Cultivar Classification of Single Sweet Corn Seed Using Fourier Transform Near-Infrared Spectroscopy Combined with Discriminant Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Seed Samples
2.2. FT-NIR Spectroscopy Acquisition
2.3. Data Pre-processing
2.4. Feature Wavelengths Selection
2.5. Discriminant Analysis Algorithms
2.5.1. K-Nearest Neighbor (KNN)
2.5.2. Soft Independent Method of Class Analogy (SIMCA)
2.5.3. Partial Least-squares Discriminant Analysis (PLS-DA)
2.5.4. Support Vector Machine Discriminant Analysis (SVM-DA)
2.6. Model Validation and Evaluation
3. Results and Discussion
3.1. Data Pre-processing
3.2. Discrimination Analysis
3.2.1. Principal Component Analysis
3.2.2. Discriminant Models Based on Full-Range Wavelength Variables
3.2.3. Feature Wavelength Variables
3.2.4. Discriminant Models Based on Feature Wavelength Variables
4. Conclusions and Future Work
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lertrat, K.; Pulam, T. Breeding for Increased Sweetness in Sweet Corn. Int. J. Plant Breed. 2007, 1, 27–30. [Google Scholar]
- Zhang, R.; Huang, L.; Deng, Y.; Chi, J.; Zhang, Y.; Wei, Z.; Zhang, M. Phenolic content and antioxidant activity of eight representative sweet corn varieties grown in South China. Int. J. Food Prop. 2017, 20, 3043–3055. [Google Scholar] [CrossRef]
- Singh, I.; Langyan, S.; Yadava, P. Sweet Corn and Corn-Based Sweeteners. Sugar Tech. 2014, 16, 144–149. [Google Scholar] [CrossRef]
- Szymanek, M.; Tanaś, W.; Kassar, F.H. Kernel Carbohydrates Concentration in Sugary-1, Sugary Enhanced and Shrunken Sweet Corn Kernels. Agric. Agric. Sci. Procedia 2015, 7, 260–264. [Google Scholar] [CrossRef]
- Olsen, J.K.; Giles, J.E.; Jordan, R.A. Post-harvest carbohydrate changes and sensory quality of three sweet corn cultivars. Sci. Hortic. 1990, 44, 179–189. [Google Scholar] [CrossRef]
