Duplicate Detection of Spike Events: A Relevant Problem in Human Single-Unit Recordings
Abstract
1. Introduction
1.1. Motivation
1.1.1. Artifact Detection Methods
- 1.
- Artifact detection based on spike shape
- 2.
- Artifact detection based on spike timing
- 3.
- Manual artifact detection based on spike shape and timing
1.1.2. Characteristics of Coincident Spike Events in Human Single-Unit Recordings
2. Materials and Methods
2.1. Data and Materials
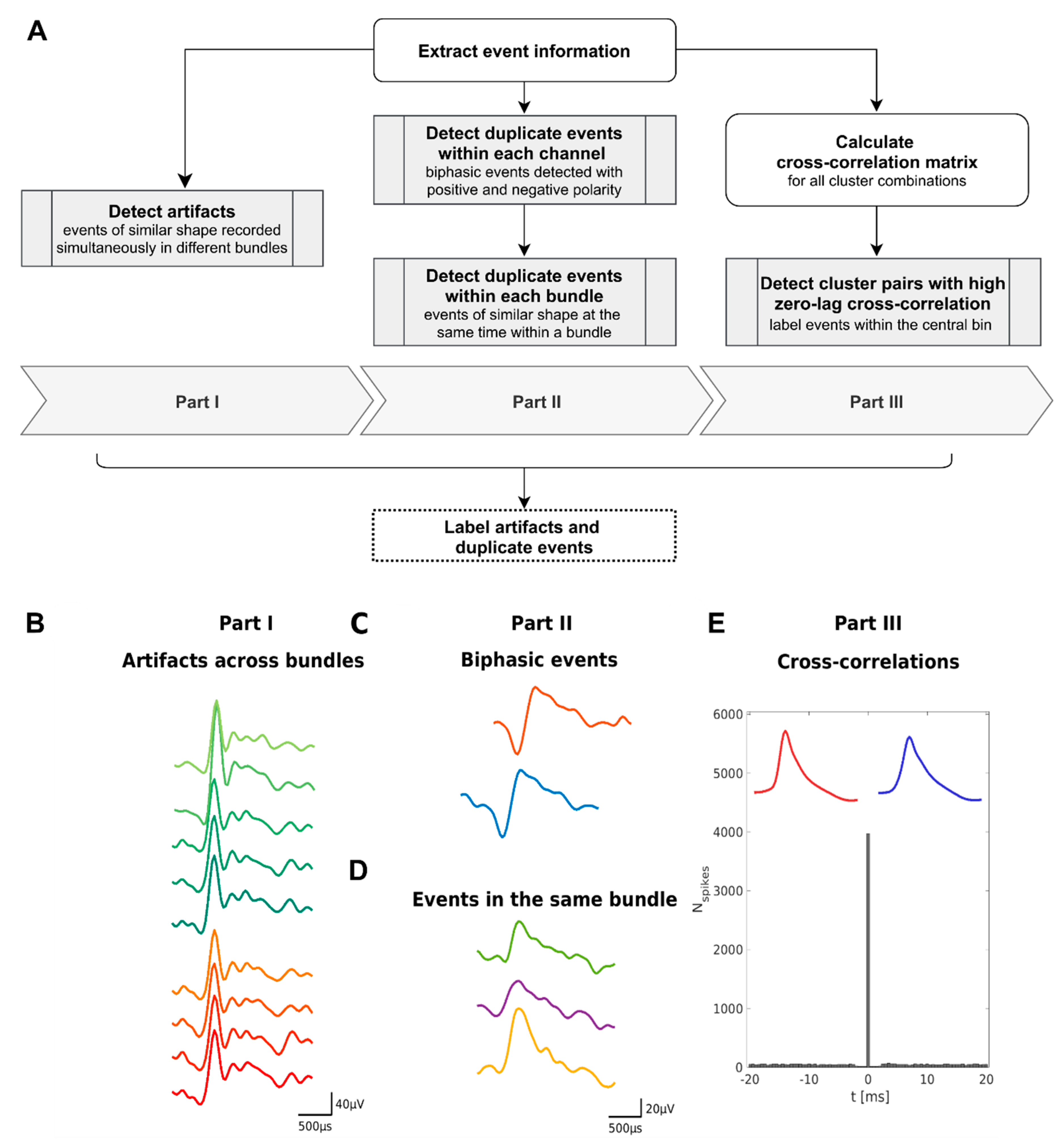
2.2. Structure of the Duplicate Event Removal Algorithm
- Part I. Detect simultaneous spike events between different bundles.
- Part II. Identify duplicate detected biphasic spike events on the same channel and simultaneous spike events on different channels within the same bundle.
- Part III. Detect duplicate spike events based on unphysiologically high zero-lag cross-correlation between clusters.
