New Parameters for Seedling Vigor Developed via Phenomics
Abstract
:1. Introduction
2. Materials and Methods
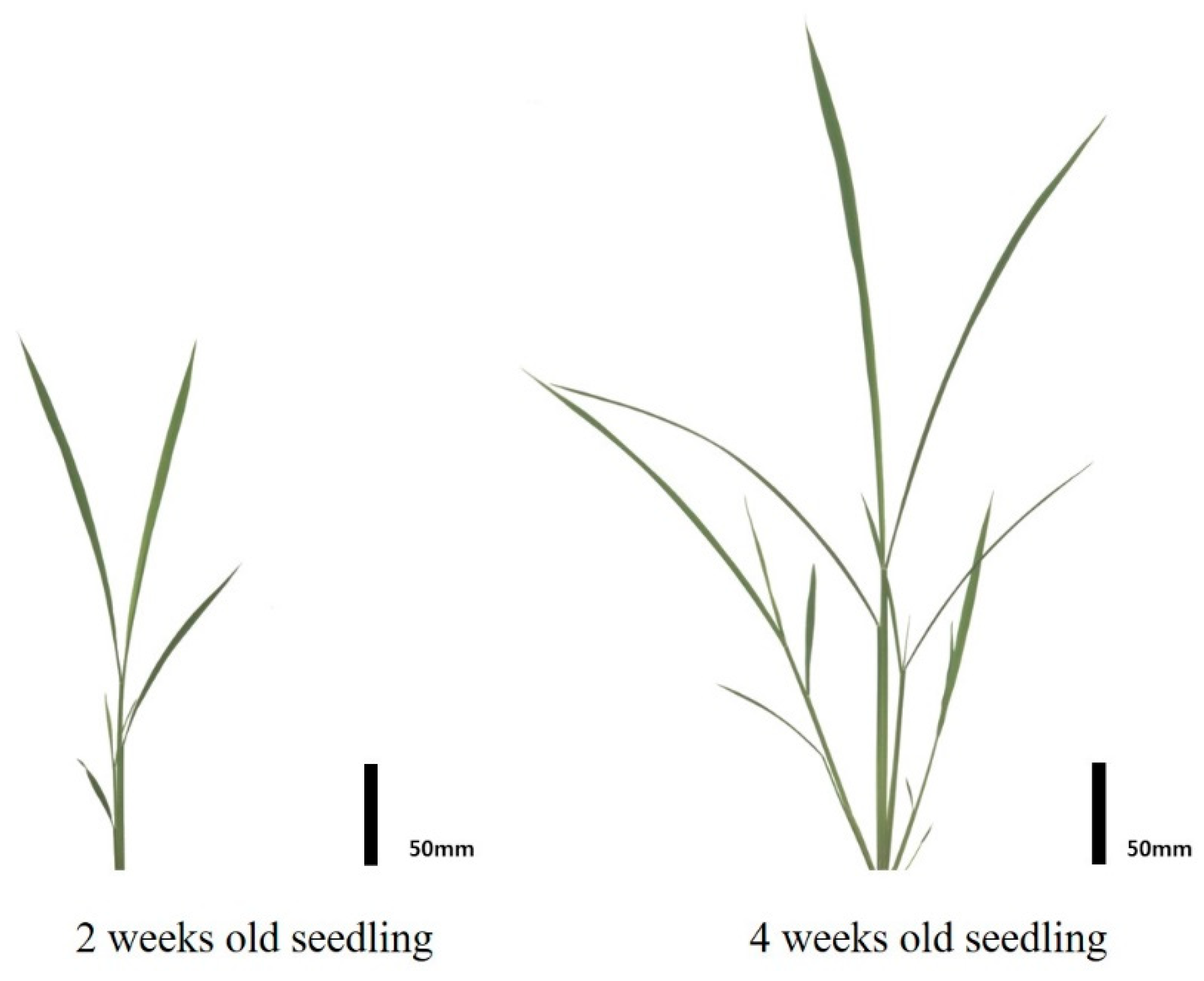
2.1. Plant Materials and Growth Condition
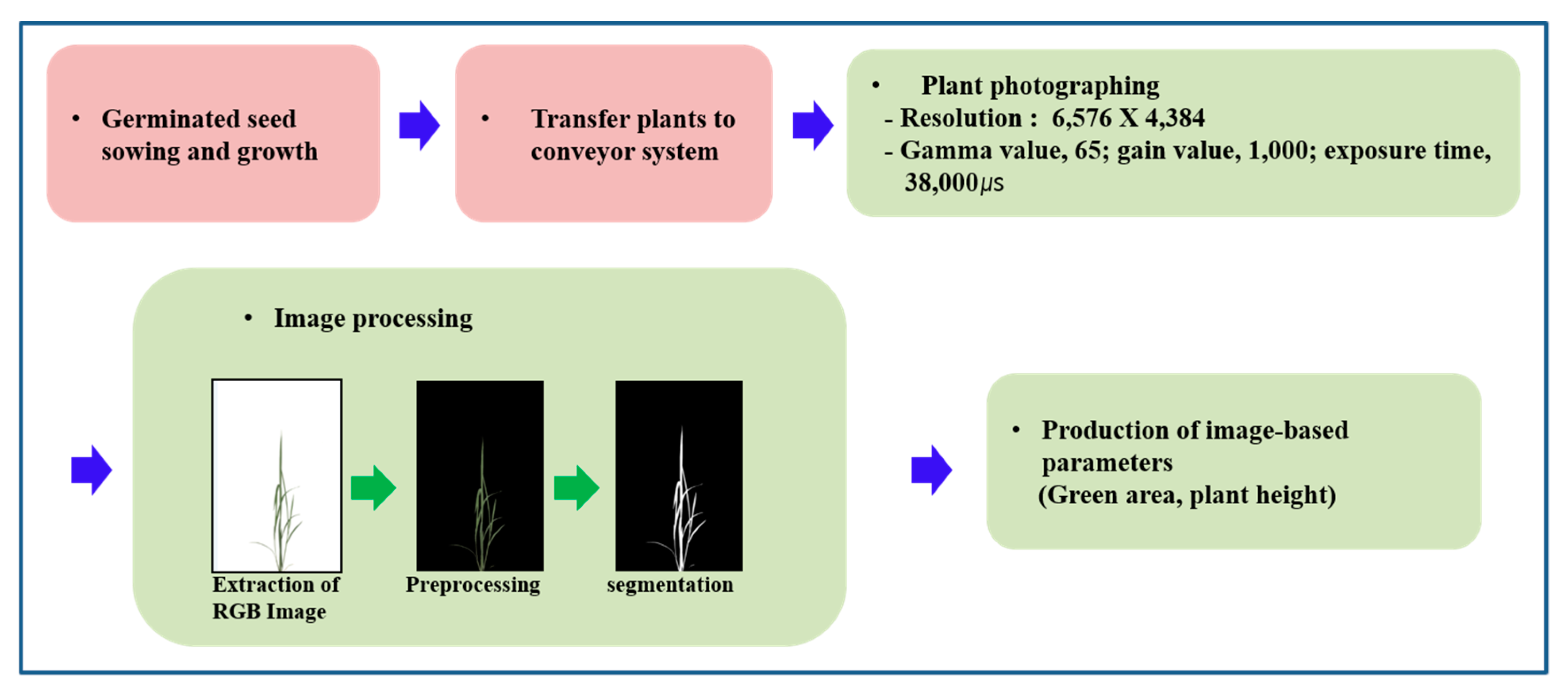
2.2. Image Acquisition
2.3. Algorithm Application in Image Analysis
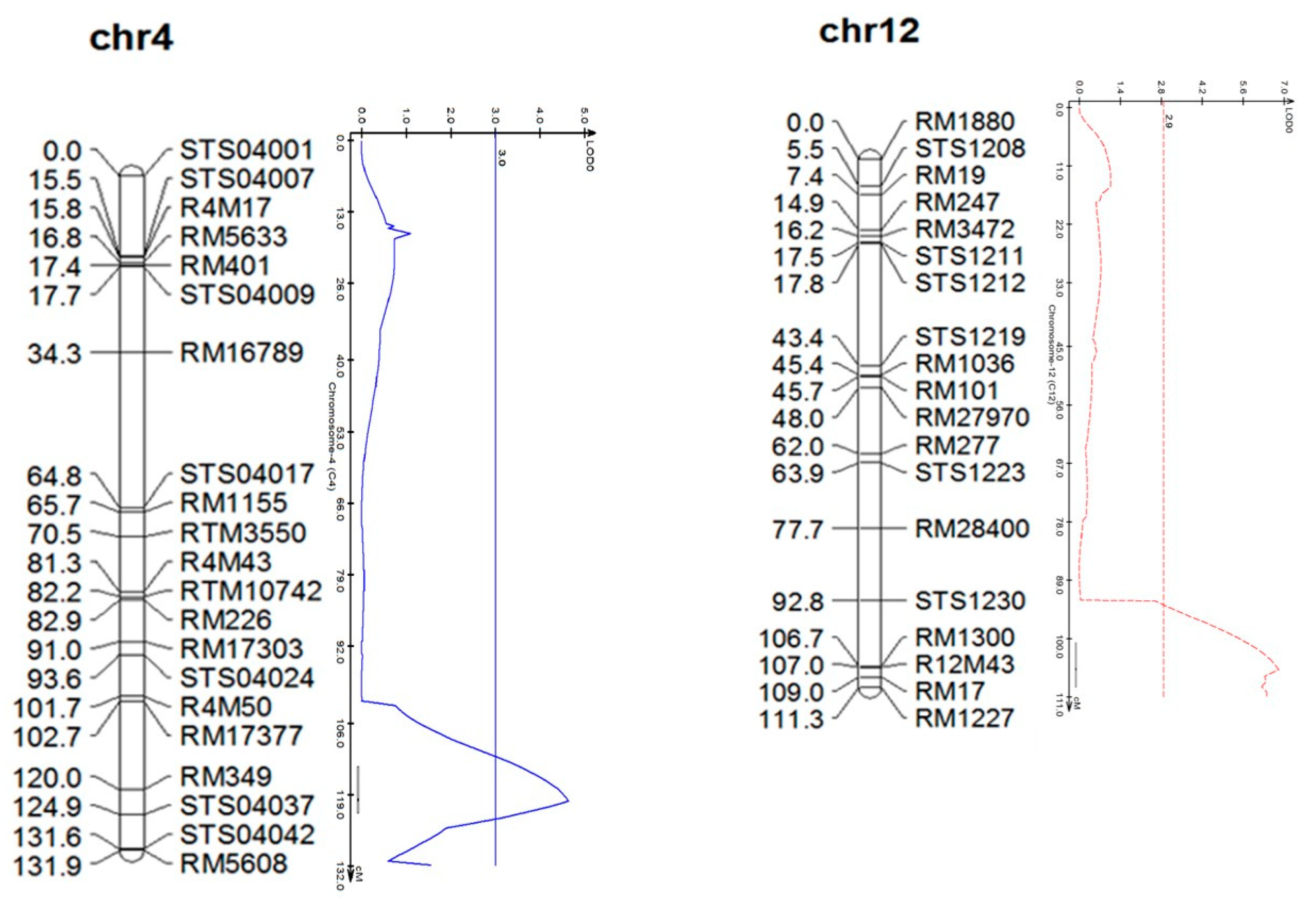
2.4. QTL Analysis for Calculating the Growth Rate
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mishra, A.; Salokhe, V.M. Rice root growth and physiological responses to sri water management and implications for crop productivity. Paddy Water Environ. 2011, 9, 41–52. [Google Scholar] [CrossRef]
- Akram, M.; Ajmal, S.U.; Munir, M. Inheritance of traits related to seedling vigor and grain yield in rice (Oryza sativa L.). Pak. J. Bot. 2007, 39, 37–45. [Google Scholar]
- Kumar, A.; Verulkar, S.; Dixit, S.; Chauhan, B.; Bernier, J.; Venuprasad, R.; Zhao, D.; Shrivastava, M. Yield and yield-attributing traits of rice (Oryza sativa L.) under lowland drought and suitability of early vigor as a selection criterion. Field Crops Res. 2009, 114, 99–107. [Google Scholar] [CrossRef]
- Biswas, J.C.; Ladha, J.K.; Dazzo, F.B.; Yanni, Y.G.; Rolfe, B.G. Rhizobial inoculation influences seedling vigor and yield of rice. Agron. J. 2000, 92, 880–886. [Google Scholar] [CrossRef]
- Cui, K.; Peng, S.; Xing, Y.; Xu, C.; Yu, S.; Zhang, Q. Molecular dissection of seedling-vigor and associated physiological traits in rice. Theor. Appl. Genet. 2002, 105, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, A.; Terao, T. Qtls for shoot length and chlorophyll content of rice seedlings grown under low-temperature conditions, using a cross between indica and japonica cultivars. Plant Prod. Sci. 2015, 18, 128–136. [Google Scholar] [CrossRef]
