Abstract
Although Ca2+ ion plays an essential role in cellular physiology, calcium-binding proteins (CaBPs) were long used for mainly as immunohistochemical markers of specific cell types in different regions of the central nervous system. They are a heterogeneous and wide-ranging group of proteins. Their function was studied intensively in the last two decades and a tremendous amount of information was gathered about them. Girard et al. compiled a comprehensive list of the gene-expression profiles of the entire EF-hand gene superfamily in the murine brain. We selected from this database those CaBPs which are related to information processing and/or neuronal signalling, have a Ca2+-buffer activity, Ca2+-sensor activity, modulator of Ca2+-channel activity, or a yet unknown function. In this way we created a gene function-based selection of the CaBPs. We cross-referenced these findings with publicly available, high-quality RNA-sequencing and in situ hybridization databases (Human Protein Atlas (HPA), Brain RNA-seq database and Allen Brain Atlas integrated into the HPA) and created gene expression heat maps of the regional and cell type-specific expression levels of the selected CaBPs. This represents a useful tool to predict and investigate different expression patterns and functions of the less-known CaBPs of the central nervous system.
1. About Ca2+ Briefly
The Ca2+ ion plays an essential role in cellular physiology. It has two major functions: as an ion it affects the membrane potential, and as a second messenger it activates several intracellular mechanisms, e.g., the contraction of the myocardium, hormone secretion by endocrine cells, enzyme activation, degranulation of various white blood cells, as well as neurotransmitter release from the synaptic terminals. Besides these rapid effects, calcium has longer-lasting effects too (like long-term potentiation), and influences even gene transcription, cell proliferation and differentiation.
In the resting state of a cell the concentration of free calcium in the cytoplasm varies between 10–100 nmol/L, more than a 1000-fold lower than that in the blood. A functionally optimal concentration is achieved by the fine-tuned balance of input-output mechanisms. In general, the intracellular increase in calcium concentration can be attributed to two main mechanisms. On one hand, the Ca2+ ions can enter the cell from the extracellular space through the ion channels of the cell membrane that can be gated by the membrane potential, ligand binding or other factors. On the other hand, Ca2+ is released from internal stores, such as the endoplasmic reticulum (named sarcoplasmic reticulum in the muscle cells), mitochondria or calcium buffers. The release of Ca2+ from the internal stores is led by Ca2+ itself or by a group of distinct messengers. Voltage-gated calcium entry takes place in electrically excitable cells whereas in non-excitable cells the intracellular calcium level increase is mainly the result of release from the internal stores. The main mechanisms for decreasing the intracellular Ca2+ concentration are the different exchangers and pumps, which remove Ca2+ from the cytosol. Their importance varies depending on the cell type. These processes transport calcium against its electro-chemical gradient and, therefore, are energy dependent. Calcium can be pumped into the organelles (intracellular Ca2+ stores), or it can be pumped to the extracellular space via plasma membrane calcium pumps or by indirect active transport achieved by exchanger proteins. The intracellular Ca2+ concentration is also dependent on the dynamic binding to different calcium-binding proteins.
An excessive amount of intracellular calcium can lead to cell damage or even cell death. By this mechanism, over-excitation of neural circuits can cause excitotoxicity. Therefore, the fine-tuning of calcium levels by several receptors, calcium-channels, calcium pumps and exchangers, calcium-buffers and numerous calcium-binding proteins (CaBPs) is of utmost importance.
2. Calcium in Neurons
Neurotransmitter release is closely correlated to calcium levels. Calcium ion channel density is high at active zones in the presynaptic terminals, opposite the post-synaptic receptors. Calcium ions do not diffuse far from their site of entry because they are immediately buffered by calcium-binding proteins [1]. Therefore, calcium influx generates a local rise in calcium concentration at the site of the active zones. In all synapses neurotransmitter release has a non-linear correlation with Ca2+ influx, meaning that a twofold increase in Ca2+ levels can produce a 16-fold increase of the amount of neurotransmitter released [2]. By fine-tuning the calcium levels in the presynaptic terminal, throughout the duration of the action potential, the neurotransmitter release is also regulated, therefore the synaptic transmission is also affected [3].
Calcium, as a second messenger, carries signals throughout the nerve cell as a reaction to membrane depolarization. Therefore it is transferring information on neuronal activity status both locally, for example in a dendritic spine or a small dendritic segment, and all over within the neuron, for instance to increase energy metabolism [4]. Changes in Ca2+ levels are often restricted to specific regions of the cytosol, allowing different processes (like exocytosis of a secretory vesicle) to happen locally, without affecting other processes elsewhere in the cell [5]. A crucial function of calcium is to regulate activity-dependent signalling. By feed-back and feed-forward mechanisms, calcium signalling reinforces relevant synaptic connections, eliminates unnecessary connections and evades overexcitation, therefore it controls neuronal excitability [6] and it is involved in the mechanisms of learning and memory.
3. The Role of Ca2+ Binding
The cytosolic Ca2+ concentration can transiently increase by 10–100 fold, due to the signal-induced release from the endoplasmic reticulum or influx from the extracellular medium by calcium channels. The sudden cytosolic change in calcium level is detected by certain Ca2+-binding proteins [5]. These calcium binding proteins regulate Ca2+ concentration but they are also regulated by the intracellular amount of calcium. The concentration of free cytosolic calcium in a resting cell is low due to the uptake into the endoplasmic reticulum and/or the continuous transport out of the cell, orchestrated by ATP-fuelled pumps. Complementing these main input–output mechanisms, when calcium buffers become loaded with calcium, they serve as fine-tuning agents of the spatial and temporal properties of calcium signals. These proteins influence both the amplitude and the recovery time of calcium transients [7]. Furthermore, CaBPs possibly directly or indirectly facilitate sensitization or desensitization of calcium channels and may cut off additional calcium entry into the cell [8].
Ca2+ binding proteins are a heterogeneous and extensive group of proteins (see Figure 1, based on Elies et al. [9], that are engaged in many cellular and extracellular functions from calcium homeostasis to calcium signalling pathways [10]. The calcium signal is decoded by various calcium-binding motifs. These are present in the specialised calcium-buffer and calcium-sensor proteins that couple changes in calcium concentration to a wide variety of functions depending on their disposition and calcium source [11]. Despite the fact that they have various structures and properties, they selectively and reversibly bind calcium in specific domains, the kinetics of this interaction frequently being very fast [12].