2.3. Part I—Detection of Artifacts across Bundles
2.3.1. Detection of Simultaneous Artifacts
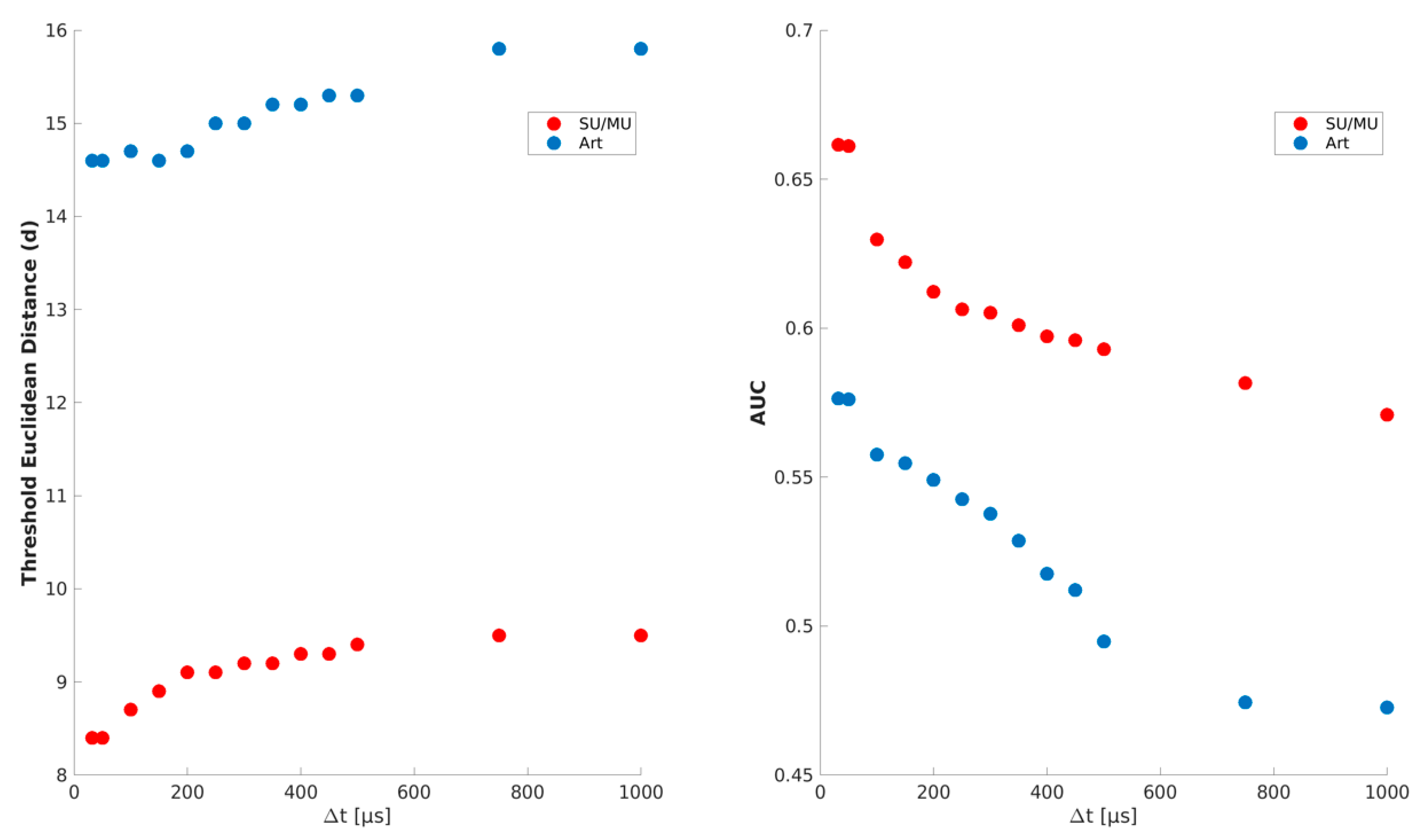
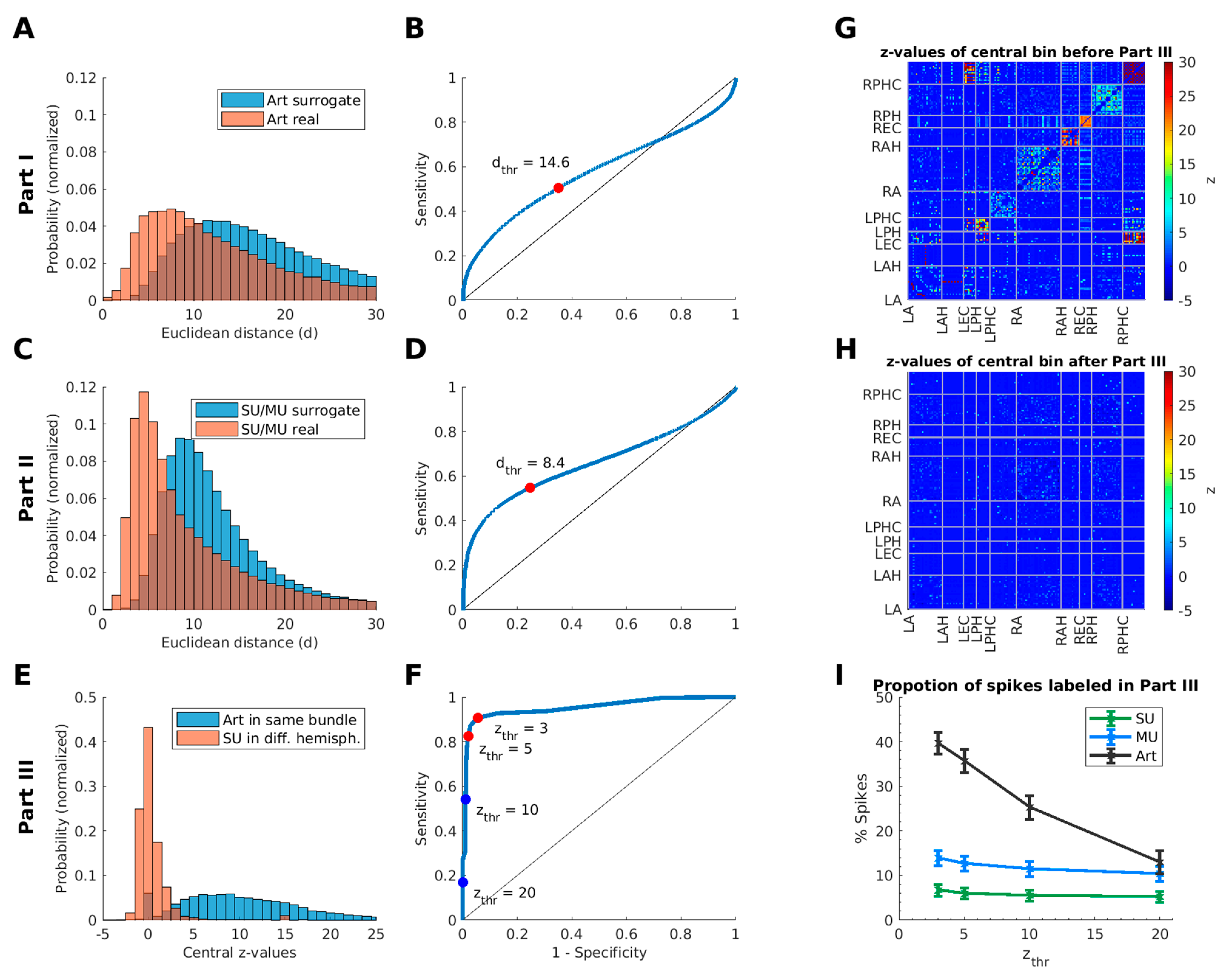
2.3.2. Definition of Thresholds of the Euclidean Distance and the Time Window
2.4. Part II—Duplicate Events in the Same Bundle
2.4.1. Same Channel
- If one of the two duplicate events was labeled as an artifact by an automated clustering algorithm or by manual reclustering, this event is labeled as artificial (Table 2, Case 1).
