Effects of Amendments on Soil Microbial Diversity, Enzyme Activity and Nutrient Accumulation after Assisted Phytostabilization of an Extremely Acidic Metalliferous Mine Soil
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. DNA Extraction, 16S rRNA Polymerase Chain Reaction (PCR) and Bacterial Community T-Restriction Fragment Length Polymorphism (RFLP) Profiles Analysis
2.3. Microbial Activity and Microbial Biomass carbon Analysis
2.4. Soil Enzyme Activities Analysis
2.5. Soil Nutrients Analysis
2.6. Statistical Analysis
3. Results
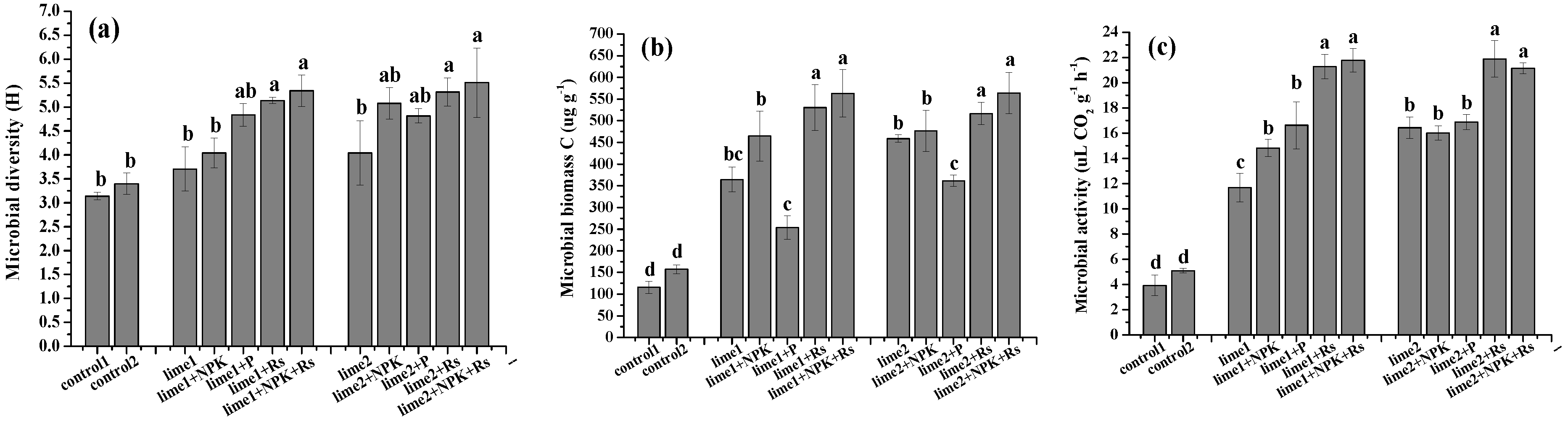
3.1. Effects of the Amendments on Microbial Diversity, Community Structure and Function
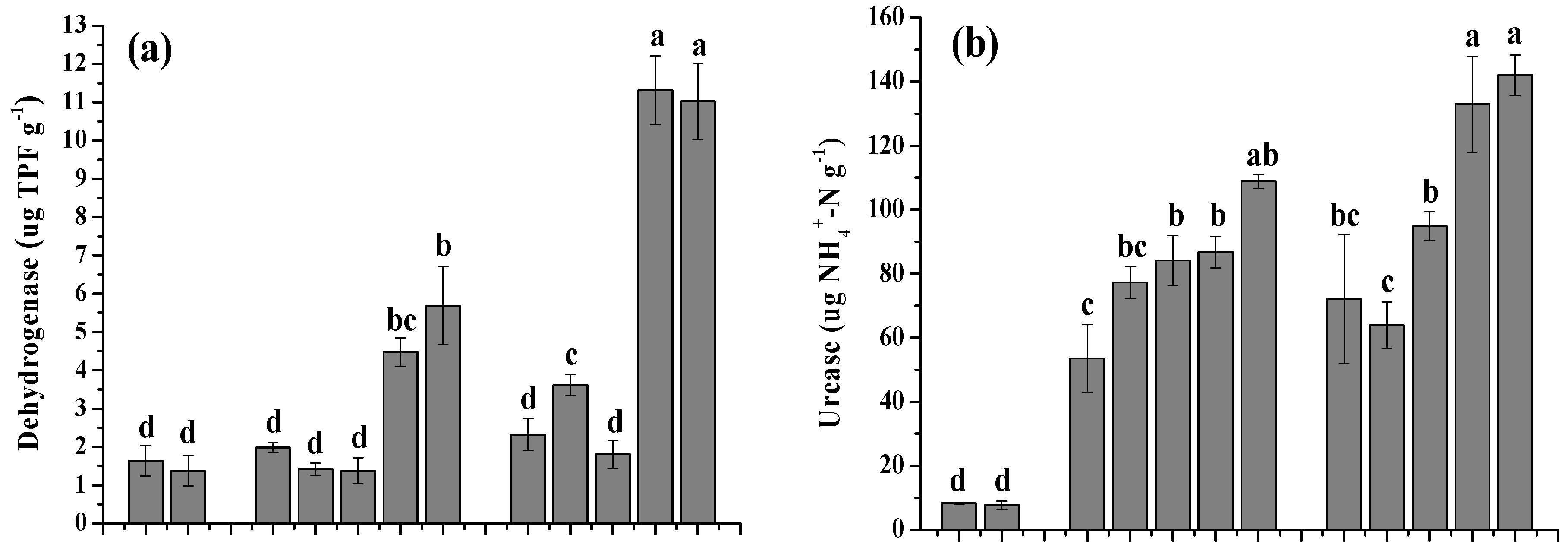
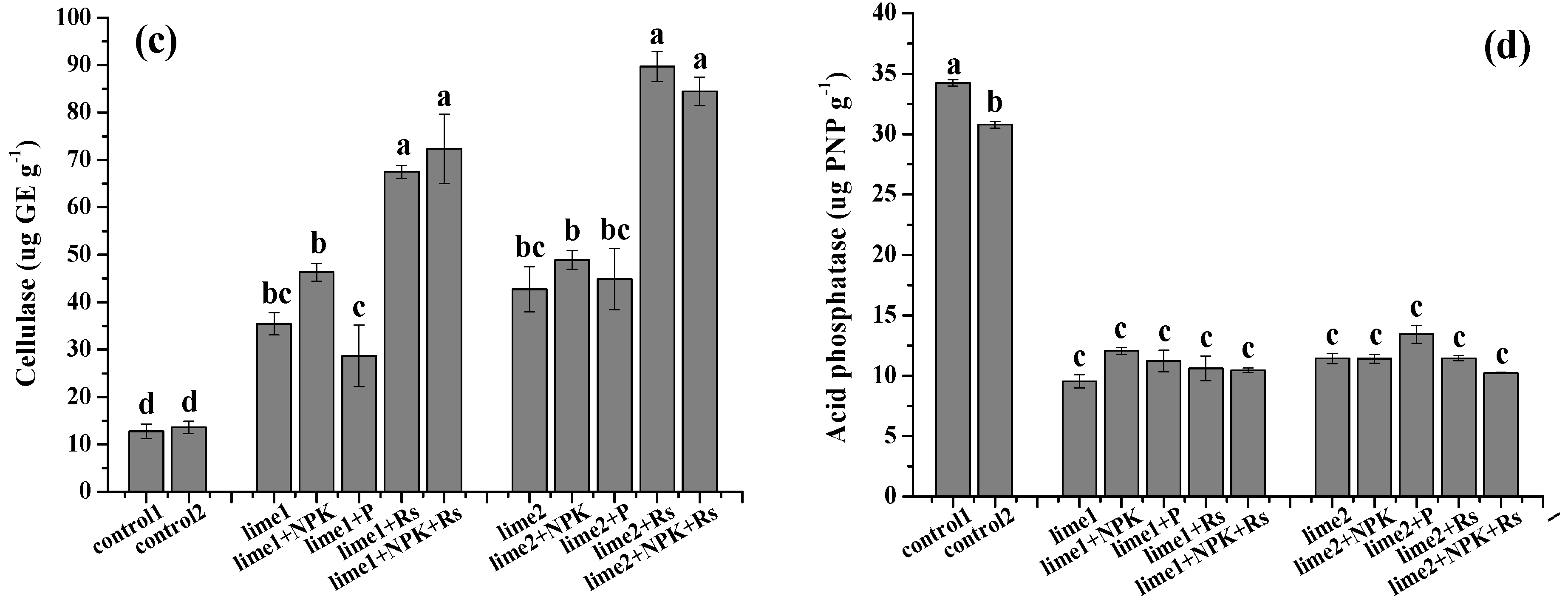
3.2. Effects of the Amendments on Soil Enzyme Activity
3.3. Effects of the Amendments on Soil Nutrient Elements
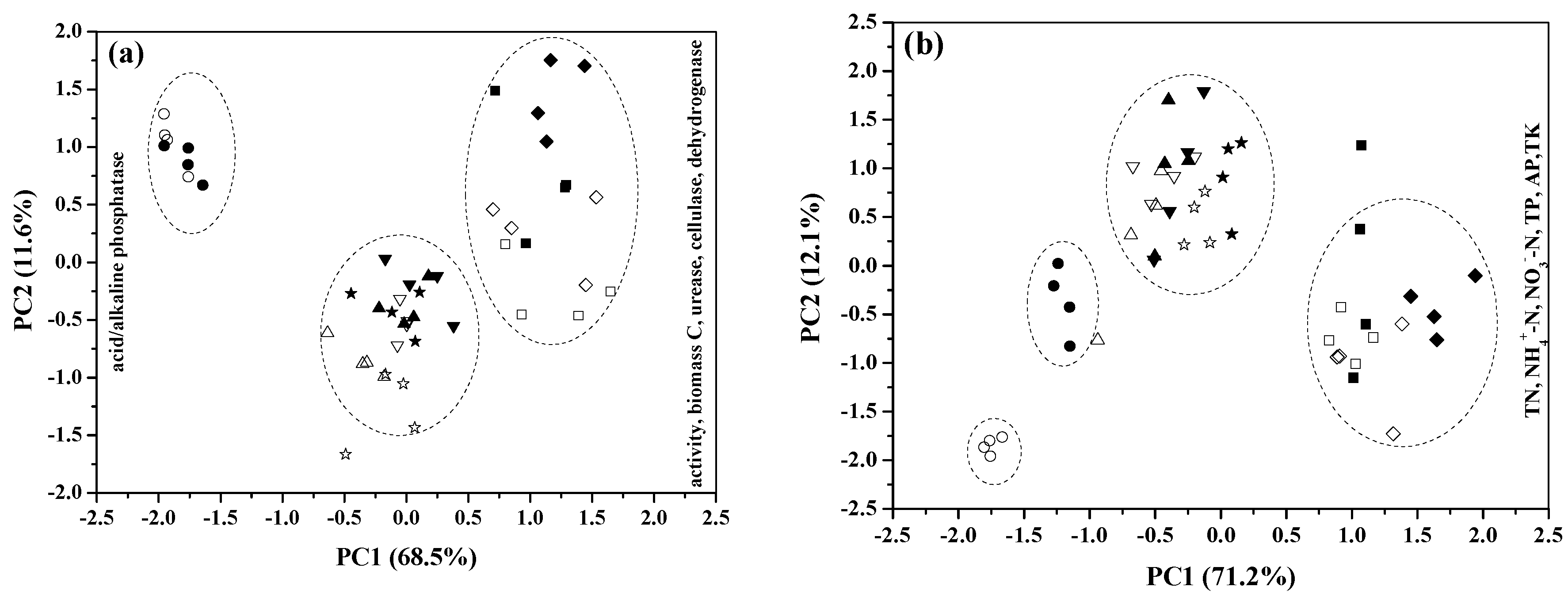
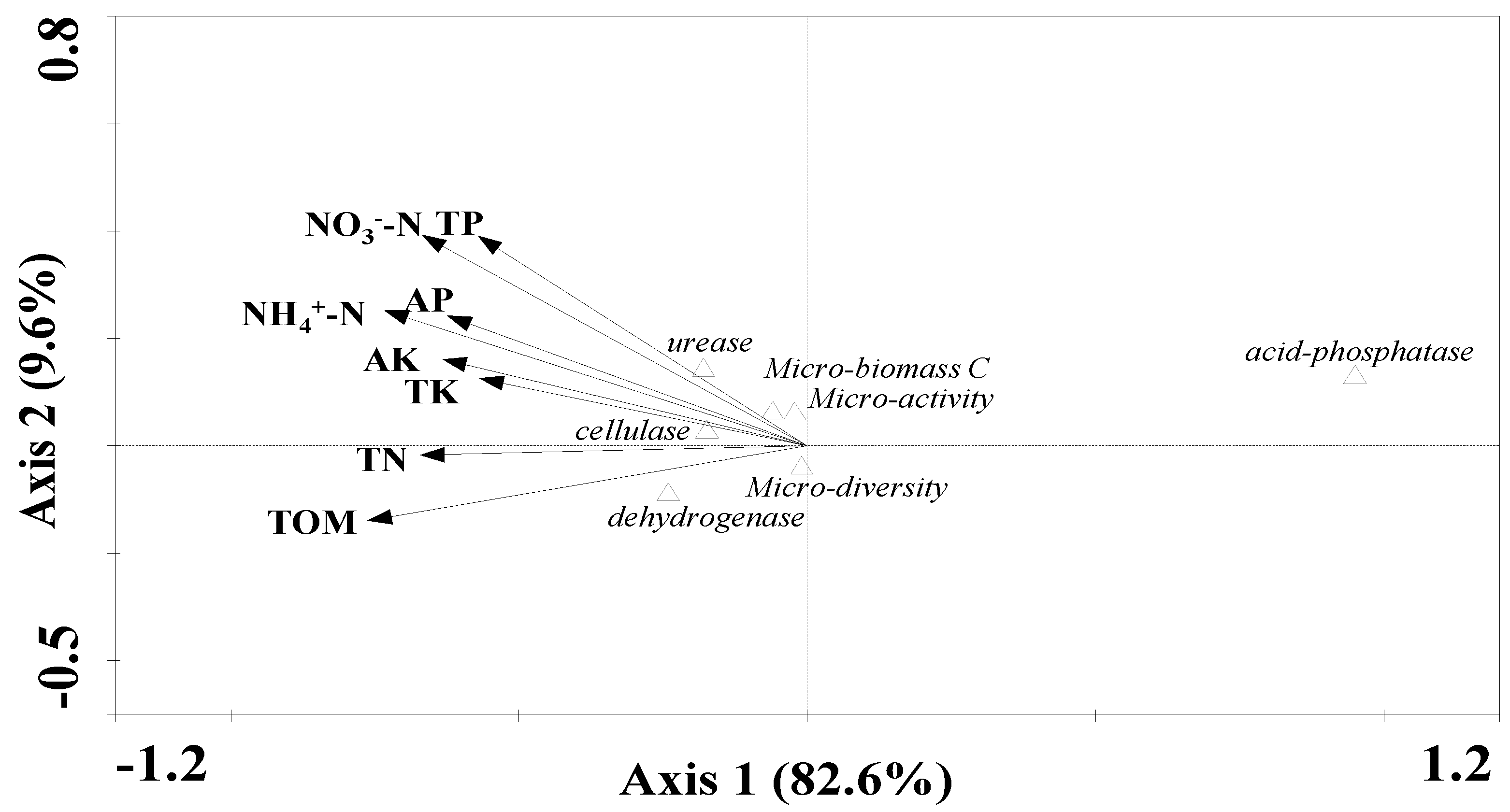
3.4. Evaluation of the Differences and Similarities of Amendment Effects using PCA
3.5. Relationships between Soil Microbial Properties and Nutrient Parameters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Karaca, O.; Cameselle, C.; Reddy, K.R. Mine tailings disposal sites: Contamination problems, remedial options and phytocaps for sustainable remediation. Rev. Environ. Sci. Biol. 2018, 17, 205–228. [Google Scholar] [CrossRef]
- Li, Y.; Jia, Z.; Sun, Q.; Zhan, J.; Yang, Y.; Wang, D. Ecological restoration alters microbial communities in mine tailings profiles. Sci. Rep. 2016, 6, 25193. [Google Scholar] [CrossRef]
- Wang, L.; Ji, B.; Hu, Y.H.; Liu, R.Q.; Sun, W. A review on in situ phytoremediation of mine tailings. Chemosphere 2017, 184, 594–600. [Google Scholar] [CrossRef]