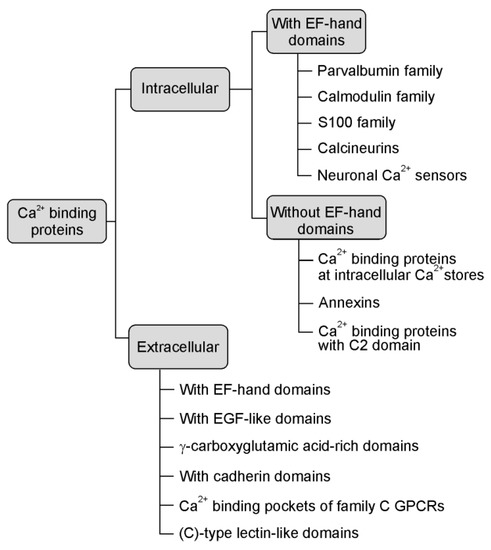
Figure 1.
The main types of calcium-binding proteins (CaBPs) [9].
The rise in cytosolic calcium is sensed especially by EF-hand family CaBPs. The large family of EF-hand domain-containing proteins comprises of various important and ubiquitously expressed CaBPs, like calmodulin [12,13], but also several proteins with functions that we are just beginning to understand.
4. The EF-Hand Calcium-Binding Proteins
Among the different types of calcium-binding proteins, the EF-hand CaBPs represent the largest family, for example, humans express approximately 250 EF-hand containing proteins [14]. This family includes proteins consisting of one or more EF-hand domains and possessing various functions like calcium buffering in the cytosol or signal transduction between cellular compartments. The term EF-hand is a descriptive one. It not only refers to the molecular structure of this calcium-binding domain but also to its motion that the binding of calcium can generate [15]. The EF-hand motif itself is composed of a highly conserved sequence of 12 amino acids which can chelate a single Ca2+ ion, surrounded by two α-helices. These two helices are bound by the short loop in such a distinct way, that the helix-loop-helix motif can be seen as the spread thumb and forefinger of a human hand [5] (see Figure 2). The calcium ions are coordinated within the loop and the affinity for Ca2+ is a determining factor for the function of the protein. In some cases the loop can accommodate Mg2+ as well [15].
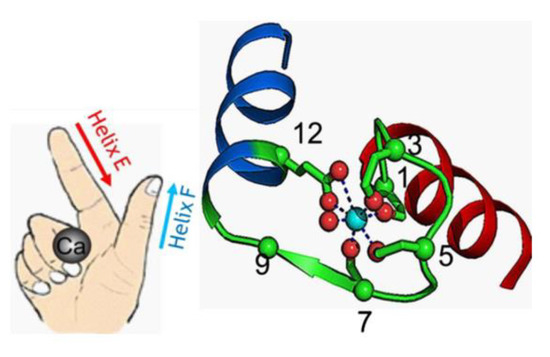
Figure 2.
The helix-loop-helix EF-hand Ca2+-binding motif based on Zhou et al., 2009 (Wikimedia commons licence). The conformation of the helixes resembles a spread thumb and forefinger of a human hand. In the representative 3D model of a typical canonical EF-hand motif from calmodulin, the Ca2+ ion is integrated within the loop, which includes seven oxygen atoms from the sidechain carboxyl or hydroxyl groups (loop sequence positions 1, 3, 5, 12), a main chain carbonyl group (position 7), and a bridged water (via position 9) [16]. Figure copyright permission obtained.
Frequently, EF-hand motifs occur in adjoining pairs, for instance, calmodulin and troponin C contain 4 EF-hand motifs, whereas parvalbumin contains only 2 EF-hand motifs [12]. Calcium ions bind to EF-hand domains with different affinities in a range of 10−6 M to 10−3 M, which allows them to fulfil their diverse biological functions [11,17].
From a functional point of view calcium-binding proteins can be divided into two major groups. First, there are the Ca2+ sensor proteins/signal modulators, which have a dissociation constant (Kd) Kd > 0.1 μM (e.g., calmodulin family, S100 families, synaptotagmin). These proteins have relatively low affinity to calcium and undergo significant conformational changes upon binding calcium in order to further interact with specific downstream targets, thereby switching their activities on or off [18]. A great deal of what we know about the mechanism of action of Ca2+ sensors is based on the structural analysis of calmodulin. If calcium sensors are present in high concentration, they can act as calcium buffers as well [19].
The second main group comprises of the Ca2+ buffers which have high affinity, Kd < 0.1 μM, that are thought to mainly chelate calcium (e.g., parvalbumin) [18,20]. In this case the binding of calcium and, therefore, the dispersion of local calcium does not cause major modification of the protein structure. They do not directly influence the activity of other macromolecules [21], instead, they moderate calcium transients. They modulate the shape and/or duration of Ca2+ signals and help maintain Ca2+ homeostasis. Their main function is to control free calcium in the cell. During resting-state calcium buffers are mainly free of calcium, but if calcium levels rise, these proteins will modulate the spatiotemporal aspects of calcium signals. The way in which calcium transients are affected is also influenced by the intracellular distribution and mobility of these calcium-buffers [22]. By modifying the calcium concentration, calcium buffers can indirectly affect neuronal excitability and synaptic plasticity [21].
There is no distinct boundary between the function of calcium sensors and buffers, seeing that the activity of these proteins is conditioned by their local concentration and the availability of interacting partners in signalling networks [23].
5. The Regional Arrangement of the EF-Hand CaBPs Throughout the Central Nervous System (CNS)
The distribution of EF-hand CaBPs throughout the central nervous system (CNS) is not homogeneous. Some of them are expressed ubiquitously, whereas others have distinct expression patterns, they can be limited to specific brain regions, subtypes of neurons or glial cells or they can appear in limited time intervals during the development of the CNS. Members of the calmodulin calcium-binding family are frequently mentioned as ubiquitously expressed proteins in all eukaryote cells. As an opposite example, we can mention the specific expression pattern of CaBPs like secretagogin, which is expressed by amacrine cells and rod photoreceptors of the retina [24], and neurons of the olfactory bulb [25,26]. Recently other CaBPs, the N-terminal EF-hand calcium-binding proteins (NECAB) 1 and 2 were shown to be expressed predominantly by CB1/CCK-positive interneurons throughout the isocortex, hippocampal formation and basolateral amygdala complex. Furthermore, they exhibit different subcellular distribution in the CB1/CCK-positive GABAergic interneurons [27].