- If the two events have different unit labels (i.e., one is a single- and the other a multi-unit), we keep the spike event from the single-unit and mark the other one as an artifact (Table 2, Case 3).
- If both spike events are of the same unit class (both single- or multi-unit), we calculate the signal-to-noise ratio (SNR, peak amplitude/spike extraction threshold) of each event and keep the event corresponding to the higher value (Table 2, Case 4). For further details about the thresholds of spike extraction, see [27,29].
2.4.2. Same Bundle
2.5. Part III—Cross-Correlations
2.5.1. Calculation of all Cross-Correlations
2.5.2. Detection of Suspicious Cross-Correlations
2.5.3. Threshold for the Central Bin of the Cross-Correlogram
3. Results
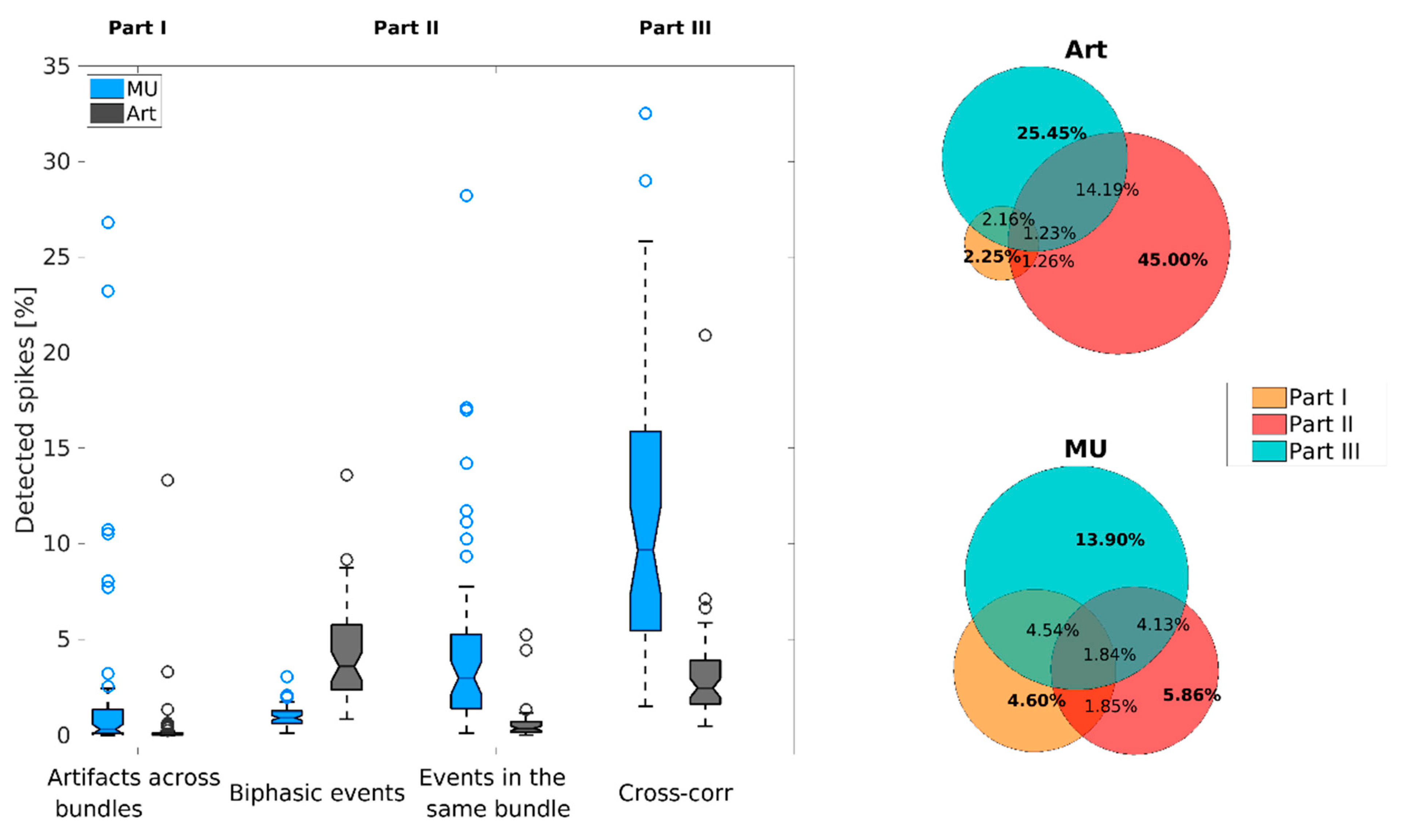
3.1. Examples of Improved Data Quality
3.2. Overall Performance of the DER Algorithm
3.3. Estimation of False-Positive Rate
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

| Software | Reference | Source |
|---|---|---|
| DER | This paper | https://github.com/Geaht/DER, accessed on 20 May 2021 |
| MATLAB 9.8 | MathWorks | https://www.mathworks.com, accessed on 08 April 2020 |
| Statistics and Machine Learning Toolbox 11.7 | ||
| Wavelet Toolbox 5.4 | ||
| Octave | GNU | https://www.gnu.org/software/octave, accessed on 1 December 2014 |
| Psychtoolbox | Brainard, 1997 [63] | http://psychtoolbox.org/, accessed on 11 June 2013 |
| Cheetah software | Neuralynx Inc. | https://neuralynx.com/software/cheetah, accessed on 27 August 2012 |
| Combinato Spike Sorting | Niediek et al., 2016 [29] | https://github.com/jniediek/combinato/, accessed on 11 May 2017 |
| Wave_clus 3 | Chaure et al., 2018 [41] | https://github.com/csn-le/wave_clus, accessed on 24 October 2020 |
| tightfig.m | Kim Dohyun (2021) [64] | https://de.mathworks.com/matlabcentral/fileexchange/73644-tightfig, accessed on 22 January 2021 |
| Venn.m | Darik (2021) [65] | https://de.mathworks.com/matlabcentral/fileexchange/22282-venn, accessed on 30 November 2020 |
References
- Kreiman, G.; Koch, C.; Fried, I. Category-Specific Visual Responses of Single Neurons in the Human Medial Temporal Lobe. Nat. Neurosci. 2000, 3, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Quiroga, R.Q.; Reddy, L.; Kreiman, G.; Koch, C.; Fried, I. Invariant Visual Representation by Single Neurons in the Human Brain. Nature 2005, 435, 1102–1107. [Google Scholar] [CrossRef]
- Mormann, F.; Dubois, J.; Kornblith, S.; Milosavljevic, M.; Cerf, M.; Ison, M.; Tsuchiya, N.; Kraskov, A.; Quiroga, R.Q.; Adolphs, R.; et al. A Category-Specific Response to Animals in the Right Human Amygdala. Nat. Neurosci. 2011, 14, 1247–1249. [Google Scholar] [CrossRef]
- Rutishauser, U.; Ye, S.; Koroma, M.; Tudusciuc, O.; Ross, I.B.; Chung, J.M.; Mamelak, A.N. Representation of Retrieval Confidence by Single Neurons in the Human Medial Temporal Lobe. Nat. Neurosci. 2015, 18, 1041–1050. [Google Scholar] [CrossRef] [PubMed]
- Mormann, F.; Niediek, J.; Tudusciuc, O.; Quesada, C.M.; Coenen, V.A.; Elger, C.E.; Adolphs, R. Neurons in the Human Amygdala Encode Face Identity, but Not Gaze Direction. Nat. Neurosci. 2015, 18, 1568–1570. [Google Scholar] [CrossRef]
