A R-Script for Generating Multiple Sclerosis Lesion Pattern Discrimination Plots
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Data
2.2. Software
2.3. Basics of the MS-Lesion Pattern Discrimination Plot
3. Results (Developed Code)
3.1. LDPgenerator.r: Program and Data Flow
- Section 1 (code lines 11–12):
- Section 2 (code lines 14–29):
- Section 3 (code lines 32–36): Input and output files are selected; LDPgenerator.r uses the straightforward graphical user interface (GUI) of base R.
- Section 4 (code lines 46–207): In the central processing loop, for each input file, the following operations are performed:
- Section 4a (code lines 51–69): RNifti extracts a voxel array and associated geometry data (number of voxels in x, y, z direction, xyz dimension per voxel).
- Section 4b (code lines 71–76): The extracted voxel array is thresholded, yielding a classified result (1 = MS-lesion, 0 = rest).
- Section 4c (code lines 78–139): Three individual empirical variograms are calculated—one per voxel array x, y, z direction, for lags 1 to Max_lag. Lag values and pair counts are stored.
- Section 4d (code lines 141–175): An exponential variogram model is separately fit to each x, y, z empirical variograms, using the R nls function with starting estimates Guess_a, Guess_C. Derived model parameters ax, Cx, ay, Cy, az, Cz, and mean a, mean c values are stored for x, y, z directions.
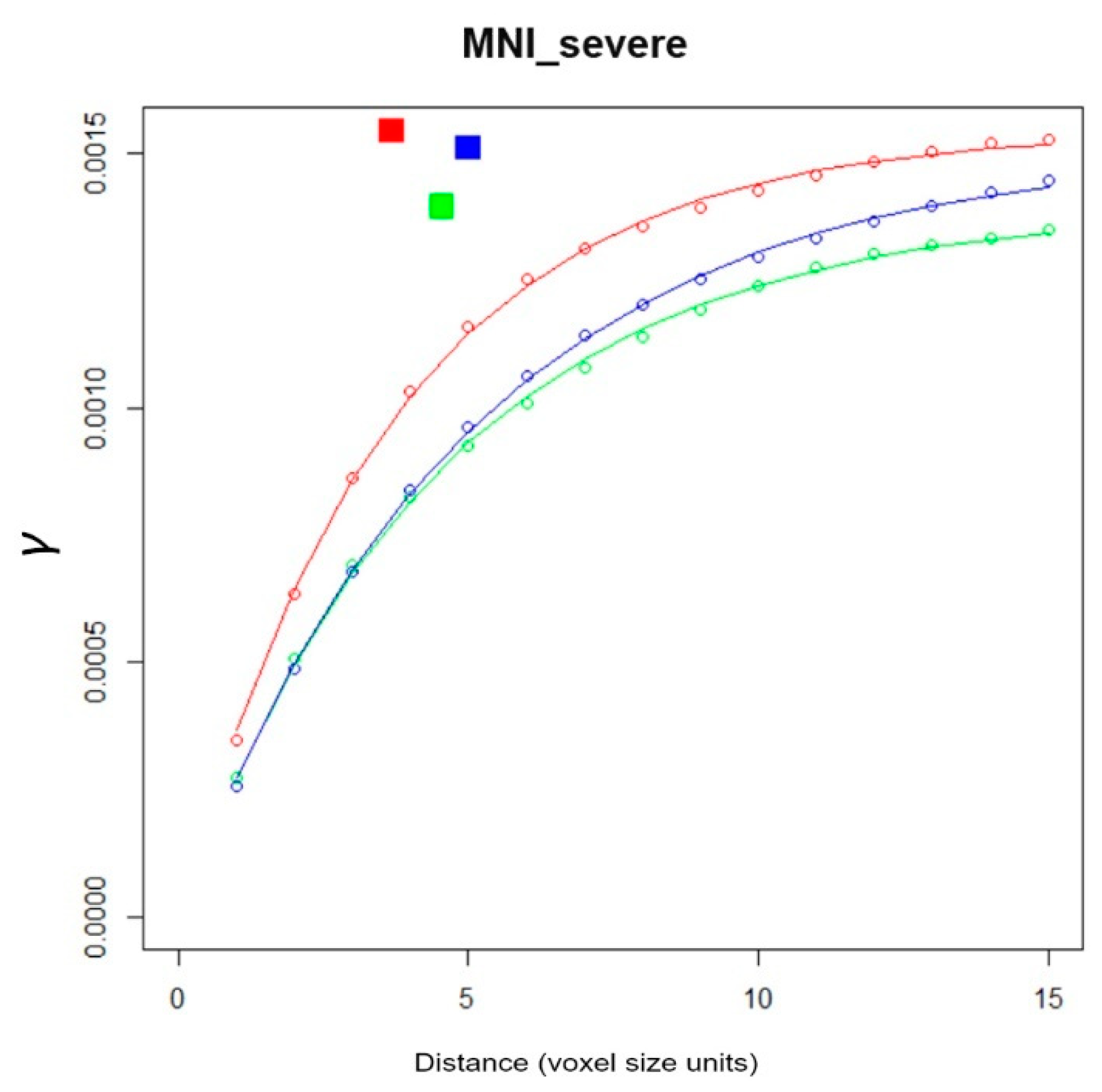
- Section 4e (code lines 177–200): Per input file, empirical variogram graphs and associated exponential variogram model functions for individual x, y, z directions are displayed and stored in png format, with associated filenames.