- Kabas, S.; Faz, A.; Acosta, J.A.; Zomoza, R.; Martinez-Martinez, S.; Camona, D.M.; Bech, J. Effect of marble waste and pig slurry on the growth of native vegetation and heavy metal mobility in a mine tailing pond. J. Geochem. Explor. 2012, 123, 69–76. [Google Scholar] [CrossRef]
- Pardo, T.; Bernal, M.P.; Clemente, R. Efficiency of soil organic and inorganic amendments on the remediation of a contaminated mine soil: I. Effects on trace elements and nutrients solubility and leaching risk. Chemosphere 2014, 107, 121–128. [Google Scholar] [CrossRef]
- Li, J.J.; Zhou, X.M.; Yan, J.X.; Li, H.J.; He, J.Z. Effects of regenerating vegetation on soil enzyme activity and microbial structure in reclaimed soils on a surface coal mine site. Appl. Soil Ecol. 2015, 87, 56–62. [Google Scholar] [CrossRef]
- Thavamani, P.; Samkumar, R.A.; Satheesh, V.; Subashchandrabose, S.R.; Ramadass, K.; Naidu, R.; Venkateswarlu, K.; Megharaj, M. Microbes from mined sites: Harnessing their potential for reclamation of derelict mine sites. Environ. Pollut. 2017, 230, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Zornoza, R.; Acosta, J.A.; Faz, A.; Bååth, E. Microbial growth and community structure in acid mine soils after addition of different amendments for soil reclamation. Geoderma 2016, 272, 64–72. [Google Scholar] [CrossRef]
- Li, X.F.; You, F.; Bond, P.L.; Huang, L.B. Establishing microbial diversity and functions in weathered and neutral Cu-Pb-Zn tailings with native soil addition. Geoderma 2015, 247–248, 108–116. [Google Scholar] [CrossRef]
- Izquierdo, I.; Caravaca, F.; Alguacil, M.M.; Hernández, G.; Roldán, A. Use of microbiological indicators for evaluating success in soil restoration after revegetation of a mining area under subtropical conditions. Appl. Soil Ecol. 2005, 30, 3–10. [Google Scholar] [CrossRef]