- Reber, T.P.; Faber, J.; Niediek, J.; Boström, J.; Elger, C.E.; Mormann, F. Single-Neuron Correlates of Conscious Perception in the Human Medial Temporal Lobe. Curr. Biol. 2017, 27, 2991–2998.e2. [Google Scholar] [CrossRef] [PubMed]
- Mormann, F.; Kornblith, S.; Cerf, M.; Ison, M.J.; Kraskov, A.; Tran, M.; Knieling, S.; Quiroga, R.Q.; Koch, C.; Fried, I. Scene-Selective Coding by Single Neurons in the Human Parahippocampal Cortex. Proc. Natl. Acad. Sci. USA 2017, 114, 1153–1158. [Google Scholar] [CrossRef] [PubMed]
- Reber, T.P.; Bausch, M.; Mackay, S.; Boström, J.; Elger, C.E.; Mormann, F. Representation of Abstract Semantic Knowledge in Populations of Human Single Neurons in the Medial Temporal Lobe. PLoS Biol. 2019, 17, e3000290. [Google Scholar] [CrossRef]
- Rey, H.G.; Gori, B.; Chaure, F.J.; Collavini, S.; Blenkmann, A.O.; Seoane, P.; Seoane, E.; Kochen, S.; Quian Quiroga, R. Single Neuron Coding of Identity in the Human Hippocampal Formation. Curr. Biol. 2020, 30, 1152–1159.e3. [Google Scholar] [CrossRef]
- Rutishauser, U.; Mamelak, A.N.; Schuman, E.M. Single-Trial Learning of Novel Stimuli by Individual Neurons of the Human Hippocampus-Amygdala Complex. Neuron 2006, 49, 805–813. [Google Scholar] [CrossRef]
- Staresina, B.P.; Reber, T.P.; Niediek, J.; Boström, J.; Elger, C.E.; Mormann, F. Recollection in the Human Hippocampal-Entorhinal Cell Circuitry. Nat. Commun. 2019, 10, 1503. [Google Scholar] [CrossRef]
- Vaz, A.P.; Wittig, J.H.; Inati, S.K.; Zaghloul, K.A. Replay of Cortical Spiking Sequences during Human Memory Retrieval. Science 2020, 367, 1131–1134. [Google Scholar] [CrossRef]
- Rutishauser, U.; Reddy, L.; Mormann, F.; Sarnthein, J. The Architecture of Human Memory: Insights from Human Single-Neuron Recordings. J. Neurosci. 2020, 41, 883–890. [Google Scholar] [CrossRef] [PubMed]
- Quian Quiroga, R. No Pattern Separation in the Human Hippocampus. Trends Cogn. Sci. 2020, 24, 994–1007. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, H.; Adolphs, R.; Oya, H.; Kovach, C.; Damasio, H.; Kaufman, O.; Howard, M. Analysis of Single-Unit Responses to Emotional Scenes in Human Ventromedial Prefrontal Cortex. J. Cogn. Neurosci. 2005, 17, 1509–1518. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Tudusciuc, O.; Mamelak, A.N.; Ross, I.B.; Adolphs, R.; Rutishauser, U. Neurons in the Human Amygdala Selective for Perceived Emotion. Proc. Natl. Acad. Sci. USA 2014, 111, E3110–E3119. [Google Scholar] [CrossRef]
- Mormann, F.; Bausch, M.; Knieling, S.; Fried, I. Neurons in the Human Left Amygdala Automatically Encode Subjective Value Irrespective of Task. Cereb. Cortex 2019, 29, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Unruh-Pinheiro, A.; Hill, M.R.; Weber, B.; Boström, J.; Elger, C.E.; Mormann, F. Single-Neuron Correlates of Decision Confidence in the Human Medial Temporal Lobe. Curr. Biol. 2020, 30, 4722–4732.e5. [Google Scholar] [CrossRef]
- Crandall, P.H.; Walter, R.D.; Rand, R.W. Clinical Applications of Studies on Stereotactically Implanted Electrodes in Temporal-Lobe Epilepsy. J. Neurosurg. 1963, 20, 827–840. [Google Scholar] [CrossRef]
- Bertrand, G.; Jasper, H. Microelectrode Recording of Unit Activity in the Human Thalamus. Stereotact. Funct. Neurosurg. 1965, 26, 205–208. [Google Scholar] [CrossRef]