- Lu, X.-L.; Niu, A.-L.; Cai, H.-Y.; Zhao, Y.; Liu, J.-W.; Zhu, Y.-G.; Zhang, Z.-H. Genetic dissection of seedling and early vigor in a recombinant inbred line population of rice. Plant Sci. 2007, 172, 212–220. [Google Scholar] [CrossRef]
- Redona, E.; Mackill, D. Genetic variation for seedling vigor traits in rice. Crop Sci. 1996, 36, 285–290. [Google Scholar] [CrossRef]
- Redona, E.; Mackill, D. Mapping quantitative trait loci for seedling vigor in rice using rflps. Theor. Appl. Genet. 1996, 92, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Yano, K.; Takashi, T.; Nagamatsu, S.; Kojima, M.; Sakakibara, H.; Kitano, H.; Matsuoka, M.; Aya, K. Efficacy of microarray profiling data combined with qtl mapping for the identification of a qtl gene controlling the initial growth rate in rice. Plant Cell Physiol. 2012, 53, 729–739. [Google Scholar] [CrossRef]
- Zhang, Z.-H.; Qu, X.-S.; Wan, S.; Chen, L.-H.; Zhu, Y.-G. Comparison of qtl controlling seedling vigour under different temperature conditions using recombinant inbred lines in rice (Oryza sativa). Ann. Bot. 2004, 95, 423–429. [Google Scholar] [CrossRef]
- Zhang, Z.-H.; Yu, S.-B.; Yu, T.; Huang, Z.; Zhu, Y.-G. Mapping quantitative trait loci (qtls) for seedling-vigor using recombinant inbred lines of rice (Oryza sativa L.). Field Crops Res. 2005, 91, 161–170. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, J.-K.; Yi, Q.; Wang, Y.-Z.; Zhu, Y.-G.; Zhang, Z.-H. Quantitative trait loci for seedling vigor in rice under field conditions. Field Crops Res. 2007, 100, 294–301. [Google Scholar] [CrossRef]
- Richards, R.A.; Rebetzke, G.J.; Watt, M.; Condon, A.T.; Spielmeyer, W.; Dolferus, R. Breeding for improved water productivity in temperate cereals: Phenotyping, quantitative trait loci, markers and the selection environment. Funct. Plant Biol. 2010, 37, 85–97. [Google Scholar] [CrossRef]