6. The “Classical” CaBPs of the CNS
CaBPs were regarded initially as immunohistochemical markers of specific cell types in different regions of the CNS. Parvalbumin, calbindin and calretinin are the most often used CaBPs for this purpose, especially for the identification of inhibitory local circuit neurons [28,29]. Sometimes their presence can predict cellular excitability and the fashion of neurotransmitter release following electrical stimuli. As these proteins have an essential role in cellular physiology, they deserve our attention, not just as cellular markers. Their complex and often interlaced roles in calcium homeostasis, in intracellular signalling, and also their alterations in brain diseases should be elucidated.
In the last two decades, a tremendous amount of information has been published about CaBPs. It is beyond the scope of our review to give a comprehensive overview of all CaBPs and Ca2+-dependent signalling cascades. We will focus on the neuronal CaBPs, their function and distribution in the central nervous system by systematically analysing publicly available, high-quality RNA-sequencing and in situ hybridization (ISH) databases. This allows us to better understand the specific spatiotemporal distribution and function of genes expressed in the brain. By summarising quantitative regional and cell-specific data in a visually clear way, our review aims to find relevant information regarding the functionally important CaBPs in the brain.
7. Search for New CaBPs. Detailed Analysis of RNA-Seq and ISH Databases
Through extensive examination of the Uniprot (www.uniprot.org accessed on 12 May 2021) and National Center for Biotechnology Information (NCBI Gene, www.ncbi.nlm.nih.gov/gene accessed on 12 May 2021) databases, Girard et al. found that the EF-hand family comprises in mouse at least 249 putative members [30]. With the help of Uniprot database and the Allen Brain Atlas (ABA, www.brain-map.org accessed on 12 May 2021) they created a meticulous list of the EF-hand super-family in the mouse genome. The functions of these genes were given using Gene Ontology, including roles like participation in muscle homeostasis, leucocyte adhesion, regulation of pH, apoptosis, etc. For the complete list of their integrated functions according to Gene Ontology terms see Table 1, adapted from Girard et al.

Table 1.
Putative functions of calcium-binding proteins (CaBPs) involved in neuronal signalling in the central nervous system (CNS).
Out of the pool of various functions we carefully selected those CaBPs that are connected to information processing and/or neuronal signalling, have Ca2+-buffer activity, Ca2+- sensor activity, modulator of Ca2+ channel activity or yet unknown function (Table 1), furthermore they show detectable gene expression in the brain. In this way we created a gene function-based selection of the calcium-binding proteins.
We cross-referenced our calcium-binding protein list with several other databases. First, we used the Human Protein Atlas (HPA, www.proteinatlas.org, accessed on 29 April 2020, [31,32]) Brain Atlas transcriptomics analysis performed on multiple brain regions (Figure 3). In this dataset, transcript expression levels are summarized per gene in 13 brain regions based on RNA-seq transcripts per million (TPM) and protein-coding transcripts per million (pTPM). The TPM values were TMM (trimmed means of M values) normalized between all samples respectively, and then each gene was Pareto scaled. The normalized expression (NX) calculation is based on pTPM of the individual samples. The Human Protein Atlas version 19.3 and Ensembl version 92.38 were used for analysis. To shine a light on the regional expression levels of the selected CaBPs we created the following gene expression heat maps.
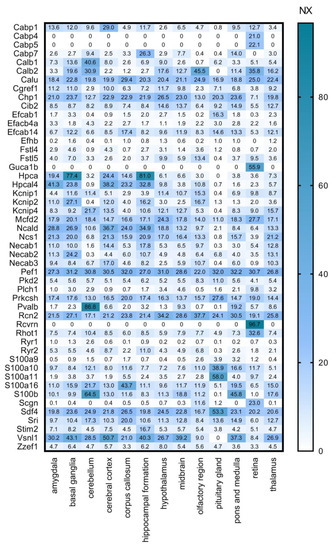
Figure 3.
Regional expression levels of the selected CaBP genes. Mouse gene data from Human Protein Atlas (HPA) transcriptomics analysis is shown as NX values in different brain regions. Numerical data is presented in Supplementary Materials Table S1.
As a next step we used the RNA Allen mouse brain region gene data (obtained from the Allen Brain Atlas, ABA) integrated into the HPA, which contains transcript expression levels summarized per gene in 10 brain regions based on in situ hybridisation (ISH) in the adult mouse brain. This ISH data provides spatial expression data on a single-cell level (Figure 4). The expression energy shown in Figure 4, based on ISH, is defined as the sum of expression pixel intensity divided by the sum of all pixels in division [33].
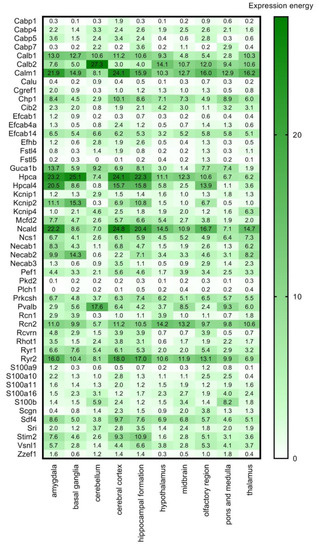
Figure 4.
Transcript expression levels in 10 brain regions based on in situ hybridisation. RNA Allen mouse brain region gene data of the selected CaBPs from in situ hybridization (ISH) analysis is represented by the expression energy value. Numerical data are presented in Supplementary Materials Table S2.
High overlap and complementarity can be observed between the HPA transcriptomics and the ISH databases (Figure 3 and Figure 4).
In order to obtain more specific information about the distinct expression patterns of the aforementioned proteins, we used a cell-type-specific analysis, RNA-seq results of selected cells using immunopanning from the Brain RNA-seq database (www.brainrnaseq.org, accessed on 12 May 2020). Whilst creating this database, Ye Zhang et al. (2014) [34] purified the major cell types in the adult mouse brain, namely neurons, astrocytes, various maturation states of oligodendrocytes, microglia and endothelial cells. For all cell types, one biological replicate consisted of pooled cells from a litter of 3–12 mice. To create a transcriptome database by RNA sequencing, two replicates of pooled animals for each cell type were sequenced. Expression level estimation is reported as fragments per kilobase of transcript sequence per million mapped fragments (FPKM) value together with confidence intervals for each sample.
