoxication in Mice. Neuropsychopharmacology 2021, 46, 500–508. [Google Scholar] [CrossRef] [PubMed]
- Mihara, M.; Hashizume, M.; Yoshida, H.; Suzuki, M.; Shiina, M. IL-6/IL-6 Receptor System and Its Role in Physiological and Pathological Conditions. Clin. Sci. 2012, 122, 143–159. [Google Scholar] [CrossRef]
- Kontogiorgis, C.; Papaioannou, P.; Hadjipavlou-Litina, D. Matrix Metalloproteinase Inhibitors: A Review on Pharmacophore Mapping and (Q)Sars Results. Curr. Med. Chem. 2005, 12, 339–355. [Google Scholar] [CrossRef]
- Abbott, N.J.; Rönnbäck, L.; Hansson, E. Astrocyte-Endothelial Interactions at the Blood-Brain Barrier. Nat. Rev. Neurosci. 2006, 7, 41–53. [Google Scholar] [CrossRef]
- Kwok, J.C.F.; Dick, G.; Wang, D.; Fawcett, J.W. Extracellular Matrix and Perineuronal Nets in CNS Repair. Dev. Neurobiol. 2011, 71, 1073–1089. [Google Scholar] [CrossRef]
- Van Lint, P.; Libert, C. Chemokine and Cytokine Processing by Matrix Metalloproteinases and Its Effect on Leukocyte Migration and Inflammation. J. Leukoc. Biol. 2007, 82, 1375–1381. [Google Scholar] [CrossRef]
- Verma, R.P.; Hansch, C. Matrix Metalloproteinases (MMPs): Chemical-Biological Functions and (Q)SARs. Bioorganic Med. Chem. 2007, 15, 2223–2268. [Google Scholar] [CrossRef]
- El Hajj, E.C.; El Hajj, M.C.; Voloshenyuk, T.G.; Mouton, A.J.; Khoutorova, E.; Molina, P.E.; Gilpin, N.W.; Gardner, J.D. Alcohol Modulation of Cardiac Matrix Metalloproteinases (MMPs) and Tissue Inhibitors of MMPs Favors Collagen Accumulation. Alcohol. Clin. Exp. Res. 2014, 38, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Stefaniuk, M.; Beroun, A.; Lebitko, T.; Markina, O.; Leski, S.; Meyza, K.; Grzywacz, A.; Samochowiec, J.; Samochowiec, A.; Radwanska, K.; et al. Archival Report Matrix Metalloproteinase-9 and Synaptic Plasticity in the Central Amygdala in Control of Alcohol-Seeking Behavior. Biol. Psychiatry 2017, 81, 907–917. [Google Scholar] [CrossRef]
- Langenfurth, A.; Rinnenthal, J.L.; Vinnakota, K.; Prinz, V.; Carlo, A.-S.; Stadelmann, C.; Siffrin, V.; Peaschke, S.; Endres, M.; Heppner, F.; et al. Membrane-Type 1 Metalloproteinase Is Upregulated in Microglia/Brain Macrophages in Neurodegenerative and Neuroinflammatory Diseases. J. Neurosci. Res. 2014, 92, 275–286. [Google Scholar] [CrossRef]
- Fu, Y.; Nagy, J.A.; Brown, L.F.; Shih, S.C.; Johnson, P.Y.; Chan, C.K.; Dvorak, H.F.; Wight, T.N. Proteolytic Cleavage of Versican and Involvement of ADAMTS-1 in VEGF-A/VPF-Induced Pathological Angiogenesis. J. Histochem. Cytochem. 2011, 59, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Schlomann, U.; Rathke-Hartlieb, S.; Yamamoto, S.; Jockusch, H.; Bartsch, J.W. Tumor Necrosis Factor α Induces a Metalloprotease-Disintegrin, ADAM8 (CD 156): Implications for Neuron-Glia Interactions during Neurodegeneration. J. Neurosci. 2000, 20, 7964–7971. [Google Scholar] [CrossRef] [PubMed]
- Hoseth, E.Z.; Ueland, T.; Dieset, I.; Birnbaum, R.; Shin, J.H.; Kleinman, J.E.; Hyde, T.M.; Mørch, R.H.; Hope, S.; Lekva, T.; et al. A Study of TNF Pathway Activation in Schizophrenia and Bipolar Disorder in Plasma and Brain Tissue. Schizophr. Bull. 2017, 43, 881–890. [Google Scholar] [CrossRef]