- Furbank, R.T.; Tester, M. Phenomics–technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 2011, 16, 635–644. [Google Scholar] [CrossRef]
- Yang, W.; Guo, Z.; Huang, C.; Duan, L.; Chen, G.; Jiang, N.; Fang, W.; Feng, H.; Xie, W.; Lian, X. Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat. Commun. 2014, 5, 5087. [Google Scholar] [CrossRef] [Green Version]
- Tanger, P.; Klassen, S.; Mojica, J.P.; Lovell, J.T.; Moyers, B.T.; Baraoidan, M.; Naredo, M.E.B.; McNally, K.L.; Poland, J.; Bush, D.R. Field-based high throughput phenotyping rapidly identifies genomic regions controlling yield components in rice. Sci. Rep. 2017, 7, 42839. [Google Scholar] [CrossRef] [Green Version]
- Hittalmani, S.; Huang, N.; Courtois, B.; Venuprasad, R.; Shashidhar, H.; Zhuang, J.; Zheng, K.; Liu, G.; Wang, G.; Sidhu, J. Identification of qtl for growth-and grain yield-related traits in rice across nine locations of asia. Theor. Appl. Genet. 2003, 107, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Kim, H.; Lee, G.-S.; Yoon, U.-H.; Kim, T.-H.; Seol, Y.-J.; Yun, D.-W.; Koh, H.-J.; Eun, M.-Y. Development of rice molecular genetic and physical map using pcr-based DNA markers with the recombinant inbred population derived from milyang23/gihobyeo cross. Korean J. Breed. Sci. 2012, 44. [Google Scholar]
- Lee, J.S. Digital image smoothing and the sigma filter. Comput. Vis. Graph. Image Proc. 1983, 24, 255–269. [Google Scholar] [CrossRef]
- Chen, L.; Hui, L.; Shutao, L. Depth Image Enhancement for Kinect Using Region Growing and Bilateral Filter. In Proceedings of the 21st International Conference on Pattern Recognition (ICPR2012), Tsukuba, Japan, 11–15 November 2012; IEEE: Piscataway, NJ, USA, 2012. [Google Scholar]