This dataset allows us to visualize the expression levels of the selected CaBPs in the major cell types in the adult mouse brain (Figure 5).
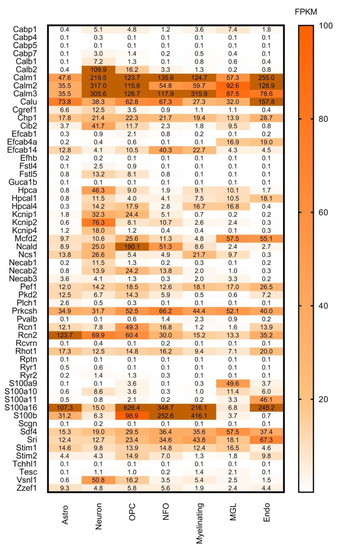
Figure 5.
Cell-type specific gene expression levels of selected CaBPs from the Brain RNA-seq database. Expression level estimation is reported as fragments per kilobase of transcript sequence per million mapped fragments (FPKM). Astro: astrocytes, OPC: oligodendrocyte progenitor cells, NFO: newly formed oligodendrocytes, Myelinating: myelinating oligodendrocyte, MGL: microglia/macrophage, Endo: endothelial. Numerical data are presented in Supplementary Materials Table S3.
In the analysis of Girard et al. some genes are mentioned as ‘no expression detected in the brain (n.e.d.)’ or ’no data available in the ABA database (n.d.)’, but when we cross referenced these genes with the HPA database, several of them were found. These genes are presented in Table 2 and Figure 6, Figure 7 and Figure 8.

Table 2.
Putative functions of calcium-binding proteins with lower expression in the central nervous system based on the Girard database.
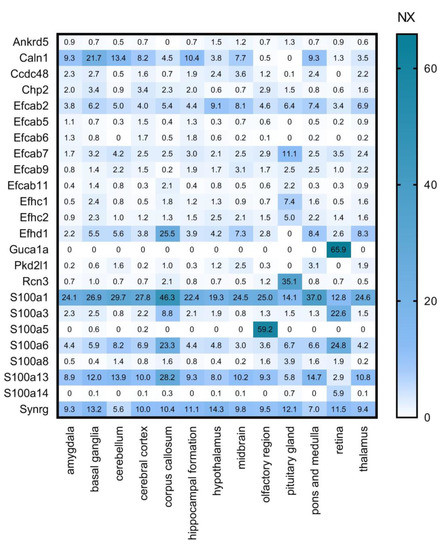
Figure 6.
Mouse regional gene data of CaBPs with lower expression in the CNS by Girard database. Transcriptomics analysis from HPA showing normalized expression (NX) values of genes in different brain regions. Numerical data are presented in Supplementary Materials Table S4.
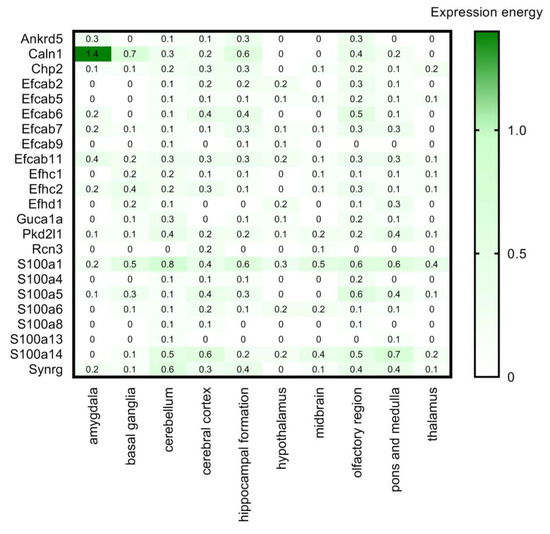
Figure 7.
Transcript expression levels of CaBPs with lower expression in the CNS by Girard database in 10 brain regions, based on in situ hybridisation. RNA Allen mouse brain region gene data is represented by the expression energy value. Note the different color scale from Figure 4. Numerical data are presented in Supplementary Materials Table S5.
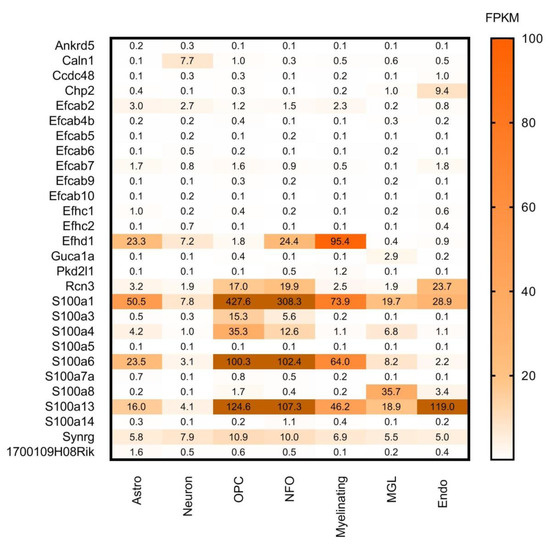
Figure 8.
Cell-type specific gene expression levels of CaBPs with lower expression in the CNS by Girard database. Expression level estimation from Brain RNA-seq database is reported as fragments per kilobase of transcript sequence per million mapped fragments (FPKM). Astro: astrocytes, OPC: oligodendrocyte progenitor cells, NFO: newly formed oligodendrocytes, Myelinating: myelinating oligodendrocyte, MGL: microglia/macrophage, Endo: endothelial. Numerical data are presented in Supplementary Materials Table S6.
The aim of our work was to provide a condensed database and visual presentation of data scattered over several primary sources. Using the color-coded figures, it is easier to spot relevant expression patterns of different CaBP genes. These figures can help to find which genes have the highest expression level in a given region, and which proteins occur in the same place. It is also possible to combine information on their distribution patterns in different brain regions and cell types. Next we present a few examples regarding the application of our databases and data visualisation.