- Gil-Sotres, F.; Trasar-Cepeda, C.; Leirós, M.C.; Seoane, S. Different approaches to evaluating soil quality using biochemical properties. Soil Biol. Biochem. 2005, 37, 877–887. [Google Scholar] [CrossRef]
- Tejada, M.; Hernandez, M.T.; Garcia, C. Application of two organic amendments on soil restoration: Effects on the soil biological properties. J. Environ. Qual. 2006, 35, 1010–1017. [Google Scholar] [CrossRef]
- Alvarenga, P.; Palma, P.; Gonçalves, A.P.; Fernandes, R.M.; de Varennes, A.; Vallini, G.; Duarte, E.; Cunha-Queda, A.C. Organic residues as immobilizing agents in aided phytostabilization: (II) Effects on soil biochemical and ecotoxicological characteristics. Chemosphere 2009, 74, 1301–1308. [Google Scholar] [CrossRef]
- Yang, S.X.; Cao, J.B.; Li, F.M.; Peng, X.Z.; Yang, Z.H.; Chai, L.Y. Field evaluation of the effectiveness of three industrial by-products as organic amendments for aided phytostabilization of a Pb/Zn mine tailings. Environ. Sci. Proc. Impacts 2016, 18, 95–103. [Google Scholar] [CrossRef]
- Finkenbein, P.; Kretschmer, K.; Kulka, K.; Klotz, S.; Heilmeier, H. Soil enzyme activities as bioindicators for substrate quality in revegetation of a subtropical coal mining dump. Soil Biol. Biochem. 2013, 56, 87–89. [Google Scholar] [CrossRef]
- Li, X.F.; Bond, P.L.; Van Nostrand, J.D.; Zhou, J.Z.; Huang, L.B. From lithotroph- to organotroph-dominant: Directional shift of microbial community in sulphidic tailings during phytostabilization. Sci. Rep. 2015, 5, 12978. [Google Scholar] [CrossRef]
- Yang, S.X.; Li, J.T.; Yang, B.; Liao, B.; Zhang, J.T.; Shu, W.S. Effectiveness of amendments on re-acidification and heavy metal immobilization in an extremely acidic mine soil. J. Environ. Monitor. 2011, 13, 1876–1883. [Google Scholar] [CrossRef]
