Neuronal Localization of SENP Proteins with Super Resolution Microscopy
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Primary Cultures
2.3. Immunofluorescence Experiments
2.4. Confocal and Structured Illumination Microscopy
2.5. Statistical Analysis
3. Results
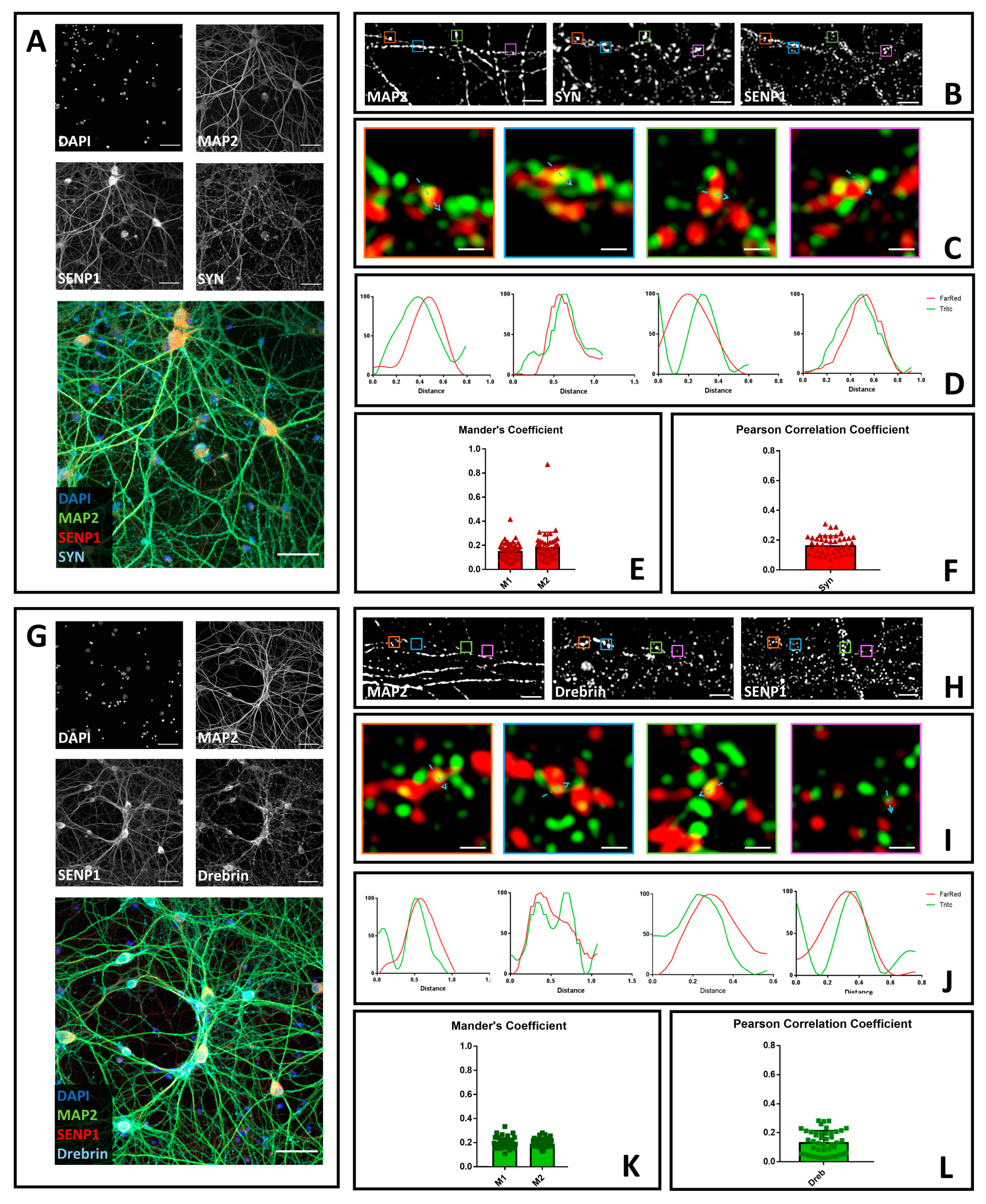
3.1. SENP1 Partially Colocalizes with the Presynaptic Marker Synaptophysin and with the Postsynaptic Marker Drebrin
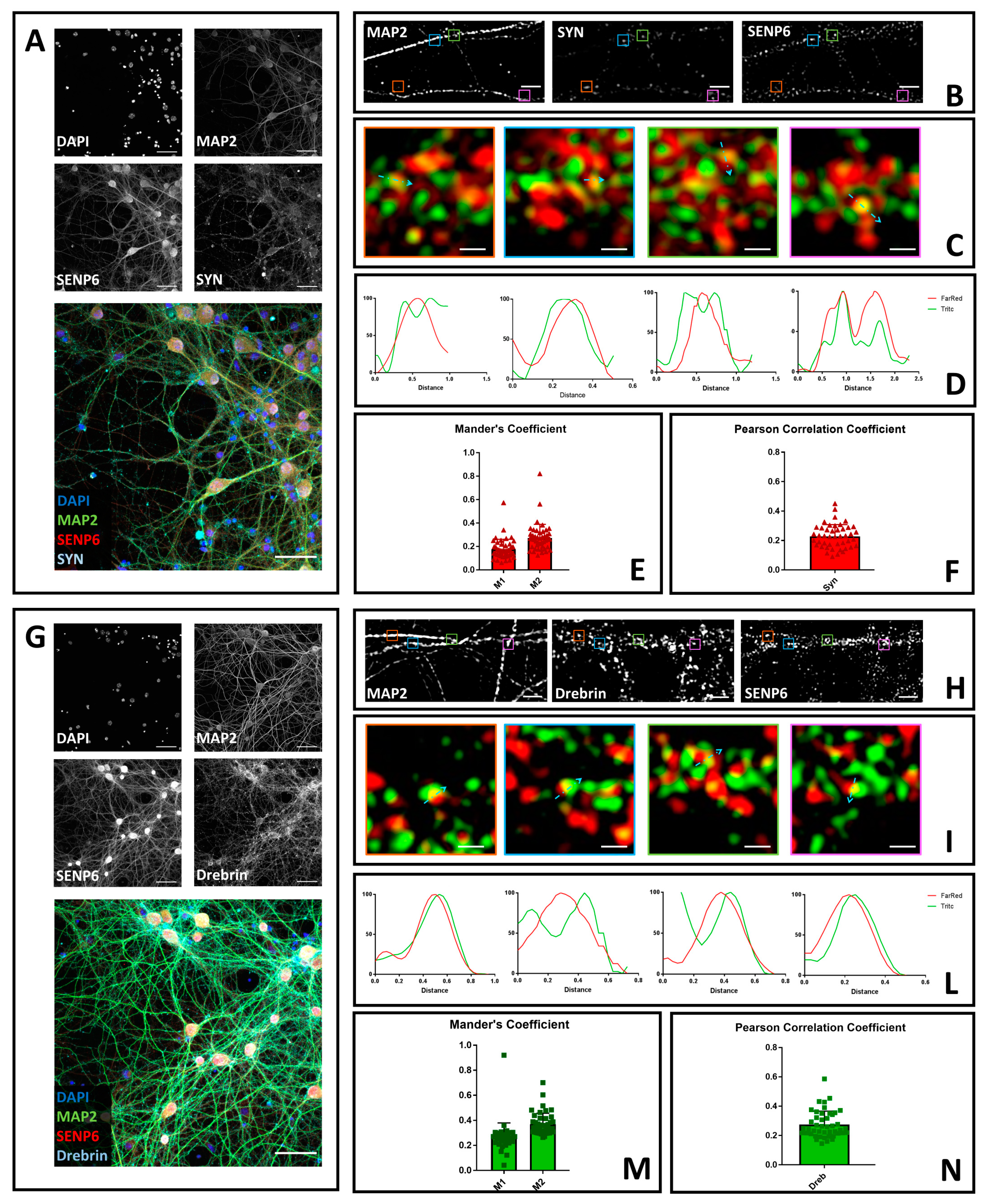
3.2. SENP6 Partially Colocalizes with the Presynaptic Marker Synaptophysin and with the Postsynaptic Marker Drebrin
3.3. SENP7 Partially Colocalizes with the Presynaptic Marker Synaptophysin and with the Postsynaptic Marker Drebrin
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Citri, A.; Malenka, R.C. Synaptic Plasticity: Multiple Forms, Functions, and Mechanisms. Neuropsychopharmacology 2008, 33, 18–41. [Google Scholar] [CrossRef]
- Rosenberg, T.; Gal-Ben-Ari, S.; Dieterich, D.C.; Kreutz, M.R.; Ziv, N.E.; Gundelfinger, E.D.; Rosenblum, K. The roles of protein expression in synaptic plasticity and memory consolidation. Front. Mol. Neurosci. 2014, 7. [Google Scholar] [CrossRef]
- Sunyer, B.; Diao, W.; Lubec, G. The role of post-translational modifications for learning and memory formation. Electrophoresis 2008, 29, 2593–2602. [Google Scholar] [CrossRef]
- Routtenberg, A. Anatomical localization of phosphoprotein and glycoprotein substrates of memory. Prog. Neurobiol. 1979, 12, 85–113. [Google Scholar] [CrossRef]
- Routtenberg, A.; Rekart, J.L. Post-translational protein modification as the substrate for long-lasting memory. Trends Neurosci. 2005, 28, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Sweatt, J.D. Memory mechanisms: The yin and yang of protein phosphorylation. Curr. Biol. 2001, 11, R391–R394. [Google Scholar] [CrossRef][Green Version]
- Gareau, J.R.; Lima, C.D. The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 2010, 11, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.; Wilkinson, K.A.; Nishimune, A.; Henley, J.M. Emerging extranuclear roles of protein SUMOylation in neuronal function and dysfunction. Nat. Rev. Neurosci. 2007, 8, 948–959. [Google Scholar] [CrossRef] [PubMed]