In Figure 3 it is striking that Cabp4, Cabp5, Guca1b, Rcvrn are expressed exclusively in the retina, and also Plch1, Rhot1 and Scgn have a much stronger expression here than in other parts of the CNS. This group can, therefore, be considered as important retinal calcium-binding proteins. This observation is consistent with the literature: CaBP4 is localised in photoreceptor synaptic terminals and is essential for neurotransmission between photoreceptors and bipolar cells as part of the Cav1.4 channel complex in the retina [35]; CaBP5 in mice is expressed in type 5 ON-cone bipolar cells, and in type 3 OFF-cone bipolar cells as well as in rod bipolar cells [36,37,38]; Guca1b is important for rod cell recovery after light exposure, by stimulation of guanylate cyclases in these photoreceptors [39]; Secretagogin plays an essential role in synapse maturation and in mouse, rat, and rabbit retina it is expressed in subtypes of cone bipolar cells, but cannot be detected in the rod bipolar cells [40]; Recoverin is thought to be a calcium sensor in retinal rod cells that can control the lifetime of photoexcited rhodopsin by inhibiting rhodopsin kinase [41,42]; and last but not least Plch1 has been shown to be expressed in mouse and human retina as well [43].
Proteins like Calm1, Calm2, Calm3, Calu, Chp1, Pef1, Prkcsh, Rcn2, Sdf4, S100a16, and S100b are highly and ubiquitously expressed by all the examined cell types: astrocytes, neurons, oligodendrocyte progenitor cells, newly formed oligodendrocytes, myelinating oligodendrocyte, microglia/macrophage, endothelial cells (Figure 5). If these proteins are expressed by all cell types, we expect that they should be present in all regions of the brain. Cross-referencing these findings with Figure 3 and Figure 4, it can be observed that these proteins are present indeed in all the representative brain regions and almost at the same expression level. Some of these genes (Calm1, Calm2, Calm3, Prkcsh, S100a16 and S100b) code proteins that have Ca2+ sensor/buffer activity. The function of the others (Calu, Chp1, Pef1, Rcn2, Sdf4) is yet unknown, but since they are similarly scattered through the brain, we may conclude that they might have fundamental functions in the cell’s calcium homeostasis as well.
In Figure 5 it is conspicuous that hippocalcin (Hpca), voltage-gated potassium channel interacting protein (Kcnip2) and visinin-like protein 1 (Vsnl1) are mainly expressed by neurons. Kcnip2 shows high expression levels in the basal ganglia, hippocampal formation, amygdala and cerebral cortex but it is less expressed in the cerebellum. This Ca2+ sensor protein is one of the four K+ channel interacting proteins (Kcnips1–4), which interacts with and modulates the activity and trafficking of Kv4 potassium channels [44]. It is a fundamental component of the endogenous A-type Kv channel complex and being an auxiliary subunit of this channel it is confined to intracellular membranes and the cell membrane [45,46,47]. In the hippocampus and neocortex Kcnip2 was found in association with the Kv4.2 subunit of the Kv4 potassium channel, mainly expressed in the apical and basal dendrites of glutamatergic pyramidal neurons [47,48,49]. The co-presence of Kcnip2 and Kv4 subunits indicates that this CaBP is likely to play a major role as modulator of somatodendritic excitability [47]. Furthermore, Kcnip2 expression is linked to the overall survival of glioblastoma patients, thereby gaining clinical significance [50].
Hippocalcin is a member of the neuronal calcium sensor protein family, dominantly expressed in the pyramidal cell layer of the hippocampus [51], moderately in the dentate granule cells and the pyramidal cells of cerebral cortex layers II–VI and weakly in the large neuronal cells of the caudate-putamen. It is mainly localized in the cytoplasm and plasma membrane of dendrites and soma [52]. Palmer et al. described its important calcium-sensing role in NMDAR (N-methyl-D-aspartate receptor)-mediated hippocampal LTD (long-term depression), where it couples NMDAR activation to the endocytosis of AMPARs (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor) [53]. It was shown that hippocalcin as a diffusible calcium sensor is the key intermediate between calcium and potassium channels that mediate a slow afterhyperpolarization current [54]. This suggests that Hpca and Kcnip2 might have complementary roles in pyramidal cell excitability.
Visinin like protein 1 (Vsnl1) is also mainly expressed by neurons and mostly in the cerebral cortex, hippocampal formation, amygdala, hypothalamus, thalamus, midbrain and cerebellum, which is consistent with the observation that VSNLs show a widespread but distinct expression pattern primarily in nerve cells [55]. More detailed studies show strong expression levels in subpopulations of calbindin-D28K and calretinin- positive GABAergic interneurons in all hippocampal regions of the rat brain, but Vsnl1 was detected in principal cells as well [56]. Vsnl1 is a Ca2+ sensing protein and it influences dendritic growth, cyclic nucleotide signalling, and nicotinic modulation of neuronal network activity, and therefore it regulates synaptic plasticity [57]. It also plays a functional role in integrating the cytosolic calcium concentration and the oxidative status of the cell [58]. Visinin-like protein 1 has been identified as a biomarker for Alzheimer’s disease [59], and it has been shown to play an important part in the development of amyotrophic lateral sclerosis [58,60].
These few examples show that the databases we created can be used to further predict and investigate different expression patterns of the little known CaBPs. The various kinetic and buffering properties of CaBPs help modulate the synaptic responses and the excitability of neurons, and therefore they are adapted to the specific needs of the cell populations that express them. The kinetics of calcium binding and the dissociation constant (Kd) varies between different CaBPs. For instance, the calcium buffering capacity of motoneurons and adrenal chromaffin cells is many folds lower than that of the Purkinje neurons. The low buffering capacity of motoneurons allows them to generate rapid Ca2+ signals, but this aspect makes them more sensitive to excitotoxicity, that in turn could promote motoneuron disease [61].