- Yang, X.; Hong, H.; You, Z.; Cheng, F. Spectral and Image Integrated Analysis of Hyperspectral Data for Waxy Corn Seed Variety Classification. Sensors 2015, 15, 15578–15594. [Google Scholar] [CrossRef]
- Wang, L.; Sun, D.; Pu, H.; Zhu, Z. Application of Hyperspectral Imaging to Discriminate the Variety of Maize Seeds. Food Anal. Methods 2016, 9, 225–234. [Google Scholar] [CrossRef]
- Huang, M.; Tang, J.; Yang, B.; Zhu, Q. Classification of maize seeds of different years based on hyperspectral imaging and model updating. Comput. Electron. Agric. 2016, 122, 139–145. [Google Scholar] [CrossRef]
- Cui, Y.; Xu, L.; An, D.; Liu, Z.; Gu, J.; Li, S.; Zhang, X.; Zhu, D. Identification of maize seed varieties based on near infrared reflectance spectroscopy and chemometrics. Int. J. Agric. Biol. Eng. 2018, 11, 177–183. [Google Scholar] [CrossRef]
- Xie, C.; He, Y. Modeling for mung bean variety classification using visible and near-infrared hyperspectral imaging. Int. J. Agric. Biol. Eng. 2018, 11, 187–191. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, F.; He, Y.; Li, X. Application of Hyperspectral Imaging and Chemometric Calibrations for Variety Discrimination of Maize Seeds. Sensors 2012, 12, 17234–17246. [Google Scholar] [CrossRef]
- Kong, W.; Zhang, C.; Liu, F.; Nie, P.; He, Y. Rice Seed Cultivar Identification Using Near-Infrared Hyperspectral Imaging and Multivariate Data Analysis. Sensors 2013, 13, 8916–8927. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhu, S.; Zhang, C.; Feng, X.; Feng, L.; He, Y. Application of hyperspectral imaging and chemometrics for variety classification of maize seeds. RSC Adv. 2018, 8, 1337–1345. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, C.; Zhu, S.; Gao, P.; Feng, L.; He, Y. Non-Destructive and Rapid Variety Discrimination and Visualization of Single Grape Seed Using Near-Infrared Hyperspectral Imaging Technique and Multivariate Analysis. Molecules 2018, 23, 1352. [Google Scholar] [CrossRef]