- Schorova, L.; Martin, S. Sumoylation in Synaptic Function and Dysfunction. Front. Synaptic. Neurosci. 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Henley, J.M.; Carmichael, R.E.; Wilkinson, K.A. Extranuclear SUMOylation in Neurons. Trends Neurosci. 2018, 41, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Cappadocia, L.; Lima, C.D. Ubiquitin-like Protein Conjugation: Structures, Chemistry, and Mechanism. Chem. Rev. 2018, 118, 889–918. [Google Scholar] [CrossRef] [PubMed]
- Hay, R.T. SUMO: A History of Modification. Mol. Cell 2005, 18, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, D.; Dasso, M. Modification in reverse: The SUMO proteases. Trends Biochem. Sci. 2007, 32, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Nayak, A.; Müller, S. SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol. 2014, 15, 422. [Google Scholar] [CrossRef]
- Hickey, C.M.; Wilson, N.R.; Hochstrasser, M. Function and Regulation of SUMO Proteases. Nat. Rev. Mol. Cell Biol. 2012, 13, 755–766. [Google Scholar] [CrossRef]
- Kunz, K.; Piller, T.; Müller, S. SUMO-specific proteases and isopeptidases of the SENP family at a glance. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef]
- Kolli, N.; Mikolajczyk, J.; Drag, M.; Mukhopadhyay, D.; Moffatt, N.; Dasso, M.; Salvesen, G.; Wilkinson, K.D. Distribution and paralogue specificity of mammalian deSUMOylating enzymes. Biochem. J. 2010, 430, 335–344. [Google Scholar] [CrossRef]
- Mikolajczyk, J.; Drag, M.; Békés, M.; Cao, J.T.; Ronai, Z.; Salvesen, G.S. Small ubiquitin-related modifier (SUMO)-specific proteases: Profiling the specificities and activities of human SENPs. J. Biol. Chem. 2007, 282, 26217–26224. [Google Scholar] [CrossRef]
- Békés, M.; Prudden, J.; Srikumar, T.; Raught, B.; Boddy, M.N.; Salvesen, G.S. The dynamics and mechanism of SUMO chain deconjugation by SUMO-specific proteases. J. Biol. Chem. 2011, 286, 10238–10247. [Google Scholar] [CrossRef]