Due to the lack of data, extensive study of the kinetics, the affinity and calcium binding capacity of the presented CaBPs are beyond the scope of our review. However, considering that there is a strong correlation between the calcium-binding properties of certain CaBPs and the electrophysiological and metabolic behaviour of the neurons that express them, further studies are necessary to better understand the functions of these proteins.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/brainsci11050634/s1, Table S1: Numerical data of regional expression levels of the selected CaBP genes. Mouse gene data from Human Protein Atlas (www.proteinatlas.org, accessed on 29 April 2020) transcriptomics analysis is shown as NX values in different brain regions. Heat map format is presented in Figure 3; Table S2: Numerical data of transcript expression levels in 10 brain regions based on in situ hybridisation. RNA Allen mouse brain region gene data of the selected CaBPs from ISH analysis is represented by the expression energy value. Heat map format is presented in Figure 4; Table S3: Numerical data of cell-type specific gene expression levels of selected CaBPs from the Brain RNA-seq database (www.brainrnaseq.org, accessed on 12 May 2020). Expression level estimation is reported as fragments per kilobase of transcript sequence per million mapped fragments (FPKM). Heat map format is presented in Figure 5; Table S4: Numerical data of mouse regional gene data of CaBPs with lower expression in the CNS by Girard database. Transcriptomics analysis from Human Protein Atlas (www.proteinatlas.org, accessed on 29 April 2020) showing NX values of genes in different brain regions. Heat map format is presented in Figure 6; Table S5: Numerical data of transcript expression levels of CaBPs with lower expression in the CNS by Girard database in 10 brain regions, based on in situ hybridisation. RNA Allen mouse brain region gene data is represented by the expression energy value. Heat map format is presented in Figure 7; Table S6: Numerical data of cell-type specific gene expression levels of CaBPs with lower expression in the CNS by Girard database. Expression level estimation from Brain RNA-seq database (www.brainrnaseq.org, accessed on 12 May 2020) is reported as fragments per kilobase of transcript sequence per million mapped fragments (FPKM). The heat map format is presented in Figure 8.
Author Contributions
Conceptualization, K.K. and T.S.; methodology, K.K. and T.S.; validation, K.K.; formal analysis, K.K.; data curation, K.K. and T.S.; writing—original draft preparation, K.K.; writing—review and editing, T.S.; visualization, K.K. and T.S.; supervision, T.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study (www.proteinatlas.org accessed in 29 April 2020, www.brainrnaseq.org accessed in 12 May 2020). The compiled databases presented in the article are available as Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Blaustein, M.P. Calcium transport and buffering in neurons. Trends Neurosci. 1988, 11, 438–443. [Google Scholar] [CrossRef]
- Kasai, H. Comparative biology of Ca2+-dependent exocytosis: Implications of kinetic diversity for secretory function. Trends Neurosci. 1999, 22, 88–93. [Google Scholar] [CrossRef]
- Kandel, E.R.; Schwartz, J.H.; Jessell, T.M.; Siegelbaum, S.A.; Hudspeth, A.J. Principles of Neural Science, 5th ed.; Health Professions Division; McGraw-Hill: New York, NY, USA, 2013. [Google Scholar]
- Augustine, G.J.; Santamaria, F.; Tanaka, K. Local Calcium Signaling in Neurons. Neuron 2003, 40, 331–346. [Google Scholar] [CrossRef]
- Lodish, H. Molecular Cell Biology, 8th ed.; W.H. Freeman: New York, NY, USA, 2016. [Google Scholar]
- Gleichmann, M.; Mattson, M.P. Neuronal Calcium Homeostasis and Dysregulation. Antioxid. Redox Signal. 2011, 14, 1261–1273. [Google Scholar] [CrossRef] [PubMed]
- Berridge, M.J.; Bootman, M.D.; Roderick, H.L. Calcium signalling: Dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 2003, 4, 517–529. [Google Scholar] [CrossRef]
- Bastianelli, E. Distribution of calcium-binding proteins in the cerebellum. Cerebellum 2003, 2, 242–262. [Google Scholar] [CrossRef] [PubMed]
- Elíes, J.; Yáñez, M.; Pereira, T.M.C.; Gil-Longo, J.; MacDougall, D.A.; Campos-Toimil, M. An Update to Calcium Binding Proteins. Adv. Exp. Med. Biol. 2019, 1131, 183–213. [Google Scholar]
- Xu, J.-H.; Tang, F.-R. Voltage-Dependent Calcium Channels, Calcium Binding Proteins, and Their Interaction in the Pathological Process of Epilepsy. Int. J. Mol. Sci. 2018, 19, 2735. [Google Scholar] [CrossRef] [PubMed]
- Bagur, R.; Hajnóczky, G. Intracellular Ca2+ Sensing: Its Role in Calcium Homeostasis and Signaling. Mol. Cell 2017, 66, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Yáñez, M.; Gil-Longo, J.; Campos-Toimil, M. Calcium Binding Proteins. In Advances in Experimental Medicine and Biology; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2012; Volume 740, pp. 461–482. [Google Scholar]
- Tang, S.; Deng, X.; Jiang, J.; Kirberger, M.; Yang, J.J. Design of Calcium-Binding Proteins to Sense Calcium. Molecules 2020, 25, 2148. [Google Scholar] [CrossRef] [PubMed]
- Schwaller, B. The continuing disappearance of “pure” Ca2+ buffers. Cell. Mol. Life Sci. 2009, 66, 275–300. [Google Scholar] [CrossRef]