- Section 4f (code lines 202–207): The index number (1 ... n) of processed MS-WML, the natural logarithm of model parameters mean(ax, ay, az), mean(Cx, Cy, Cz) and directional components ax, Cx, ay, Cy, az, Cz, and the respective input file name are appended to the LDP container file (ASCII). File contents are a good starting point for postprocessing geostatistical data on MS-WML (Table 1). MS-WML index numbers are displayed in MS-LDP and Component MS-LDP graphics to reference input file names while not overloading graphics.
- Section 5 (code lines 209–248): MS-LDP and Component MS-LDP graphics are displayed and exported in png format. LDPgenerator.r terminates.
3.2. A Worked Example in 5 Steps
- Step 1: From standard R, launch LDPgenerator.r (no changes of parameters necessary in the script);
- Step 2: Define input files: Navigate to the relevant directory and select MNI_mild.hdr, MNI_moderate.hdr and MNI_severe.hdr from the file list;
- Step 3: Define the LDP container output file: In the highlighted input box, type MNI.var (Supplementary Materials, see Appendix A for description);
- Step 4: Define the LDP graphics output file: In the highlighted input box, type MNI_LDP.png (Supplementary Materials, see Appendix A for description);
- Step 5: Define the Component LDP graphics output file: In the highlighted input box, type MNI_LDP_xyz.png (Supplementary Materials, see Appendix A for description).
4. Discussion
- Adding a 3D viewer: The recent version of RNifti (1.1) contains a basic viewer for easy implementation of Nifti/Analyze viewing. Providing interactive viewing of MS-WML would facilitate setting the correct binarization threshold values, via enabling visual checking of resulting lesion probability maps.
- Using a GUI package, e.g., https://r4stats.com/articles/software-reviews/r-gui-comparison/.
- Availability of state-of-art GUI elements like customizable buttons, input boxes or spinners would enable easy tuning of variography and graphics parameters.