- Bylebyl, G.R.; Belichenko, I.; Johnson, E.S. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J. Biol. Chem. 2003, 278, 44113–44120. [Google Scholar] [CrossRef]
- Lima, C.D.; Reverter, D. Structure of the Human SENP7 Catalytic Domain and Poly-SUMO Deconjugation Activities for SENP6 and SENP7. J. Biol. Chem. 2008, 283, 32045–32055. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.N.; Geoffroy, M.C.; Jaffray, E.G.; Hay, R.T. Characterization of SENP7, a SUMO-2/-3 specific isopeptidase. Biochem. Biochem. J. 2009, 421, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Shin, E.J.; Shin, H.M.; Nam, E.; Kim, W.S.; Kim, J.-H.; Oh, B.-H.; Yun, Y. DeSUMOylating isopeptidase: A second class of SUMO protease. EMBO Rep. 2012, 13, 339–346. [Google Scholar] [CrossRef]
- Schulz, S.; Chachami, G.; Kozaczkiewicz, L.; Winter, U.; Stankovic-Valentin, N.; Haas, P.; Hofmann, K.; Urlaub, H.; Ovaa, H.; Wittbrodt, J.; et al. Ubiquitin-specific protease-like 1 (USPL1) is a SUMO isopeptidase with essential, non-catalytic functions. EMBO Rep. 2012, 13, 930–938. [Google Scholar] [CrossRef] [PubMed]
- Henley, J.M.; Seager, R.; Nakamura, Y.; Talandyte, K.; Nair, J.; Wilkinson, K.A. SUMOylation of synaptic and synapse-associated proteins: An update. J. Neurochem. 2020. [Google Scholar] [CrossRef]
- Henley, J.M.; Craig, T.J.; Wilkinson, K.A. Neuronal SUMOylation: Mechanisms, physiology, and roles in neuronal dysfunction. Physiol. Rev. 2014, 94, 1249–1285. [Google Scholar] [CrossRef]
- Feligioni, M.; Nishimune, A.; Henley, J.M. Protein SUMOylation modulates calcium influx and glutamate release from presynaptic terminals. Eur. J. Neurosci. 2009, 29, 1348–1356. [Google Scholar] [CrossRef]
- Martin, S.; Nishimune, A.; Mellor, J.R.; Henley, J.M. SUMOylation regulates kainate-receptor-mediated synaptic transmission. Nature 2007, 447, 321–325. [Google Scholar] [CrossRef]
- Tang, L.T.H.; Craig, T.J.; Henley, J.M. SUMOylation of synapsin Ia maintains synaptic vesicle availability and is reduced in an autism mutation. Nat. Commun. 2015, 6, 7728. [Google Scholar] [CrossRef]