- Lewit-Bentley, A.; Réty, S. EF-hand calcium-binding proteins. Curr. Opin. Struct. Biol. 2000, 10, 637–643. [Google Scholar] [CrossRef]
- Zhou, Y.; Frey, T.K.; Yang, J.J. Viral calciomics: Interplays between Ca2+ and virus. Cell Calcium 2009, 46, 1–17. [Google Scholar] [CrossRef]
- Gifford, J.L.; Walsh, M.P.; Vogel, H.J. Structures and metal-ion-binding properties of the Ca2+-binding helix–loop–helix EF-hand motifs. Biochem. J. 2007, 405, 199–221. [Google Scholar] [CrossRef] [PubMed]
- Andressen, C.; Blümcke, I.; Celio, M.R. Calcium-binding proteins: Selective markers of nerve cells. Cell Tissue Rev. Artic. 1993, 271, 181–208. [Google Scholar] [CrossRef]
- McMahon, S.M.; Jackson, M.B. An Inconvenient Truth: Calcium Sensors Are Calcium Buffers. Trends Neurosci. 2018, 41, 880–884. [Google Scholar] [CrossRef] [PubMed]
- Kreutz, M.R.; Naranjo, J.R.; Koch, K.; Schwaller, B. The Neuronal Functions of EF-hand Ca(2+)-binding Proteins. Front. Mol. Neurosci. 2012, 5, 92. [Google Scholar] [CrossRef]
- Gattoni, G.; Bernocchi, G. Calcium-Binding Proteins in the Nervous System during Hibernation: Neuroprotective Strategies in Hypometabolic Conditions? Int. J. Mol. Sci. 2019, 20, 2364. [Google Scholar] [CrossRef] [PubMed]
- Schwaller, B. Emerging Functions of the “Ca2+ Buffers” Parvalbumin, Calbindin D-28k and Calretinin in the Brain. In Handbook of Neurochemistry and Molecular Neurobiology; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2007; pp. 197–221. [Google Scholar]
- Alpár, A.; Attems, J.; Mulder, J.; Hökfelt, T.; Harkany, T. The renaissance of Ca2+-binding proteins in the nervous system: Secretagogin takes center stage. Cell. Signal. 2012, 24, 378–387. [Google Scholar] [CrossRef]
- Kántor, O.; Mezey, S.; Adeghate, J.; Naumann, A.; Nitschke, R.; Énzsöly, A.; Szabó, A.; Lukáts, Á.; Németh, J.; Somogyvári, Z.; et al. Calcium buffer proteins are specific markers of human retinal neurons. Cell Tissue Res. 2016, 365, 29–50. [Google Scholar] [CrossRef]
- Mulder, J.; Zilberter, M.; Spence, L.; Tortoriello, G.; Uhlén, M.; Yanagawa, Y.; Aujard, F.; Hökfelt, T.; Harkany, T. Secretagogin is a Ca2+-binding protein specifying subpopulations of telencephalic neurons. Proc. Natl. Acad. Sci. USA 2009, 106, 22492–22497. [Google Scholar] [CrossRef]
- Fairless, R.; Williams, S.K.; Diem, R. Calcium-Binding Proteins as Determinants of Central Nervous System Neuronal Vulnerability to Disease. Int. J. Mol. Sci. 2019, 20, 2146. [Google Scholar] [CrossRef]
- Miczán, V.; Kelemen, K.; Glavinics, J.R.; László, Z.I.; Barti, B.; Kenesei, K.; Kisfali, M.; Katona, I. NECAB1 and NECAB2 are Prevalent Calcium-Binding Proteins of CB1/CCK-Positive GABAergic Interneurons. Cereb. Cortex 2021, 31, 1786–1806. [Google Scholar] [CrossRef] [PubMed]
- Klausberger, T.; Somogyi, P. Neuronal Diversity and Temporal Dynamics: The Unity of Hippocampal Circuit Operations. Science 2008, 321, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Pelkey, K.A.; Chittajallu, R.; Craig, M.T.; Tricoire, L.; Wester, J.C.; McBain, C.J. Hippocampal GABAergic inhibitory interneurons. Physiol. Rev. 2017, 97, 1619–1747. [Google Scholar] [CrossRef] [PubMed]
- Girard, F.; Venail, J.; Schwaller, B.; Celio, M. The EF-hand Ca2+-binding protein super-family: A genome-wide analysis of gene expression patterns in the adult mouse brain. Neuroscience 2015, 294, 116–155. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S.; et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [Google Scholar] [CrossRef] [PubMed]
- Sjöstedt, E.; Zhong, W.; Fagerberg, L.; Karlsson, M.; Mitsios, N.; Adori, C.; Oksvold, P.; Edfors, F.; Limiszewska, A.; Hikmet, F.; et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 2020, 367, eaay5947. [Google Scholar] [CrossRef] [PubMed]
- Ezaldivar, A.; Krichmar, J.L. Allen Brain Atlas-Driven Visualizations: A web-based gene expression energy visualization tool. Front. Aging Neurosci. 2014, 8, 51. [Google Scholar]
- Zhang, Y.; Chen, K.; Sloan, S.A.; Bennett, M.L.; Scholze, A.R.; O’Keeffe, S.; Phatnani, H.P.; Guarnieri, P.; Caneda, C.; Ruderisch, N.; et al. An RNA-Sequencing Transcriptome and Splicing Database of Glia, Neurons, and Vascular Cells of the Cerebral Cortex. J. Neurosci. 2014, 34, 11929–11947. [Google Scholar] [CrossRef]
- Haeseleer, F.; Imanishi, Y.; Maeda, T.; E Possin, D.; Maeda, A.; Lee, A.; Rieke, F.; Palczewski, K. Essential role of Ca2+-binding protein 4, a Cav1.4 channel regulator, in photoreceptor synaptic function. Nat. Neurosci. 2004, 7, 1079–1087. [Google Scholar] [CrossRef]
- Haeseleer, F.; Sokal, I.; Verlinde, C.L.M.J.; Erdjument-Bromage, H.; Tempst, P.; Pronin, A.N.; Benovic, J.L.; Fariss, R.N.; Palczewski, K. Five Members of a Novel Ca2+-binding Protein (CABP) Subfamily with Similarity to Calmodulin. J. Biol. Chem. 2000, 275, 1247–1260. [Google Scholar] [CrossRef]
- Haverkamp, S.; Ghosh, K.K.; Hirano, A.A.; Wässle, H. Immunocytochemical description of five bipolar cell types of the mouse retina. J. Comp. Neurol. 2002, 455, 463–476. [Google Scholar] [CrossRef]
- Ghosh, K.K.; Bujan, S.; Haverkamp, S.; Feigenspan, A.; Wässle, H. Types of bipolar cells in the mouse retina. J. Comp. Neurol. 2003, 469, 70–82. [Google Scholar] [CrossRef]