- Gillingham, J. Forty-Five Years of Stereotactic Surgery for Parkinson’s Disease: A Review. Stereotact. Funct. Neurosurg. 2000, 74, 95–98. [Google Scholar] [CrossRef]
- Holtzheimer, P.E.; Husain, M.M.; Lisanby, S.H.; Taylor, S.F.; Whitworth, L.A.; McClintock, S.; Slavin, K.V.; Berman, J.; McKhann, G.M.; Patil, P.G.; et al. Subcallosal Cingulate Deep Brain Stimulation for Treatment-Resistant Depression: A Multisite, Randomised, Sham-Controlled Trial. Lancet Psychiatry 2017, 4, 839–849. [Google Scholar] [CrossRef]
- Misra, A.; Burke, J.F.; Ramayya, A.G.; Jacobs, J.; Sperling, M.R.; Moxon, K.A.; Kahana, M.J.; Evans, J.J.; Sharan, A.D. Methods for Implantation of Micro-Wire Bundles and Optimization of Single/Multi-Unit Recordings from Human Mesial Temporal Lobe. J. Neural Eng. 2014, 11, 026013. [Google Scholar] [CrossRef] [PubMed]
- McNaughton, B.L.; O’Keefe, J.; Barnes, C.A. The Stereotrode: A New Technique for Simultaneous Isolation of Several Single Units in the Central Nervous System from Multiple Unit Records. J. Neurosci. Methods 1983, 8, 391–397. [Google Scholar] [CrossRef]
- Gray, C.M.; Maldonado, P.E.; Wilson, M.; McNaughton, B. Tetrodes Markedly Improve the Reliability and Yield of Multiple Single-Unit Isolation from Multi-Unit Recordings in Cat Striate Cortex. J. Neurosci. Methods 1995, 63, 43–54. [Google Scholar] [CrossRef]
- Harris, K.D.; Henze, D.A.; Csicsvari, J.; Hirase, H.; Buzsáki, G. Accuracy of Tetrode Spike Separation as Determined by Simultaneous Intracellular and Extracellular Measurements. J. Neurophysiol. 2000, 84, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Quiroga, R.Q.; Nadasdy, Z.; Ben-Shaul, Y. Unsupervised Spike Detection and Sorting with Wavelets and Superparamagnetic Clustering. Neural Comput. 2004, 16, 1661–1687. [Google Scholar] [CrossRef] [PubMed]
- Rutishauser, U.; Schuman, E.M.; Mamelak, A.N. Online Detection and Sorting of Extracellularly Recorded Action Potentials in Human Medial Temporal Lobe Recordings, in Vivo. J. Neurosci. Methods 2006, 154, 204–224. [Google Scholar] [CrossRef]
- Niediek, J.; Boström, J.; Elger, C.E.; Mormann, F. Reliable Analysis of Single-Unit Recordings from the Human Brain under Noisy Conditions: Tracking Neurons over Hours. PLoS ONE 2016, 11, e0166598. [Google Scholar] [CrossRef] [PubMed]
- Rossant, C.; Kadir, S.N.; Goodman, D.F.M.; Schulman, J.; Hunter, M.L.D.; Saleem, A.B.; Grosmark, A.; Belluscio, M.; Denfield, G.H.; Ecker, A.S.; et al. Spike Sorting for Large, Dense Electrode Arrays. Nat. Neurosci. 2016, 19, 634–641. [Google Scholar] [CrossRef]
- Chung, J.E.; Magland, J.F.; Barnett, A.H.; Tolosa, V.M.; Tooker, A.C.; Lee, K.Y.; Shah, K.G.; Felix, S.H.; Frank, L.M.; Greengard, L.F. A Fully Automated Approach to Spike Sorting. Neuron 2017, 95, 1381–1394.e6. [Google Scholar] [CrossRef]
- Keshtkaran, M.R.; Yang, Z. Noise-Robust Unsupervised Spike Sorting Based on Discriminative Subspace Learning with Outlier Handling. J. Neural Eng. 2017, 14, 036003. [Google Scholar] [CrossRef] [PubMed]
- Jun, J.J.; Mitelut, C.; Lai, C.; Gratiy, S.L.; Anastassiou, C.A.; Harris, T.D. Real-Time Spike Sorting Platform for High-Density Extracellular Probes with Ground-Truth Validation and Drift Correction. BioRxiv 2017, 101030. [Google Scholar]