- Oh, J.; Takahashi, R.; Kondo, S.; Mizoguchi, A.; Adachi, E.; Sasahara, R.M.; Nishimura, S.; Imamura, Y.; Kitayama, H.; Alexander, D.B.; et al. The Membrane-Anchored MMP Inhibitor RECK Is a Key Regulator of Extracellular Matrix Integrity and Angiogenesis. Cell 2001, 107, 789–800. [Google Scholar] [CrossRef]
- Takahashi, C.; Sheng, Z.; Horan, T.P.; Kitayama, H.; Maki, M.; Hitomi, K.; Kitaura, Y.; Takai, S.; Sasahara, R.M.; Horimoto, A.; et al. Regulation of Matrix Metalloproteinase-9 and Inhibition of Tumor Invasion by the Membrane-Anchored Glycoprotein RECK. Proc. Natl. Acad. Sci. USA 1998, 95, 13221–13226. [Google Scholar] [CrossRef]
- Wang, H.; Imamura, Y.; Ishibashi, R.; Chandana, E.P.S.; Yamamoto, M.; Noda, M. The Reck Tumor Suppressor Protein Alleviates Tissue Damage and Promotes Functional Recovery after Transient Cerebral Ischemia in Mice. J. Neurochem. 2010, 115, 385–398. [Google Scholar] [CrossRef]
- Gould, D.B.; Phalan, F.C.; Breedveld, G.J.; Van Mil, S.E.; Smith, R.S.; Schimenti, J.C.; Aguglia, U.; Van Der Knaap, M.S.; Heutink, P.; John, S.W.M. Mutations in Col4a1 Cause Perinatal Cerebral Hemorrhage and Porencephaly. Science 2005, 308, 1167–1171. [Google Scholar] [CrossRef]
- van Agtmael, T.; Bailey, M.A.; Schlötzer-Schrehardt, U.; Craigie, E.; Jackson, I.J.; Brownstein, D.G.; Megson, I.L.; Mullins, J.J. Col4a1 Mutation in Mice Causes Defects in Vascular Function and Low Blood Pressure Associated with Reduced Red Blood Cell Volume. Hum. Mol. Genet. 2010, 19, 1119–1128. [Google Scholar] [CrossRef]
- Mouton, A.J.; Ninh, V.K.; El Hajj, E.C.; El Hajj, M.C.; Gilpin, N.W.; Gardner, J.D. Exposure to Chronic Alcohol Accelerates Development of Wall Stress and Eccentric Remodeling in Rats with Volume Overload. J. Mol. Cell. Cardiol. 2016, 97, 15–23. [Google Scholar] [CrossRef]
- Haider, S.; Pal, R. Integrated Analysis of Transcriptomic and Proteomic Data. Curr. Genom. 2013, 14, 91–110. [Google Scholar] [CrossRef]
- Laurent, J.M.; Vogel, C.; Kwon, T.; Craig, S.A.; Boutz, D.R.; Huse, H.K.; Nozue, K.; Walia, H.; Whiteley, M.; Ronald, P.C.; et al. Protein Abundances Are More Conserved than MRNA Abundances across Diverse Taxa. Proteomics 2010, 10, 4209–4212. [Google Scholar] [CrossRef]
- Perl, K.; Ushakov, K.; Pozniak, Y.; Yizhar-Barnea, O.; Bhonker, Y.; Shivatzki, S.; Geiger, T.; Avraham, K.B.; Shamir, R. Reduced Changes in Protein Compared to MRNA Levels across Non-Proliferating Tissues. BMC Genom. 2017, 18, 305. [Google Scholar] [CrossRef]
- Kaur, S.; Li, J.; Stenzel-Poore, M.P.; Ryabinin, A.E. Corticotropin-Releasing Factor Acting on Corticotropin-Releasing Factor Receptor Type 1 Is Critical for Binge Alcohol Drinking in Mice. Alcohol. Clin. Exp. Res. 2012, 36, 369–376. [Google Scholar] [CrossRef]
- Rinker, J.A.; Marshall, S.A.; Mazzone, C.M.; Lowery-Gionta, E.G.; Gulati, V.; Pleil, K.E.; Kash, T.L.; Navarro, M.; Thiele, T.E. Extended Amygdala to Ventral Tegmental Area Corticotropin-Releasing Factor Circuit Controls Binge Ethanol Intake. Biol. Psychiatry 2017, 81, 930–940. [Google Scholar] [CrossRef]
- Itoga, C.A.; Roltsch Hellard, E.A.; Whitaker, A.M.; Lu, Y.L.; Schreiber, A.L.; Baynes, B.B.; Baiamonte, B.A.; Richardson, H.N.; Gilpin, N.W. Traumatic Stress Promotes Hyperalgesia via Corticotropin-Releasing Factor-1 Receptor (CRFR1) Signaling in Central Amygdala. Neuropsychopharmacology 2016, 41, 2463–2472. [Google Scholar] [CrossRef]