- Qiu, Z.; Chen, J.; Zhao, Y.; Zhu, S.; He, Y.; Zhang, C. Variety Identification of Single Rice Seed Using Hyperspectral Imaging Combined with Convolutional Neural Network. Appl. Sci. 2018, 8, 212. [Google Scholar] [CrossRef]
- Kumar, S.; Andy, A. Fourier transform-near infrared reflectance spectroscopy calibration development for screening of oil content of intact safflower seeds. Int. Food Res. J. 2013, 20, 759. [Google Scholar]
- Xiao, H.; Sun, K.; Sun, Y.; Wei, K.; Tu, K.; Pan, L. Comparison of Benchtop Fourier-Transform (FT) and Portable Grating Scanning Spectrometers for Determination of Total Soluble Solid Contents in Single Grape Berry (Vitis vinifera L.) and Calibration Transfer. Sensors 2017, 17, 2693. [Google Scholar] [CrossRef]
- Gislum, R.; Nikneshan, P.; Shrestha, S.; Tadayyon, A.; Deleuran, L.; Boelt, B. Characterisation of Castor (Ricinus communis L.) Seed Quality Using Fourier Transform Near-Infrared Spectroscopy in Combination with Multivariate Data Analysis. Agriculture 2018, 8, 59. [Google Scholar] [CrossRef]
- Ahn, C.K.; Cho, B.K.; Kang, J.S.; Lee, K.J. Study on non-destructive sorting technique for lettuce seed using fourier transform near-Infrared spectrometer. J. Agric. Sci. 2012, 39, 111–116. [Google Scholar]
- Lohumi, S.; Mo, C.; Kang, J.S.; Hong, S.J.; Cho, B.K. Nondestructive Evaluation for the Viability of Watermelon (Citrullus lanatus) Seeds Using Fourier Transform Near Infrared Spectroscopy. J. Biosyst. Eng. 2013, 38, 312–317. [Google Scholar] [CrossRef]
- Ambrose, A.; Lohumi, S.; Lee, W.; Cho, B.K. Comparative nondestructive measurement of corn seed viability using Fourier transform near-infrared (FT-NIR) and Raman spectroscopy. Sens. Actuators B Chem. 2016, 224, 500–506. [Google Scholar] [CrossRef]
- Qiu, G.; Lü, E.; Lu, H.; Xu, S.; Zeng, F.; Shui, Q. Single-Kernel FT-NIR Spectroscopy for Detecting Supersweet Corn (Zea mays L. Saccharata Sturt) Seed Viability with Multivariate Data Analysis. Sensors 2018, 18, 1010. [Google Scholar] [CrossRef]