- Saif-ur-Rehman, M.; Lienkämper, R.; Parpaley, Y.; Wellmer, J.; Liu, C.; Lee, B.; Kellis, S.; Andersen, R.; Iossifidis, I.; Glasmachers, T.; et al. SpikeDeeptector: A Deep-Learning Based Method for Detection of Neural Spiking Activity. J. Neural Eng. 2019, 16, 056003. [Google Scholar] [CrossRef]
- Pachitariu, M.; Steinmetz, N.A.; Kadir, S.N.; Carandini, M.; Harris, K.D. Fast and Accurate Spike Sorting of High-Channel Count Probes with KiloSort. In Advances in Neural Information Processing Systems 29 (NIPS 2016); NIPS Proceedings: Barcelona, Spain, 2016. [Google Scholar]
- Pouzat, C.; Mazor, O.; Laurent, G. Using Noise Signature to Optimize Spike-Sorting and to Assess Neuronal Classification Quality. J. Neurosci. Methods 2002, 122, 43–57. [Google Scholar] [CrossRef]
- Schmitzer-Torbert, N.; Jackson, J.; Henze, D.; Harris, K.; Redish, A.D. Quantitative Measures of Cluster Quality for Use in Extracellular Recordings. Neuroscience 2005, 131, 1–11. [Google Scholar] [CrossRef]
- Knieling, S.; Sridharan, K.S.; Belardinelli, P.; Naros, G.; Weiss, D.; Mormann, F.; Gharabaghi, A. An Unsupervised Online Spike-Sorting Framework. Int. J. Neural Syst. 2016, 26, 1550042. [Google Scholar] [CrossRef]
- Hilgen, G.; Sorbaro, M.; Pirmoradian, S.; Muthmann, J.-O.; Kepiro, I.E.; Ullo, S.; Ramirez, C.J.; Puente Encinas, A.; Maccione, A.; Berdondini, L.; et al. Unsupervised Spike Sorting for Large-Scale, High-Density Multielectrode Arrays. Cell Rep. 2017, 18, 2521–2532. [Google Scholar] [CrossRef]
- Lee, J.; Carlson, D.; Shokri, H.; Yao, W.; Goetz, G.; Hagen, E.; Batty, E.; Chichilnisky, E.; Einevoll, G.; Paninski, L. YASS: Yet Another Spike Sorter. BioRxiv 2017, 151928. [Google Scholar] [CrossRef]
- Chaure, F.J.; Rey, H.G.; Quian Quiroga, R. A Novel and Fully Automatic Spike-Sorting Implementation with Variable Number of Features. J. Neurophysiol. 2018, 120, 1859–1871. [Google Scholar] [CrossRef]
- Yger, P.; Spampinato, G.L.; Esposito, E.; Lefebvre, B.; Deny, S.; Gardella, C.; Stimberg, M.; Jetter, F.; Zeck, G.; Picaud, S.; et al. A Spike Sorting Toolbox for up to Thousands of Electrodes Validated with Ground Truth Recordings in Vitro and in Vivo. eLife 2018, 7, e34518. [Google Scholar] [CrossRef]
- Rácz, M.; Liber, C.; Németh, E.; Fiáth, R.; Rokai, J.; Harmati, I.; Ulbert, I.; Márton, G. Spike Detection and Sorting with Deep Learning. J. Neural Eng. 2020, 17, 016038. [Google Scholar] [CrossRef]
- Park, I.Y.; Eom, J.; Jang, H.; Kim, S.; Park, S.; Huh, Y.; Hwang, D. Deep Learning-Based Template Matching Spike Classification for Extracellular Recordings. Appl. Sci. 2020, 10, 301. [Google Scholar] [CrossRef]
- Chapeton, J.I.; Haque, R.; Wittig, J.H.; Inati, S.K.; Zaghloul, K.A. Large-Scale Communication in the Human Brain Is Rhythmically Modulated through Alpha Coherence. Curr. Biol. 2019, 29, 2801–2811.e5. [Google Scholar] [CrossRef] [PubMed]
- Henze, D.A.; Borhegyi, Z.; Csicsvari, J.; Mamiya, A.; Harris, K.D.; Buzsáki, G. Intracellular Features Predicted by Extracellular Recordings in the Hippocampus In Vivo. J. Neurophysiol. 2000, 84, 390–400. [Google Scholar] [CrossRef]
- Buzsáki, G. Large-Scale Recording of Neuronal Ensembles. Nat. Neurosci. 2004, 7, 446–451. [Google Scholar] [CrossRef]