- Haralick, R.M.; Stanley, R.S.; Xinhua, Z. Image analysis using mathematical morphology. IEEE Trans. Pattern Anal. Mach. Intell. 1987, 9, 532–550. [Google Scholar] [CrossRef]
- Yin, W.; Cheng, X.; Cui, H.; Li, D.; Zhou, L. Adaptive and accurate color edge extraction method for one-shot shape acquisition. Opt. Eng. 2016, 55, 094104. [Google Scholar] [CrossRef]
- Bai, X.; Wu, B.; Xing, Y. Yield-related qtls and their applications in rice genetic improvement f. J. Integr. Plant Biol. 2012, 54, 300–311. [Google Scholar] [CrossRef]
- Khattab, D.; Ebied, H.M.; Hussein, A.S.; Tolba, M.F. Color image segmentation based on different color space models using automatic grabcut. Sci. World J. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Khoshroo, A.; Arefi, A.; Khodaei, J. Detection of red tomato on plants using image processing techniques. Agric. Commun. 2014, 2, 9–15. [Google Scholar]
- Phakade, S.; Flora, D.; Malashree, H.; Rashmi, J. Automatic fruit defect detection using hsv and rgb color space model. Int. J. Innov. Res. Comput. Sci. Technol. 2014, 3, 67–73. [Google Scholar]
- Lorieux, M. Mapdisto: Fast and efficient computation of genetic linkage maps. Mol. Breed. 2012, 30, 1231–1235. [Google Scholar] [CrossRef]
- Voorrips, R. Mapchart: Software for the Graphical Presentation of Linkage Maps and Qtls. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Wang, S. Windows QTL Cartographer 2.5; Department of Statistics, North Carolina State University: Raleigh, NC, USA, 2006. [Google Scholar]
- Hemamalini, G.; Shashidhar, H.; Hittalmani, S. Molecular marker assisted tagging of morphological and physiological traits under two contrasting moisture regimes at peak vegetative stage in rice (Oryza sativa L.). Euphytica 2000, 112, 69–78. [Google Scholar] [CrossRef]
- Moldenhauer, K.; Slaton, N. Rice Growth and Development. In Rice Production Handbook; Africa Rice Center: Bouaké, Côte d’Ivoire, 2001; pp. 7–14. [Google Scholar]