- Using an improved graphics package—see: https://cran.r-project.org/web/views/Graphics.html. More sophisticated annotation elements like labels, scalable arrows, or improved legend elements would facilitate explorative data analysis of time series portrayed in the MS-LDP, e.g., data from follow-up MRI.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
- R Script
- ◦
- LDPgenerator.R
- Sample data for testing LDPgenerator.R
- ◦
- MNI_mild.hdr
- ◦
- MNI_mild.img
- ◦
- MNI_moderate.hdr
- ◦
- MNI_moderate.img
- ◦
- MNI_severe.hdr
- ◦
- MNI_severe.img
- Associated LDPgenerator.R result files from above sample data
- ◦
- MNI_mild.hdr_variograms.png
- ◦
- MNI_moderate.hdr_variograms.png
- ◦
- MNI_severe.hdr_variograms.png
- ◦
- MNI.var
- ◦
- MNI_LDP_xyz.png
- ◦
- MNI_LDP.png
- LDP with data from Clinical Study [7] (Marschallinger et al., 2018), created by LDPgenerator.r
- ◦
- LDP_Supplement.jpg
References
- Compston, A.; Coles, A. Multiple sclerosis. Lancet 2008, 372, 1502–1517. [Google Scholar] [CrossRef]
- Thompson, A.J.; Banwell, B.L.; Barkhof, F.; Carroll, W.M.; Coetzee, T.; Comi, G.; Correale, J.; Fazekas, F.; Filippi, M.; Freedman, M.S.; et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018, 17, 162–173. [Google Scholar] [CrossRef]
- Filippi, M.; Rocca, M.A. MR Imaging of Multiple Sclerosis. Radiology 2011, 25, 659–681. [Google Scholar] [CrossRef] [PubMed]
- Louapre, C. Conventional and advanced MRI in multiple sclerosis. Rev. Neurol. 2018, 174, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Marschallinger, R.; Golaszewski, S.M.; Kunz, A.B.; Kronbichler, M.; Ladurner, G.; Hofmann, P.; Trinka, E.; McCoy, M.; Kraus, J. Usability and potential of geostatistics for spatial discrimination of multiple sclerosis lesion patterns. J. Neuroimaging 2014, 4, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Marschallinger, R.; Schmidt, P.; Hofmann, P.; Zimmer, C.; Atkinson, P.M.; Sellner, J.; Trinka, E.; Mühlau, M. A MS-lesion pattern discrimination plot based on geostatistics. Brain Behav. 2016, 6, e00430. [Google Scholar] [CrossRef] [PubMed]
- Marschallinger, R.; Mühlau, M.; Pongratz, V.; Kirschke, J.S.; Marschallinger, S.; Schmidt, P.; Sellner, J. Geostatistical Analysis of White Matter Lesions in Multiple Sclerosis Identifies Gender Differences in Lesion Evolution. Front. Mol. Neurosci. 2018, 11, 460. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017; Available online: https://www.R-project.org/ (accessed on 7 January 2021).