- Yang, T.T.; Liu, J.; Chen, W.C.; Chen, X.; Shu, H.Y.; Jia, P.; Liao, B.; Shu, W.S.; Li, J.T. Changes in microbial community composition following phytostabilization of an extremely acidic Cu mine tailings. Soil Biol. Biochem. 2017, 114, 52–58. [Google Scholar] [CrossRef]
- Tan, G.L.; Shu, W.S.; Hallberg, K.B.; Li, F.; Lan, C.Y.; Huang, L.N. Cultivation-dependent and cultivation-independent characterization of the microbial community in acid mine drainage associated with acidic Pb/Zn mine tailings at Lechang, Guangdong, China. FEMS Microbiol. Ecol. 2007, 59, 118–126. [Google Scholar] [CrossRef]
- Dojka, M.A.; Hugenholtz, P.; Haack, S.K.; Pace, N.R. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 1998, 64, 3869–3877. [Google Scholar]
- De Leij, F.A.A.M.; Whipps, J.M.; Lynch, J.M. The use of colony development for the characterization of bacterial communities in soil and roots. Microb. Ecol. 1993, 27, 81–97. [Google Scholar] [CrossRef]
- Anderson, J.P.E. Soil respiration. In Methods of Soil Analysis, Part 2. Chemical and Microbiological Properties; Soil Science Society of America: Madison, WI, USA, 1982. [Google Scholar]
- Vance, E.D.; Brookes, P.C.; Jenkinson, D.S. An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 1987, 19, 703–707. [Google Scholar] [CrossRef]
- Thalmann, A. Zur Methodik der Bestimmung der Dehydrogenaseaktivität im Boden mittels Triphenyltetrazoliumchlorid (TTC). Landwirt. Forsch. 1968, 21, 249–258. [Google Scholar]
- Schinner, F.; von Mersi, W. Xylanase-, CM-cellulase and invertase activity in soil, an improved method. Soil Biol. Biochem. 1990, 22, 511–515. [Google Scholar] [CrossRef]