- Choi, J.H.; Park, J.Y.; Park, S.P.; Lee, H.; Han, S.; Park, K.H.; Suh, Y.H. Regulation of mGluR7 trafficking by SUMOylation in neurons. Neuropharmacology 2016, 102, 229–235. [Google Scholar] [CrossRef]
- Parker, A.R.; Forster, L.A.; Baro, D.J. Modulator-Gated, SUMOylation-Mediated, Activity-Dependent Regulation of Ionic Current Densities Contributes to Short-Term Activity Homeostasis. J. Neurosci. 2019, 39, 596–611. [Google Scholar] [CrossRef] [PubMed]
- Daniel, J.A.; Cooper, B.H.; Palvimo, J.J.; Zhang, F.P.; Brose, N.; Tirard, M. Response: Commentary: Analysis of SUMO1-conjugation at synapses. Front. Cell. Neurosci. 2018, 12. [Google Scholar] [CrossRef]
- Daniel, J.A.; Cooper, B.H.; Palvimo, J.J.; Zhang, F.P.; Brose, N.; Tirard, M. Analysis of SUMO1-conjugation at synapses. eLife 2017, 6, e26338. [Google Scholar] [CrossRef] [PubMed]
- Tirard, M.; Hsiao, H.H.; Nikolov, M.; Urlaub, H.; Melchior, F.; Brose, N. In vivo localization and identification of SUMOylated proteins in the brain of His6-HA-SUMO1 knock-in mice. Proc. Natl. Acad. Sci. USA 2012, 109, 21122–21127. [Google Scholar] [CrossRef]
- Wilkinson, K.A.; Martin, S.; Tyagarajan, S.K.; Arancio, O.; Craig, T.J.; Guo, C.; Fraser, P.E.; Goldstein, S.A.N.; Henley, J.M. Commentary: Analysis of SUMO1-conjugation at synapses. Front. Cell. Neurosci. 2017, 11. [Google Scholar] [CrossRef]
- Colnaghi, L.; Russo, L.; Natale, C.; Restelli, E.; Cagnotto, A.; Salmona, M.; Chiesa, R.; Fioriti, L. Super Resolution Microscopy of SUMO Proteins in Neurons. Front. Cell. Neurosci. 2019, 13. [Google Scholar] [CrossRef]
- Loriol, C.; Khayachi, A.; Poupon, G.; Gwizdek, C.; Martin, S. Activity-dependent regulation of the sumoylation machinery in rat hippocampal neurons. Biol. Cell 2013, 105, 30–45. [Google Scholar] [CrossRef]
- Loriol, C.; Parisot, J.; Poupon, G.; Gwizdek, C.; Martin, S. Developmental Regulation and Spatiotemporal Redistribution of the Sumoylation Machinery in the Rat Central Nervous System. PLoS ONE 2012, 7, e33757. [Google Scholar] [CrossRef] [PubMed]