- Albrechet-Souza, L.; Hwa, L.S.; Han, X.; Zhang, E.Y.; Debold, J.F.; Miczek, K.A. Corticotropin Releasing Factor Binding Protein and CRF2 Receptors in the Ventral Tegmental Area: Modulation of Ethanol Binge Drinking in C57BL/6J Mice. Alcohol. Clin. Exp. Res. 2015, 39, 1609–1618. [Google Scholar] [CrossRef]
- Haass-Koffler, C.L.; Henry, A.T.; Melkus, G.; Simms, J.A.; Naemmuddin, M.; Nielsen, C.K.; Lasek, A.W.; Magill, M.; Schwandt, M.L.; Momenan, R.; et al. Defining the Role of Corticotropin Releasing Factor Binding Protein in Alcohol Consumption. Transl. Psychiatry 2016, 6, e953. [Google Scholar] [CrossRef]
- Ketchesin, K.D.; Stinnett, G.S.; Seasholtz, A.F. Binge Drinking Decreases Corticotropin-Releasing Factor-Binding Protein Expression in the Medial Prefrontal Cortex of Mice. Alcohol. Clin. Exp. Res. 2016, 40, 1641–1650. [Google Scholar] [CrossRef]

| Biological Category | # of Genes | p Value | |
|---|---|---|---|
| Cell type | |||
| Myelinating Oligodendrocyte | 60 | 9.20 × 10−12 | |
| Endothelial Cells | 100 | 5.40 × 10−7 | |
| Astrocyte | 46 | 3.00 × 10−12 | |
| Functional Group | |||
| Extracellular Matrix (ECM) organization | 59 | 1.57 × 10−6 | |
| Ensheathment of neurons | 18 | 1.94 × 10−5 | |
| Brain development | 33 | 9.51 × 10−5 | |
| Myelination | 16 | 1.09 × 10−4 | |
| Leukocyte migration | 36 | 2.82 × 10−4 | |
| Regulation of cell adhesion | 47 | 5.28 × 10−4 | |
| Regulation of cytokine production | 62 | 5.60 × 10−4 | |
| Response to alcohol | 40 | 6.43 × 10−4 | |
| Vasculogenesis | 14 | 2.55 × 10−3 | |
| Response to oxidative stress | 39 | 2.83 × 10−3 | |
| Regulation of innate immune response | 34 | 5.41 × 10−3 | |
| Regulation of blood vessel size | 11 | 2.11 × 10−2 | |
| Molecular Pathway | |||
| NF-kappa B signaling pathway | 16 | 1.09 × 10−2 | |
| IL-6 signaling pathway | 16 | 2.90 × 10−2 | |
| IL-1 signaling pathway | 8 | 3.01 × 10−2 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kisby, B.R.; Farris, S.P.; McManus, M.M.; Varodayan, F.P.; Roberto, M.; Harris, R.A.; Ponomarev, I. Alcohol Dependence in Rats Is Associated with Global Changes in Gene Expression in the Central Amygdala. Brain Sci. 2021, 11, 1149. https://doi.org/10.3390/brainsci11091149
Kisby BR, Farris SP, McManus MM, Varodayan FP, Roberto M, Harris RA, Ponomarev I. Alcohol Dependence in Rats Is Associated with Global Changes in Gene Expression in the Central Amygdala. Brain Sciences. 2021; 11(9):1149. https://doi.org/10.3390/brainsci11091149
Chicago/Turabian StyleKisby, Brent R., Sean P. Farris, Michelle M. McManus, Florence P. Varodayan, Marisa Roberto, R. Adron Harris, and Igor Ponomarev. 2021. "Alcohol Dependence in Rats Is Associated with Global Changes in Gene Expression in the Central Amygdala" Brain Sciences 11, no. 9: 1149. https://doi.org/10.3390/brainsci11091149
APA StyleKisby, B. R., Farris, S. P., McManus, M. M., Varodayan, F. P., Roberto, M., Harris, R. A., & Ponomarev, I. (2021). Alcohol Dependence in Rats Is Associated with Global Changes in Gene Expression in the Central Amygdala. Brain Sciences, 11(9), 1149. https://doi.org/10.3390/brainsci11091149