- Makino, C.L.; Peshenko, I.V.; Wen, X.-H.; Olshevskaya, E.V.; Barrett, R.; Dizhoor, A.M. A Role for GCAP2 in Regulating the Photoresponse. J. Biol. Chem. 2008, 283, 29135–29143. [Google Scholar] [CrossRef] [PubMed]
- Puthussery, T.; Gayet-Primo, J.; Taylor, W.R. Localization of the calcium-binding protein secretagogin in cone bipolar cells of the mammalian retina. J. Comp. Neurol. 2009, 518, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Ames, J.B.; Ishima, R.; Tanaka, T.; Gordon, J.I.; Stryer, L.; Ikura, M. Molecular mechanics of calcium–myristoyl switches. Nat. Cell Biol. 1997, 389, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Borsatto, A.; Marino, V.; Abrusci, G.; Lattanzi, G.; Dell’Orco, D. Effects of Membrane and Biological Target on the Structural and Allosteric Properties of Recoverin: A Computational Approach. Int. J. Mol. Sci. 2019, 20, 5009. [Google Scholar] [CrossRef]
- Chen, Y.; Palczewski, K. Systems Pharmacology Links GPCRs with Retinal Degenerative Disorders. Annu. Rev. Pharm. Toxicol. 2016, 56, 273–298. [Google Scholar] [CrossRef]
- Xiong, H.; Kovacs, I.; Zhang, Z. Differential distribution of KChIPs mRNAs in adult mouse brain. Mol. Brain Res. 2004, 128, 103–111. [Google Scholar] [CrossRef]
- Shibata, R.; Misonou, H.; Campomanes, C.R.; Anderson, A.E.; Schrader, L.A.; Doliveira, L.C.; Carroll, K.I.; Sweatt, J.D.; Rhodes, K.J.; Trimmer, J.S. A Fundamental Role for KChIPs in Determining the Molecular Properties and Trafficking of Kv4.2 Potassium Channels. J. Biol. Chem. 2003, 278, 36445–36454. [Google Scholar] [CrossRef] [PubMed]
- Misonou, H.; Trimmer, J.S.; Misono, H. Determinants of Voltage-Gated Potassium Channel Surface Expression and Localization in Mammalian Neurons. Crit. Rev. Biochem. Mol. Biol. 2004, 39, 125–145. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, K.J.; Carroll, K.I.; Sung, M.A.; Doliveira, L.C.; Monaghan, M.M.; Burke, S.L.; Strassle, B.W.; Buchwalder, L.; Menegola, M.; Cao, J.; et al. KChIPs and Kv4 Subunits as Integral Components of A-Type Potassium Channels in Mammalian Brain. J. Neurosci. 2004, 24, 7903–7915. [Google Scholar] [CrossRef] [PubMed]
- Dabrowska, J.; Rainnie, D.G. Expression and distribution of Kv4 potassium channel subunits and potassium channel inter-acting proteins in subpopulations of interneurons in the basolateral amygdala. Neuroscience 2010, 171, 721–733. [Google Scholar] [CrossRef]
- Norris, A.J.; Foeger, N.C.; Nerbonne, J.M. Interdependent Roles for Accessory KChIP2, KChIP3, and KChIP4 Subunits in the Generation of Kv4-Encoded IA Channels in Cortical Pyramidal Neurons. J. Neurosci. 2010, 30, 13644–13655. [Google Scholar] [CrossRef]
- Néant, I.; Haiech, J.; Kilhoffer, M.-C.; Aulestia, F.J.; Moreau, M.; Leclerc, C. Ca2+-Dependent Transcriptional Repressors KCNIP and Regulation of Prognosis Genes in Glioblastoma. Front. Mol. Neurosci. 2018, 11, 472. [Google Scholar] [CrossRef]
- Takamatsu, K.; Noguchi, T. Hippocalcin: A calcium-binding protein of the EF-hand superfamily dominantly expressed in the hippocampus. Neurosci. Res. 1993, 17, 291–295. [Google Scholar] [CrossRef]
- Saitoh, S.; Takamatsu, K.; Kobayashi, M.; Noguchi, T. Distribution of hippocalcin mRNA and immunoreactivity in rat brain. Neurosci. Lett. 1993, 157, 107–110. [Google Scholar] [CrossRef]
- Palmer, C.L.; Lim, W.; Hastie, P.G.; Toward, M.; Korolchuk, V.I.; Burbidge, S.A.; Banting, G.; Collingridge, G.L.; Isaac, J.T.; Henley, J.M. Hippocalcin Functions as a Calcium Sensor in Hippocampal LTD. Neuron 2005, 47, 487–494. [Google Scholar] [CrossRef]
- Tzingounis, A.V.; Kobayashi, M.; Takamatsu, K.; Nicoll, R.A. Hippocalcin Gates the Calcium Activation of the Slow Afterhyperpolarization in Hippocampal Pyramidal Cells. Neuron 2007, 53, 487–493. [Google Scholar] [CrossRef]
- Braunewell, K.-H.; Gundelfinger, E.D. Intracellular neuronal calcium sensor proteins: A family of EF-hand calcium-binding proteins in search of a function. Cell Tissue Res. 1999, 295, 1–12. [Google Scholar] [CrossRef]
- Zhao, C.; Braunewell, K.-H. Expression of the neuronal calcium sensor visinin-like protein-1 in the rat hippocampus. Neuroscience 2008, 153, 1202–1212. [Google Scholar] [CrossRef] [PubMed]
- Braunewell, K.H. The visinin-like proteins VILIP-1 and VILIP-3 in Alzheimer’s disease—Old wine in new bottles. Front. Mol. Neurosci. 2012, 5, 20. [Google Scholar] [CrossRef] [PubMed]
- Liebl, M.P.; Kaya, A.M.; Tenzer, S.; Mittenzwei, R.; Koziollek-Drechsler, I.; Schild, H.; Moosmann, B.; Behl, C.; Clement, A.M. Dimerization of visinin-like protein 1 is regulated by oxidative stress and calcium and is a pathological hallmark of amyotrophic lateral sclerosis. Free. Radic. Biol. Med. 2014, 72, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-W.; Chang, L.-C.; Tseng, G.C.; Kirkwood, C.M.; Sibille, E.L.; Sweet, R.A. VSNL1 Co-Expression Networks in Aging Include Calcium Signaling, Synaptic Plasticity, and Alzheimer’s Disease Pathways. Front. Psychiatry 2015, 6, 30. [Google Scholar] [CrossRef]
- Recabarren-Leiva, D.; Alarcón, M. New insights into the gene expression associated to amyotrophic lateral sclerosis. Life Sci. 2018, 193, 110–123. [Google Scholar] [CrossRef]
- Palecek, J.; Lips, M.B.; Keller, B.U. Calcium dynamics and buffering in motoneurones of the mouse spinal cord. J. Physiol. 1999, 520, 485–502. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).