- Singer, W. Neuronal Synchrony: A Versatile Code for the Definition of Relations? Neuron 1999, 24, 49–65. [Google Scholar] [CrossRef]
- Gold, C.; Henze, D.A.; Koch, C.; Buzsáki, G. On the Origin of the Extracellular Action Potential Waveform: A Modeling Study. J. Neurophysiol. 2006, 95, 3113–3128. [Google Scholar] [CrossRef]
- Eom, J.; Kim, S.; Jang, H.; Shin, H.; Hwang, J.H.; Park, S.; Huh, Y.; Choi, H.J.; Hwang, D. Neural Spike Classification via Deep Neural Network. IBRO Rep. 2019, 6, S139–S140. [Google Scholar] [CrossRef]
- Grün, S. Data-Driven Significance Estimation for Precise Spike Correlation. J. Neurophysiol. 2009, 101, 1126–1140. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Zhang, G.; Zhu, W.; Liang, Z. ROC Operating Point Selection for Classification of Imbalanced Data with Application to Computer-Aided Polyp Detection in CT Colonography. Int. J. Comput. Assist. Radiol. Surg. 2014, 9, 79–89. [Google Scholar] [CrossRef]
- Ison, M.J.; Mormann, F.; Cerf, M.; Koch, C.; Fried, I.; Quiroga, R.Q. Selectivity of Pyramidal Cells and Interneurons in the Human Medial Temporal Lobe. J. Neurophysiol. 2011, 106, 1713–1721. [Google Scholar] [CrossRef]
- Gast, H.; Niediek, J.; Schindler, K.; Boström, J.; Coenen, V.A.; Beck, H.; Elger, C.E.; Mormann, F. Burst Firing of Single Neurons in the Human Medial Temporal Lobe Changes before Epileptic Seizures. Clin. Neurophysiol. 2016, 127, 3329–3334. [Google Scholar] [CrossRef] [PubMed]
- Perkel, D.H.; Gerstein, G.L.; Moore, G.P. Neuronal Spike Trains and Stochastic Point Processes: II. Simultaneous Spike Trains. Biophys. J. 1967, 7, 419–440. [Google Scholar] [CrossRef]
- Csicsvari, J.; Hirase, H.; Czurko, A.; Buzsáki, G. Reliability and State Dependence of Pyramidal Cell–Interneuron Synapses in the Hippocampus. Neuron 1998, 21, 179–189. [Google Scholar] [CrossRef]
- Maldonado, P.E.; Friedman-Hill, S.; Gray, C.M. Dynamics of Striate Cortical Activity in the Alert Macaque: II. Fast Time Scale Synchronization. Cereb. Cortex 2000, 10, 1117–1131. [Google Scholar] [CrossRef]
- Pedreira, C.; Martinez, J.; Ison, M.J.; Quian Quiroga, R. How Many Neurons Can We See with Current Spike Sorting Algorithms? J. Neurosci. Methods 2012, 211, 58–65. [Google Scholar] [CrossRef]
- Bair, W.; Zohary, E.; Newsome, W.T. Correlated Firing in Macaque Visual Area MT: Time Scales and Relationship to Behavior. J. Neurosci. 2001, 21, 1676–1697. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, S.; Amarasingham, A.; Harrison, M.T.; Buzsáki, G. Behavior-Dependent Short-Term Assembly Dynamics in the Medial Prefrontal Cortex. Nat. Neurosci. 2008, 11, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Harris, K.D.; Hirase, H.; Leinekugel, X.; Henze, D.A.; Buzsáki, G. Temporal Interaction between Single Spikes and Complex Spike Bursts in Hippocampal Pyramidal Cells. Neuron 2001, 32, 141–149. [Google Scholar] [CrossRef]
- Ostojic, S.; Brunel, N.; Hakim, V. How Connectivity, Background Activity, and Synaptic Properties Shape the Cross-Correlation between Spike Trains. J. Neurosci. 2009, 29, 10234–10253. [Google Scholar] [CrossRef] [PubMed]