- Kandeler, E.; Gerber, H. Short-term assay of soil urease activity using colorimetric determination of ammonium. Biol. Fert. Soils 1988, 6, 68–72. [Google Scholar] [CrossRef]
- Tabatabai, A.M.; Bremner, M.J. Use of p-nitrophenyl phosphate for assay of soil phosphatase activity. Soil Biol. Biochem. 1969, 1, 301–307. [Google Scholar] [CrossRef]
- Nelson, D.W.; Sommers, L.E. Total carbon, organic carbon and organic matter. In Methods of Soil Analysis: Part 2, Agronomy Monograph, 2nd ed.; Page, A.L., Ed.; American Society of Agronomy and Soil Science Society of America: Madison, WI, USA, 1982; Volume 9. [Google Scholar]
- Bremner, J.M.; Mulvaney, C.S. Total nitrogen. In Methods of Soil Analysis: Part 2, Agronomy Monograph, 2nd ed.; Page, A.L., Ed.; American Society of Agronomy and Soil Science Society of America: Madison, WI, USA, 1982; Volume 9. [Google Scholar]
- Maynard, D.G.; Kalra, Y.P.; Crumbaugh, J.A. Nitrate and Exchangeable Ammonium Nitrogen in Soil Sampling and Methods of Analysis, 2nd ed.; CRC Press, Taylor and Francis: Boca Raton, FL, USA, 2008. [Google Scholar]
- Keeney, D.R.; Nelson, D.W. Nitrogen: Inorganic forms. In Methods of Soil Analysis: Part 2, Agronomy Monograph, 2nd ed.; Page, A.L., Ed.; American Society of Agronomy and Soil Science Society of America: Madison, WI, USA, 1982; Volume 9. [Google Scholar]
- Bray, R.H.; Kurtz, L.T. Determination of total, organic and available forms of phosphorus in soil. Soil Sci. 1945, 59, 39–45. [Google Scholar] [CrossRef]
- Lu, R.S. Soil Agricultural Chemistry Analysis; China Agricultural Technology Press: Beijing, China, 1999. [Google Scholar]
- Abollino, O.; Aceto, M.; Malandrino, M.; Mentasti, E.; Sarzanini, C. Heavy metals in agricultural soils from Piedmont, Italy. Distribution, speciation and chemometric data treatment. Chemosphere 2002, 49, 545–557. [Google Scholar] [CrossRef]