- Kusumaningrum, D.; Lee, H.; Lohumi, S.; Mo, C.; Kim, M.S.; Cho, B. Non-destructive technique for determining the viability of soybean (Glycine max) seeds using FT-NIR spectroscopy. J. Sci. Food Agric. 2018, 98, 1734–1742. [Google Scholar] [CrossRef]
- De Girolamo, A.; Cervellieri, S.; Visconti, A.; Pascale, M. Rapid Analysis of Deoxynivalenol in Durum Wheat by FT-NIR Spectroscopy. Toxins 2014, 6, 3129–3143. [Google Scholar] [CrossRef]
- Taradolsirithitikul, P.; Sirisomboon, P.; Dachoupakan Sirisomboon, C. Qualitative and quantitative analysis of ochratoxin A contamination in green coffee beans using Fourier transform near infrared spectroscopy. J. Sci. Food Agric. 2017, 97, 1260–1266. [Google Scholar] [CrossRef]
- Fu, H.; Jiang, D.; Zhou, R.; Yang, T.; Chen, F.; Li, H.; Yin, Q.; Fan, Y. Predicting Mildew Contamination and Shelf-Life of Sunflower Seeds and Soybeans by Fourier Transform Near-Infrared Spectroscopy and Chemometric Data Analysis. Food Anal. Methods 2017, 10, 1597–1608. [Google Scholar] [CrossRef]
- Attaviroj, N.; Kasemsumran, S.; Noomhorm, A. Rapid Variety Identification of Pure Rough Rice by Fourier-Transform Near-Infrared Spectroscopy. Cereal Chem. J. 2011, 88, 490–496. [Google Scholar] [CrossRef]
- Chen, Y.M.; Lin, P.; He, J.Q.; He, Y.; Li, X.L. Combination of the Manifold Dimensionality Reduction Methods with Least Squares Support vector machines for Classifying the Species of Sorghum Seeds. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Luo, Y.H.; Liang, K.Q.; Zhang, D.H.; Su, J.H.; Zhang, S.T.; Liang, Y.F. Breeding of a new supersweet corn cultivar Huameitian NO. 168. Guangdong Agric. Sci. 2008, 11, 7–9. (In Chinese) [Google Scholar]
- Zhang, S.T.; Liang, K.Q.; Zhang, D.H.; Liang, Y.F.; Su, J.H. Breeding of yellow-white supersweet corn Huameitian NO. 8. Guangdong Agric. Sci. 2010, 8, 30–31. (In Chinese) [Google Scholar]