- Rorden, C.; Brett, M. Stereotaxic display of brain lesions. Behav. Neurol. 2000, 12, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, P.; Gaser, C.; Arsic, M.; Buck, D.; Forschler, A. An automated tool for detection of FLAIR-hyperintense white-matter lesions in multiple sclerosis. NeuroImage 2012, 59, 3774–3783. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Lau, J.C.; Anderson, T.; De Kraker, J.; Collins, D.L.; Peters, T.; Khan, A.R. An accurate registration of the BigBrain dataset with the MNI PD25 and ICBM152 atlases. Sci. Data 2019, 6, 210. [Google Scholar] [CrossRef] [PubMed]
- Frackowiak, R.S.J.; Friston, K.J.; Frith, C.D.; Dolan, R.J.; Mazziotta, J.C. Human Brain Function; Academic Press: New York, NY, USA, 1997. [Google Scholar]
- Woolrich, M.W.; Jbabdi, S.; Patenaude, B.; Chappell, M.; Makni, S.; Behrens, T.; Beckmann, C.; Jenkinson, M.; Smith, S.M. Bayesian analysis of neuroimaging data in FSL. NeuroImage 2009, 45, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Wackernagel, H. Multivariate Geostatistics. An Introduction with Applications; Springer: Berlin/Heidelberg, Germany, 1995; 256p. [Google Scholar]
- Lantuejoul, C. Geostatistical Simulation. Models and Algorithms; Springer: Berlin/Heidelberg, Germany, 2002; 256p. [Google Scholar]
- Christakos, G. Modern Spatiotemporal Geostatistics; Oxford University Press: Oxford, UK, 2000; 288p. [Google Scholar]
- Krige, D.G. A statistical approach to some basic mine valuation problems on the Witwatersrand. J. Chem. Metal. Min. Soc. S. Afr. 1951, 52, 119–139. [Google Scholar]
- Blewett, M.; Kildluff, M. Geostatistical Analysis of Possible Spirochetal Involvement in Multiple Sclerosis (abstract). In Proceedings of the ESRI Users Group Conference, San Diego, CA, USA, 7–11 August 2006. [Google Scholar]
- Conan, V.; Gesbert, S.; Howard, C.V.; Jeulin, D.; Meyer, F.; Renard, D. Geostatistical and morphological methods applied to three-dimensional microscopy. J. Microsc. 1992, 166, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Marschallinger, R.; Höck, V.; Topa, D. A Method for the 3-D Reconstruction of Chemically Zoned Minerals from Microprobe Scanned Serial Sections. Microsc. Anal. 1993, 5, 5–7. [Google Scholar]
- Kourgli, A.; Belhadj-aissa, A.; Bouchemakh, A. Optimizing texture primitives description based on variography and mathematical morphology. In Image Analysis and Recognition; Campilho, A., Kamel, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2004. [Google Scholar]