- Ter Braak, C.J.F.; Šamilauer, P. CANOCO Reference Munual and CanoDraw for Windows User’s Guide: Software for Canonical Community Ordination (Version 4.5); Microcomputer Power: Ithaca, NY, USA, 2002. [Google Scholar]
- Chen, L.G.; Tsui, M.M.P.; Lam, J.C.W.; Wang, Q.; Hu, C.Y.; Wai, O.W.H.; Zhou, B.S.; Lam, P.K.S. Contamination by perfluoroalkyl substances and microbial community structure in Pearl River Delta sediments. Environ. Pollut. 2019, 245, 218–225. [Google Scholar] [CrossRef]
- Cusack, D.F.; Silver, W.L.; Torn, M.S.; Burton, S.D.; Firestone, M.K. Changes in microbial community characteristics and soil organic matter with nitrogen additions in two tropical forests. Ecology 2011, 92, 621–632. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microb. 2009, 75, 5111–5120. [Google Scholar] [CrossRef]
- Andronov, E.E.; Petrova, S.N.; Pinaev, A.G. Analysis of the structure of microbial community in soils with different degrees of salinization using T-RFLP and real-time PCR techniques. Eurasian Soil. Sci. 2012, 45, 147–156. [Google Scholar] [CrossRef]
- Zornoza, R.; Acosta, J.A.; Martínez-Martínez, S.; Faz, A.; Bååth, E. Main factors controlling microbial community structure and function after reclamation of a tailing pond with aided phytostabilization. Geoderma 2015, 245–246, 1–10. [Google Scholar] [CrossRef]
- Li, Y.; Sun, Q.Y.; Zhan, J.; Yang, Y.; Wang, D. Soil-covered strategy for ecological restoration alters the bacterial community structure and predictive energy metabolic functions in mine tailings profiles. Appl. Microbiol. Biotechnol. 2017, 101, 2549–2561. [Google Scholar] [CrossRef]
| Parameters | Mine Soil | River Sediment |
|---|---|---|