- Achata, E.; Esquerre, C.; O’Donnell, C.; Gowen, A. A Study on the Application of Near Infrared Hyperspectral Chemical Imaging for Monitoring Moisture Content and Water Activity in Low Moisture Systems. Molecules 2015, 20, 2611–2621. [Google Scholar] [CrossRef]
- Nieuwoudt, H.H.; Prior, B.A.; Pretorius, I.S.; Manley, M.; Bauer, F.F. Principal Component Analysis Applied to Fourier Transform Infrared Spectroscopy for the Design of Calibration Sets for Glycerol Prediction Models in Wine and for the Detection and Classification of Outlier Samples. J. Agric. Food Chem. 2004, 52, 3726–3735. [Google Scholar] [CrossRef]
- Agelet, E.L.; Ellis, D.D.; Duvick, S.; Goggi, A.S.; Hurburgh, C.R.; Gardner, C.A. Feasibility of near infrared spectroscopy for analyzing corn kernel damage and viability of soybean and corn kernels. J. Cereal Sci. 2012, 55, 160–165. [Google Scholar] [CrossRef]
- Isaksson, T.; Naes, T. The Effect of Multiplicative Scatter Correction (MSC) and Linearity Improvement in NIR Spectroscopy. Appl. Spectrosc. 1988, 42, 1273–1284. [Google Scholar] [CrossRef]
- Shrestha, S.; Deleuran, L.C.; Gislum, R. Separation of viable and non-viable tomato (Solanum lycopersicum L.) seeds using single seed near-infrared spectroscopy. Comput. Electron. Agric. 2017, 142, 348–355. [Google Scholar] [CrossRef]
- Nørgaard, L.; Saudland, A.; Wagner, J.; Nielsen, J.P.; Munck, L.; Engelsen, S.B. Interval Partial Least-Squares Regression (iPLS): A Comparative Chemometric Study with an Example from Near-Infrared Spectroscopy. Appl. Spectrosc. 2016, 54, 413–419. [Google Scholar] [CrossRef]
- Bangalore, A.S.; Shaffer, R.E.; Small, G.W.; Arnold, M.A. Genetic Algorithm-Based Method for Selecting Wavelengths and Model Size for Use with Partial Least-Squares Regression: Application to Near-Infrared Spectroscopy. Anal. Chem. 1996, 68, 4200–4212. [Google Scholar] [CrossRef]