- Schorova, L.; Pronot, M.; Poupon, G.; Prieto, M.; Folci, A.; Khayachi, A.; Brau, F.; Cassé, F.; Gwizdek, C.; Martin, S. The synaptic balance between sumoylation and desumoylation is maintained by the activation of metabotropic mGlu5 receptors. Cell. Mol. Life Sci. 2019, 76, 3019–3031. [Google Scholar] [CrossRef]
- Yang, C.Y.; Yu, T.H.; Wen, W.L.; Ling, P.; Hsu, K.S. Conditional Deletion of CC2D1A Reduces Hippocampal Synaptic Plasticity and Impairs Cognitive Function through Rac1 Hyperactivation. J. Neurosci. 2019, 39, 4959–4975. [Google Scholar] [CrossRef] [PubMed]
- Cordelieres, F.P.; Bolte, S. JACoP v2.0: Improving the User Experience with Co-Localization Studies; Institut Curie: Orsay, France, 2008; p. 8. [Google Scholar]
- Adler, J.; Parmryd, I. Quantifying colocalization by correlation: The Pearson correlation coefficient is superior to the Mander’s overlap coefficient. Cytom. Part A 2010, 77, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Wiedenmann, B.; Franke, W.W. Identification and localization of synaptophysin, an integral membrane glycoprotein of Mr 38,000 characteristic of presynaptic vesicles. Cell 1985, 41, 1017–1028. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Y.; Zhu, A.; Huang, D.; Deng, S.; Cheng, J.; Zhu, M.X.; Li, Y. SUMO-specific protease 1 protects neurons from apoptotic death during transient brain ischemia/reperfusion. Cell Death Dis. 2016, 7, e2484. [Google Scholar] [CrossRef]
- Gustafsson, M.G.L. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 2000, 198, 82–87. [Google Scholar] [CrossRef]
- Gustafsson, M.G.L.; Shao, L.; Carlton, P.M.; Wang, C.J.R.; Golubovskaya, I.N.; Cande, W.Z.; Agard, D.A.; Sedat, J.W. Three-Dimensional Resolution Doubling in Wide-Field Fluorescence Microscopy by Structured Illumination. Biophys. J. 2008, 94, 4957–4970. [Google Scholar] [CrossRef]
- Huang, B.; Bates, M.; Zhuang, X. Super resolution fluorescence microscopy. Annu. Rev. Biochem. 2009, 78, 993–1016. [Google Scholar] [CrossRef]
- Igarashi, M.; Nozumi, M.; Wu, L.-G.; Cella Zanacchi, F.; Katona, I.; Barna, L.; Xu, P.; Zhang, M.; Xue, F.; Boyden, E. New observations in neuroscience using superresolution microscopy. J. Neurosci. 2018, 38, 9459–9467. [Google Scholar] [CrossRef]