- Trevisani, S.; Cavalli, M.; Marchi, L. Variogram maps from LiDAR data as fingerprints of surface morphology on scree slopes. Nat. Hazards Earth Syst. Sci. 2009, 9, 129–133. [Google Scholar] [CrossRef]
- Mariethoz, G.; Caers, J. Multiple-Point Geostatistics. Stochastic Modeling with Training Images; Wiley-Blackwell: Oxford, UK, 2015; 364p. [Google Scholar]
- Deutsch, C.; Journel, A.G. Geostatistical Software Library and User’s Guide; Oxford Univ. Press: Oxford, UK, 1992; 340p. [Google Scholar]
- Gringarten, E.; Deutsch, C.V. Methodology for variogram interpretation and modeling for improved reservoir characterization. In Proceedings of the SPE Annual Technical Conference and Exhibition, Houston, TX, USA, 3–6 October 1999; pp. 1–13. [Google Scholar] [CrossRef]
- Cressie, N.A.C. Statistics for Spatial Data; Wiley: Hoboken, NJ, USA, 1993. [Google Scholar]

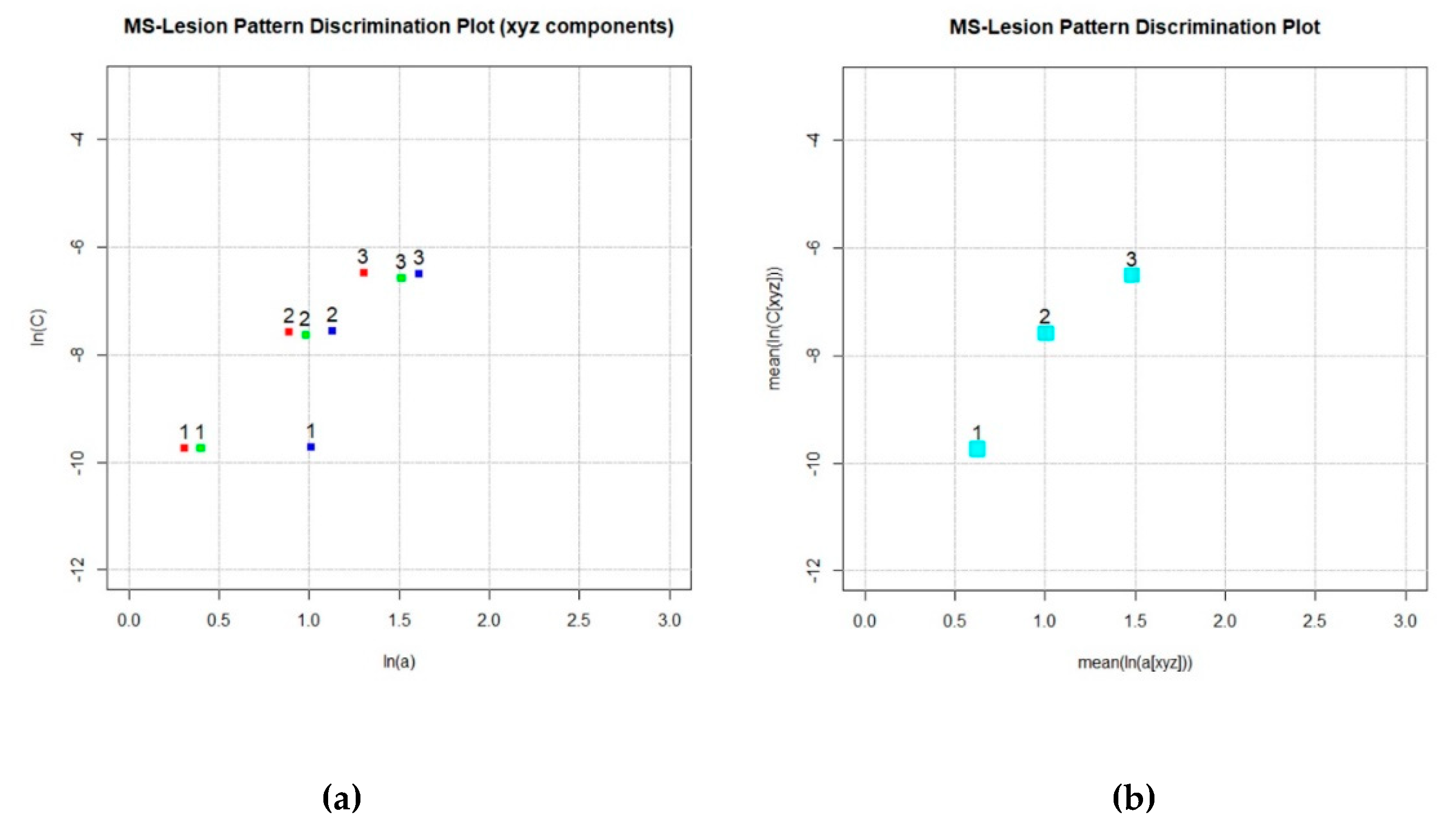

| ID | ln(avg(a[xyz])) | ln(avg(C[xyz])) | ln(aX) | ln(CX) | ln(aY) | ln(CY) | ln(az) | ln(CZ) | File |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.62610 | −9.74182 | 0.30975 | −9.74533 | 0.40066 | −9.75081 | 1.01342 | −9.72945 | MNI_mild.img |
| 2 | 1.00625 | −7.59234 | 0.89172 | −7.58171 | 0.98117 | −7.63554 | 1.13109 | −7.56123 | MNI_moderate.img |
| 3 | 1.48463 | −6.51406 | 1.30517 | −6.47390 | 1.51387 | −6.57510 | 1.61090 | −6.49598 | MNI_severe.img |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marschallinger, R.; Tur, C.; Marschallinger, H.; Sellner, J. A R-Script for Generating Multiple Sclerosis Lesion Pattern Discrimination Plots. Brain Sci. 2021, 11, 90. https://doi.org/10.3390/brainsci11010090
Marschallinger R, Tur C, Marschallinger H, Sellner J. A R-Script for Generating Multiple Sclerosis Lesion Pattern Discrimination Plots. Brain Sciences. 2021; 11(1):90. https://doi.org/10.3390/brainsci11010090
Chicago/Turabian StyleMarschallinger, Robert, Carmen Tur, Hannes Marschallinger, and Johann Sellner. 2021. "A R-Script for Generating Multiple Sclerosis Lesion Pattern Discrimination Plots" Brain Sciences 11, no. 1: 90. https://doi.org/10.3390/brainsci11010090
APA StyleMarschallinger, R., Tur, C., Marschallinger, H., & Sellner, J. (2021). A R-Script for Generating Multiple Sclerosis Lesion Pattern Discrimination Plots. Brain Sciences, 11(1), 90. https://doi.org/10.3390/brainsci11010090