- Brainard, D.H. The psychophysics toolbox. Spat. Vis. 1997, 10, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://de.mathworks.com/matlabcen-tral/fileexchange/73644-tightfig (accessed on 22 January 2021).
- Available online: https://de.mathworks.com/matlabcen-tral/fileexchange/22282-venn (accessed on 30 November 2020).



| Patients | Recording Sessions | SU | MU | Art | #Events |
|---|---|---|---|---|---|
| 13 | 51 | 2217 | 2212 | 4078 | 22,341,989 |
| Case | Cluster Combination | Spike Events Labelled as Artifacts |
|---|---|---|
| 1 | pair of artifact and (SU or MU) within the same channel | spike event in the artifact cluster |
| 2 | pair of artifact and (SU or MU) | both coincident spike events |
| 3 | SU and MU in the same bundle | coincident spikes in the MU |
| 4 | two SU or two MU in the same bundle | coincident spikes in the lower SNR |
| Case | Cluster Combination | Spike Events Labelled as Artifacts |
|---|---|---|
| 1 | pair of artifact and (SU or MU) | all coincident spike events |
| 2 | two clusters in different wire bundles | all coincident spikes |
| 3 | SU and MU in the same bundle | coincident spikes in the MU |
| 4 | two SU or two MU in the same bundle | coincident spikes in the lower SNR cluster |
| Part | Parameter | Threshold | Value |
|---|---|---|---|
| I | min. number of simultaneous spike events | nosim | 3 |
| max. time difference | ∆t | 50 µs | |
| max. Euclidean distance of spike-event shapes | dthr | 14.6 | |
| II | max. time difference in the same channel | ∆tsame channel | 650 µs |
| max. time difference in the same bundle | ∆tsame bundle | 50 µs | |
| max. Euclidean distance of spike-event shapes | dthr | 8.4 | |
| III | width of time bins in the cross-correlograms | tbin | 500 µs |
| max. z-value of central bin count in cross-correlograms | zthr | 5 |
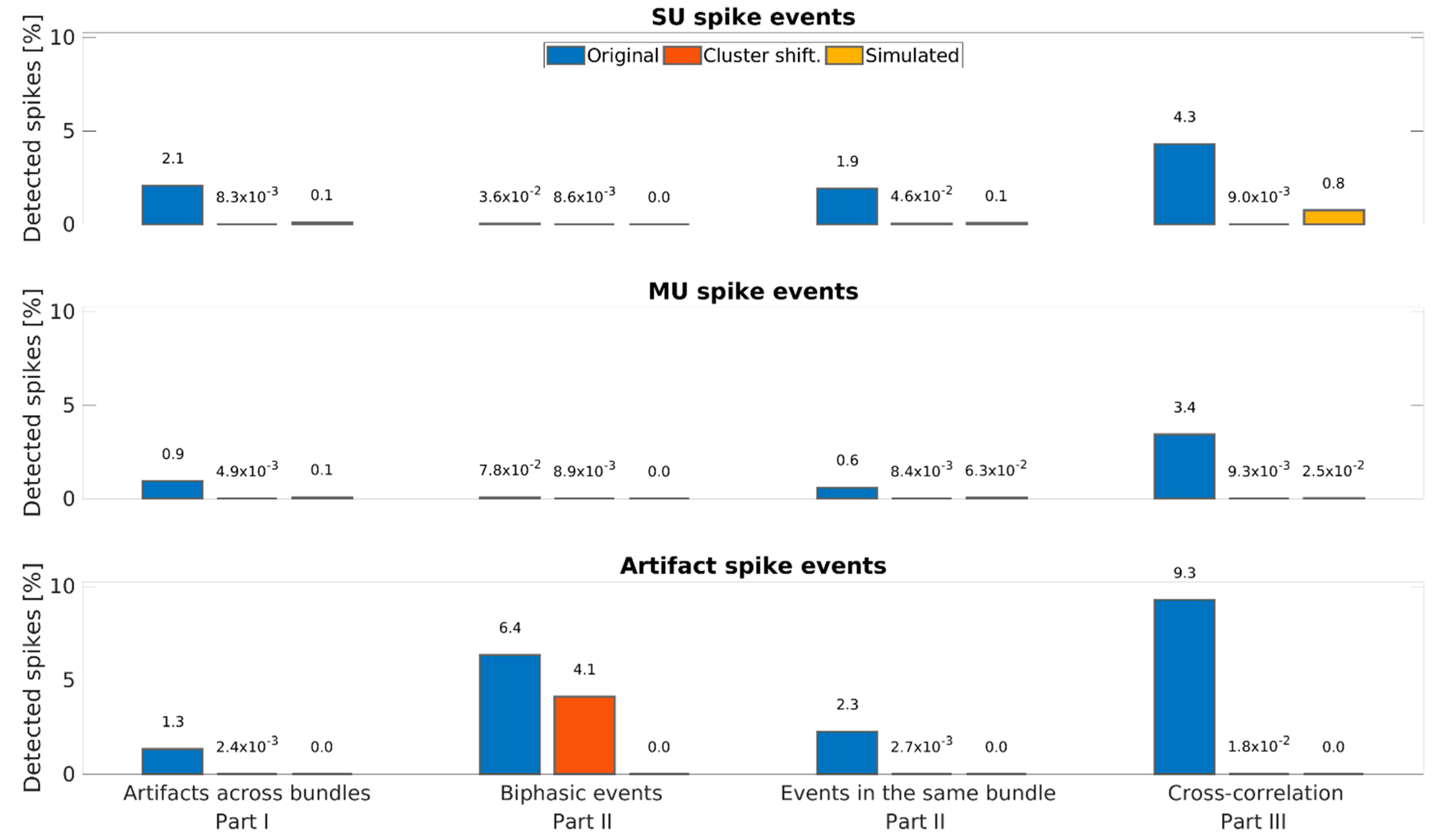
| Part I | Part II | Part III | ||
|---|---|---|---|---|
| Unit Class | Different Bundle | Same Channel | Same Bundle | Cross-Correlation |
| Artifacts | 1.34% | 6.35% | 2.26% | 9.27% |
| Multi-units | 0.94% | 0.08% | 0.61% | 3.45% |
| Single units | 2.08% | 0.04% | 1.93% | 4.30% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dehnen, G.; Kehl, M.S.; Darcher, A.; Müller, T.T.; Macke, J.H.; Borger, V.; Surges, R.; Mormann, F. Duplicate Detection of Spike Events: A Relevant Problem in Human Single-Unit Recordings. Brain Sci. 2021, 11, 761. https://doi.org/10.3390/brainsci11060761
Dehnen G, Kehl MS, Darcher A, Müller TT, Macke JH, Borger V, Surges R, Mormann F. Duplicate Detection of Spike Events: A Relevant Problem in Human Single-Unit Recordings. Brain Sciences. 2021; 11(6):761. https://doi.org/10.3390/brainsci11060761
Chicago/Turabian StyleDehnen, Gert, Marcel S. Kehl, Alana Darcher, Tamara T. Müller, Jakob H. Macke, Valeri Borger, Rainer Surges, and Florian Mormann. 2021. "Duplicate Detection of Spike Events: A Relevant Problem in Human Single-Unit Recordings" Brain Sciences 11, no. 6: 761. https://doi.org/10.3390/brainsci11060761
APA StyleDehnen, G., Kehl, M. S., Darcher, A., Müller, T. T., Macke, J. H., Borger, V., Surges, R., & Mormann, F. (2021). Duplicate Detection of Spike Events: A Relevant Problem in Human Single-Unit Recordings. Brain Sciences, 11(6), 761. https://doi.org/10.3390/brainsci11060761