- Papa, M.; Bundman, M.C.; Greenberger, V.; Segal, M. Morphological analysis of dendritic spine development in primary cultures of hippocampal neurons. J. Neurosci. Off. J. Soc. Neurosci. 1995, 15, 1–11. [Google Scholar] [CrossRef]
- Schikorski, T.; Stevens, C.F. Quantitative Ultrastructural Analysis of Hippocampal Excitatory Synapses. J. Neurosci. 1997, 17, 5858–5867. [Google Scholar] [CrossRef]
- Dunn, K.W.; Kamocka, M.M.; McDonald, J.H. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 2011, 300, C723–C742. [Google Scholar] [CrossRef]
- Aoki, C.; Sekino, Y.; Hanamura, K.; Fujisawa, S.; Mahadomrongkul, V.; Ren, Y.; Shirao, T. Drebrin A is a postsynaptic protein that localizes in vivo to the submembranous surface of dendritic sites forming excitatory synapses. J. Comp. Neurol. 2005, 483, 383–402. [Google Scholar] [CrossRef] [PubMed]
- Vertegaal, A.C.O. SUMO chains: Polymeric signals. Biochem. Soc. Trans. 2010, 38, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Fioravante, D.; Byrne, J.H. Protein degradation and memory formation. Brain Res. Bull. 2011, 85, 14–20. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colnaghi, L.; Conz, A.; Russo, L.; Musi, C.A.; Fioriti, L.; Borsello, T.; Salmona, M. Neuronal Localization of SENP Proteins with Super Resolution Microscopy. Brain Sci. 2020, 10, 778. https://doi.org/10.3390/brainsci10110778
Colnaghi L, Conz A, Russo L, Musi CA, Fioriti L, Borsello T, Salmona M. Neuronal Localization of SENP Proteins with Super Resolution Microscopy. Brain Sciences. 2020; 10(11):778. https://doi.org/10.3390/brainsci10110778
Chicago/Turabian StyleColnaghi, Luca, Andrea Conz, Luca Russo, Clara A. Musi, Luana Fioriti, Tiziana Borsello, and Mario Salmona. 2020. "Neuronal Localization of SENP Proteins with Super Resolution Microscopy" Brain Sciences 10, no. 11: 778. https://doi.org/10.3390/brainsci10110778
APA StyleColnaghi, L., Conz, A., Russo, L., Musi, C. A., Fioriti, L., Borsello, T., & Salmona, M. (2020). Neuronal Localization of SENP Proteins with Super Resolution Microscopy. Brain Sciences, 10(11), 778. https://doi.org/10.3390/brainsci10110778