| Sand (%) | 82.2 ± 9.7 | nd |
| Slit (%) | 10.1 ± 0.89 | nd |
| Clay (%) | 7.5 ± 0.43 | nd |
| pH | 2.5 ± 0.14 | 7.3 ± 0.29 |
| Electrical conductivity (dS m−1) | 3.1 ± 0.18 | 1.6 ± 0.13 |
| Organic matter (g kg−1) | 6.2 ± 0.53 | 70.6 ± 12.1 |
| Total nitrogen (mg kg−1) | 44.5 ± 3.6 | 1054 ± 88.1 |
| Total phosphate (mg kg−1) | 307 ± 25.2 | 722 ± 53.2 |
| Total potassium (mg kg−1) | 290 ± 16.4 | 2717 ± 217 |
| Zinc (Zn, mg kg−1) | 1175 ± 84.1 | 147 ± 16.8 |
| Lead (Pb, mg kg−1) | 1107 ± 67.9 | 75.1 ± 10.4 |
| Copper (Cu, mg kg−1) | 1826 ± 121 | 50.4 ± 8.8 |
| Cadmium ( Cd, mg kg−1) | 2.6 ± 0.33 | 0.76 ± 0.01 |
| Treatment types | Treatments | Amendments |
|---|---|---|
| CK | control1 | mine soil |
| control2 | mine soil + sawdust * | |
| L1 | lime1 | mine soil + lime1 (25 t ha−1) |
| lime1+NPK | mine soil + lime1 (25 t ha−1) + NPK compound fertilizer (150 kg ha−1) | |
| lime1+P | mine soil + lime1 (25 t ha−1) + phosphate fertilizer (300 kg ha−1) | |
| lime1+Rs | mine soil + lime1 (25 t ha−1) + River sediment (30%, w:w) | |
| lime1+NPK+Rs | mine soil + lime1 (25 t ha−1)+ NPK compound fertilizer (150 kg ha−1) + River sediment (30%, w:w) | |
| L2 | lime2 | mine soil + lime2 (50 t ha−1) |
| lime2+NPK | mine soil + lime2 (50 t ha−1) + NPK compound fertilizer (150 kg ha−1) | |
| lime2+P | mine soil + lime2 (50 t ha−1) + phosphate fertilizer (300 kg ha−1) | |
| lime2+Rs | mine soil + lime2 (50 t ha−1) + River sediment (30%, w:w) | |
| lime2+NPK+Rs | mine soil + lime2 (50 t ha−1) + NPK compound fertilizer (150 kg ha−1) + River sediment (30%, w:w) |
| OM (g kg−1) | TN (mg kg−1) | TP (mg kg−1) | TK (mg kg−1) |
|---|---|---|---|
| 3.7 ± 0.93c * | 72 ± 3.6d | 276 ± 2.9d | 183 ± 2.7d |
| 21 ± 2.4b | 179 ± 8.8c | 289 ± 8.2d | 229 ± 26d |