- Septiningsih, E.M.; Ignacio, J.C.I.; Sendon, P.M.; Sanchez, D.L.; Ismail, A.M.; Mackill, D.J. Qtl mapping and confirmation for tolerance of anaerobic conditions during germination derived from the rice landrace ma-zhan red. Theor. Appl. Genet. 2013, 126, 1357–1366. [Google Scholar] [CrossRef] [PubMed]
- Hattori, Y.; Miura, K.; Asano, K.; Yamamoto, E.; Mori, H.; Kitano, H.; Matsuoka, M.; Ashikari, M. A major qtl confers rapid internode elongation in response to water rise in deepwater rice. Breed. Sci. 2007, 57, 305–314. [Google Scholar] [CrossRef]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.-J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H. The ethylene response factors snorkel1 and snorkel2 allow rice to adapt to deep water. Nature 2009, 460, 1026. [Google Scholar] [CrossRef]
- Kawano, R.; Doi, K.; Yasui, H.; Mochizuki, T.; Yoshimura, A. Mapping of qtls for floating ability in rice. Breed. Sci. 2008, 58, 47–53. [Google Scholar] [CrossRef]
- Nagai, K.; Hattori, Y.; Ashikari, M. Stunt or elongate? Two opposite strategies by which rice adapts to floods. J. Plant Res. 2010, 123, 303–309. [Google Scholar] [CrossRef]
- Nagai, K.; Kondo, Y.; Kitaoka, T.; Noda, T.; Kuroha, T.; Angeles-Shim, R.B.; Yasui, H.; Yoshimura, A.; Ashikari, M. Qtl analysis of internode elongation in response to gibberellin in deepwater rice. AoB Plants 2014, 6. [Google Scholar] [CrossRef]
- Dunand, R.; Saichuk, J. Rice Plant Growth and Development. In Lousiana Rice Production Handbook; Louisiana State University Agricultural Center: Baton Rouge, LA, USA, 2014; pp. 41–53. [Google Scholar]
- Li, X.; Qian, Q.; Fu, Z.; Wang, Y.; Xiong, G.; Zeng, D.; Wang, X.; Liu, X.; Teng, S.; Hiroshi, F. Control of tillering in rice. Nature 2003, 422, 618. [Google Scholar] [CrossRef] [PubMed]
- Champoux, M.; Wang, G.; Sarkarung, S.; Mackill, D.; O’Toole, J.; Huang, N.; McCouch, S. Locating genes associated with root morphology and drought avoidance in rice via linkage to molecular markers. Theor. Appl. Genet. 1995, 90, 969–981. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Mu, P.; Zhang, H.; Chen, C.Y.; Gao, Y.; Tian, Y.; Wen, F.; Li, Z. Mapping qtls of root morphological traits at different growth stages in rice. Genetica 2008, 133, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Hittalmani, S.; Shashidhar, H.; Bagali, P.G.; Huang, N.; Sidhu, J.; Singh, V.; Khush, G. Molecular mapping of quantitative trait loci for plant growth, yield and yield related traits across three diverse locations in a doubled haploid rice population. Euphytica 2002, 125, 207–214. [Google Scholar] [CrossRef]
- Sürek, H.; Beşer, N. Correlation and path coefficient analysis for some yield-related traits in rice (Oryza sativa L.) under thrace conditions. Turk. J. Agric. For. 2003, 27, 77–83. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.L.; Chung, Y.S.; Ji, H.; Lee, H.; Choi, I.; Kim, N.; Lee, E.; Oh, J.; Kang, D.-Y.; BAEK, J.; et al. New Parameters for Seedling Vigor Developed via Phenomics. Appl. Sci. 2019, 9, 1752. https://doi.org/10.3390/app9091752
Kim SL, Chung YS, Ji H, Lee H, Choi I, Kim N, Lee E, Oh J, Kang D-Y, BAEK J, et al. New Parameters for Seedling Vigor Developed via Phenomics. Applied Sciences. 2019; 9(9):1752. https://doi.org/10.3390/app9091752
Chicago/Turabian StyleKim, Song Lim, Yong Suk Chung, Hyeonso Ji, Hongseok Lee, Inchan Choi, Nyunhee Kim, Eungyeong Lee, Jun Oh, Do-Yu Kang, JeongHo BAEK, and et al. 2019. "New Parameters for Seedling Vigor Developed via Phenomics" Applied Sciences 9, no. 9: 1752. https://doi.org/10.3390/app9091752