| 21 ± 0.76b | 235 ± 7.9b | 297 ± 10d | 286 ± 24d |
| 23 ± 1.5b | 236 ± 7.5b | 303 ± 11d | 289 ± 27d |
| 23 ± 1.4b | 232 ± 6.2b | 359 ± 11c | 386 ± 28c |
| 25 ± 0.59ab | 458 ± 12a | 446 ± 10b | 551 ± 44ab |
| 25 ± 1.7ab | 487 ± 35a | 423 ± 14b | 630 ± 52a |
| 23 ± 0.66b | 221 ± 10b | 322 ± 11cd | 266 ± 7.3d |
| 27 ± 1.4a | 237 ± 6.3b | 326 ± 5.3cd | 262 ± 6.2d |
| 27 ± 1.5a | 262 ± 4.3b | 362 ± 18c | 410 ± 41c |
| 28 ± 2.0a | 478 ± 5.3a | 418 ± 8.0b | 525 ± 14b |
| 29 ± 1.8a | 492 ± 14a | 513 ± 23a | 521 ± 24b |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.-x.; Liao, B.; Xiao, R.-b.; Li, J.-t. Effects of Amendments on Soil Microbial Diversity, Enzyme Activity and Nutrient Accumulation after Assisted Phytostabilization of an Extremely Acidic Metalliferous Mine Soil. Appl. Sci. 2019, 9, 1552. https://doi.org/10.3390/app9081552
Yang S-x, Liao B, Xiao R-b, Li J-t. Effects of Amendments on Soil Microbial Diversity, Enzyme Activity and Nutrient Accumulation after Assisted Phytostabilization of an Extremely Acidic Metalliferous Mine Soil. Applied Sciences. 2019; 9(8):1552. https://doi.org/10.3390/app9081552
Chicago/Turabian StyleYang, Sheng-xiang, Bin Liao, Rong-bo Xiao, and Jin-tian Li. 2019. "Effects of Amendments on Soil Microbial Diversity, Enzyme Activity and Nutrient Accumulation after Assisted Phytostabilization of an Extremely Acidic Metalliferous Mine Soil" Applied Sciences 9, no. 8: 1552. https://doi.org/10.3390/app9081552
APA StyleYang, S.-x., Liao, B., Xiao, R.-b., & Li, J.-t. (2019). Effects of Amendments on Soil Microbial Diversity, Enzyme Activity and Nutrient Accumulation after Assisted Phytostabilization of an Extremely Acidic Metalliferous Mine Soil. Applied Sciences, 9(8), 1552. https://doi.org/10.3390/app9081552