- Daszykowski, M.; Orzel, J.; Wrobel, M.S.; Czarnik-Matusewicz, H.; Walczak, B. Improvement of classification using robust soft classification rules for near-infrared reflectance spectral data. Chemom. Intell. Lab. Syst. 2011, 109, 86–93. [Google Scholar] [CrossRef]
- Pérez, N.F.; Ferré, J.; Boqué, R. Calculation of the reliability of classification in discriminant partial least-squares binary classification. Chemom. Intell. Lab. Syst. 2009, 95, 122–128. [Google Scholar] [CrossRef]
- Devos, O.; Ruckebusch, C.; Durand, A.; Duponchel, L.; Huvenne, J. Support vector machines (SVM) in near infrared (NIR) spectroscopy: Focus on parameters optimization and model interpretation. Chemom. Intell. Lab. Syst. 2009, 96, 27–33. [Google Scholar] [CrossRef]
- Westerhuis, J.A.; Hoefsloot, H.C.J.; Smit, S.; Vis, D.J.; Smilde, A.K.; Velzen, E.J.J.; Duijnhoven, J.P.M.; Dorsten, F.A. Assessment of PLSDA cross validation. Metabolomics 2008, 4, 81–89. [Google Scholar] [CrossRef]
- Stone, M. Cross-Validatory Choice and Assessment of Statistical Predictions. J. R. Stat. Soc. Ser. B (Methodol.) 1974, 36, 111–147. [Google Scholar] [CrossRef]
- Liu, H.; Papa, E.; Gramatica, P. QSAR Prediction of Estrogen Activity for a Large Set of Diverse Chemicals under the Guidance of OECD Principles. Chem. Res. Toxicol. 2006, 19, 1540–1548. [Google Scholar] [CrossRef]
- Aenugu, H.P.R.; Kumar, D.S.; Srisudharson; Parthiban, N.; Ghosh, S.S.; Banji, D. Near Infra Red Spectroscopy-An Overview. Int. J. ChemTech Res. 2011, 3, 825–836. [Google Scholar]
- Zhang, J.; Feng, X.; Liu, X.; He, Y. Identification of Hybrid Okra Seeds Based on Near-Infrared Hyperspectral Imaging Technology. Appl. Sci. 2018, 8, 1793. [Google Scholar] [CrossRef]
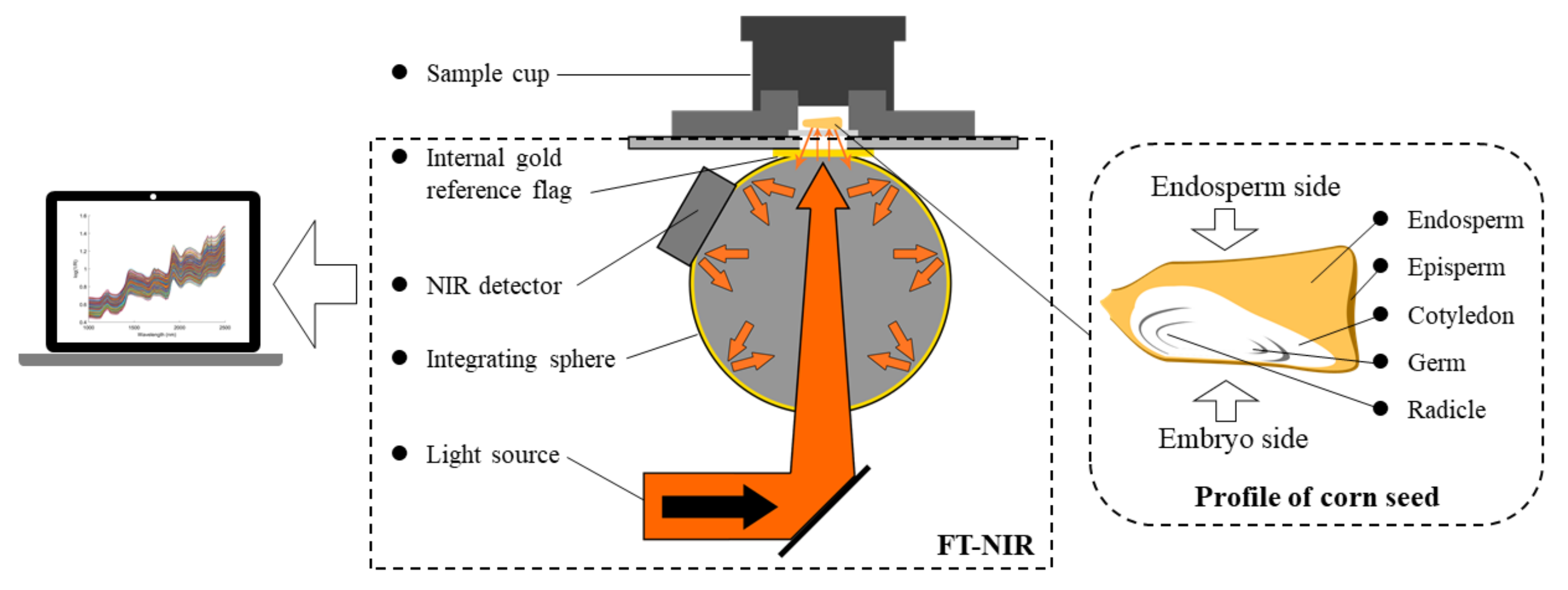
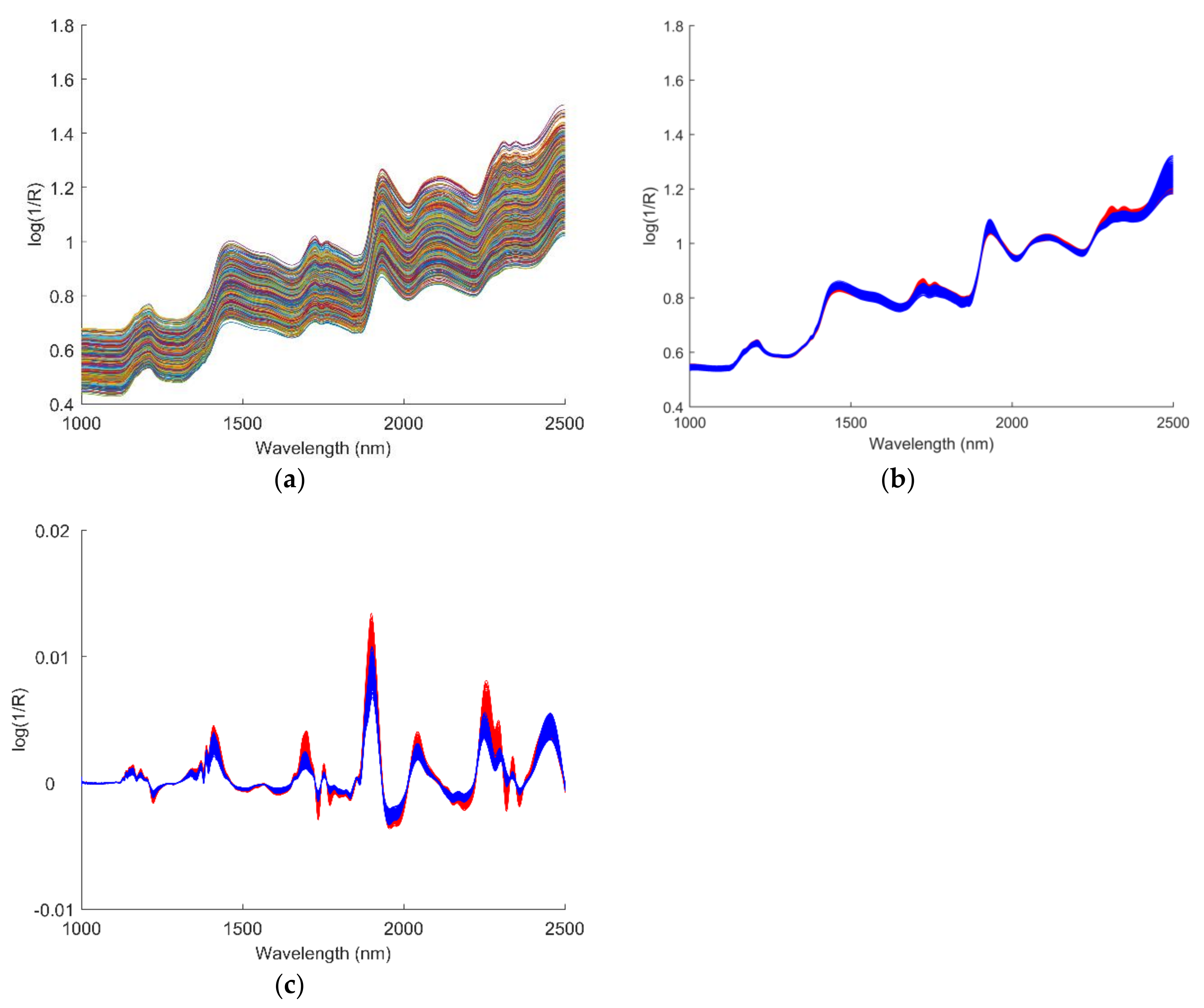
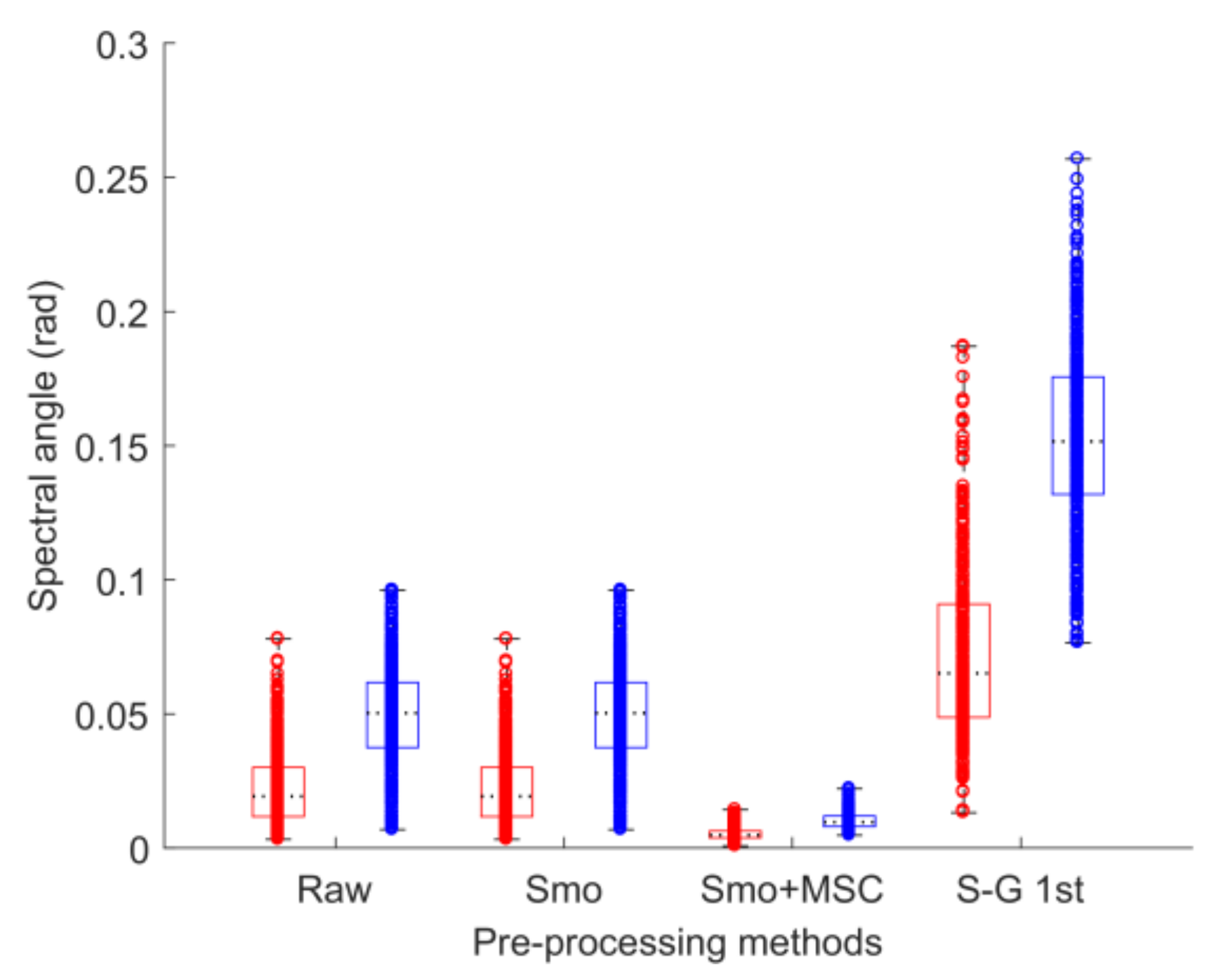

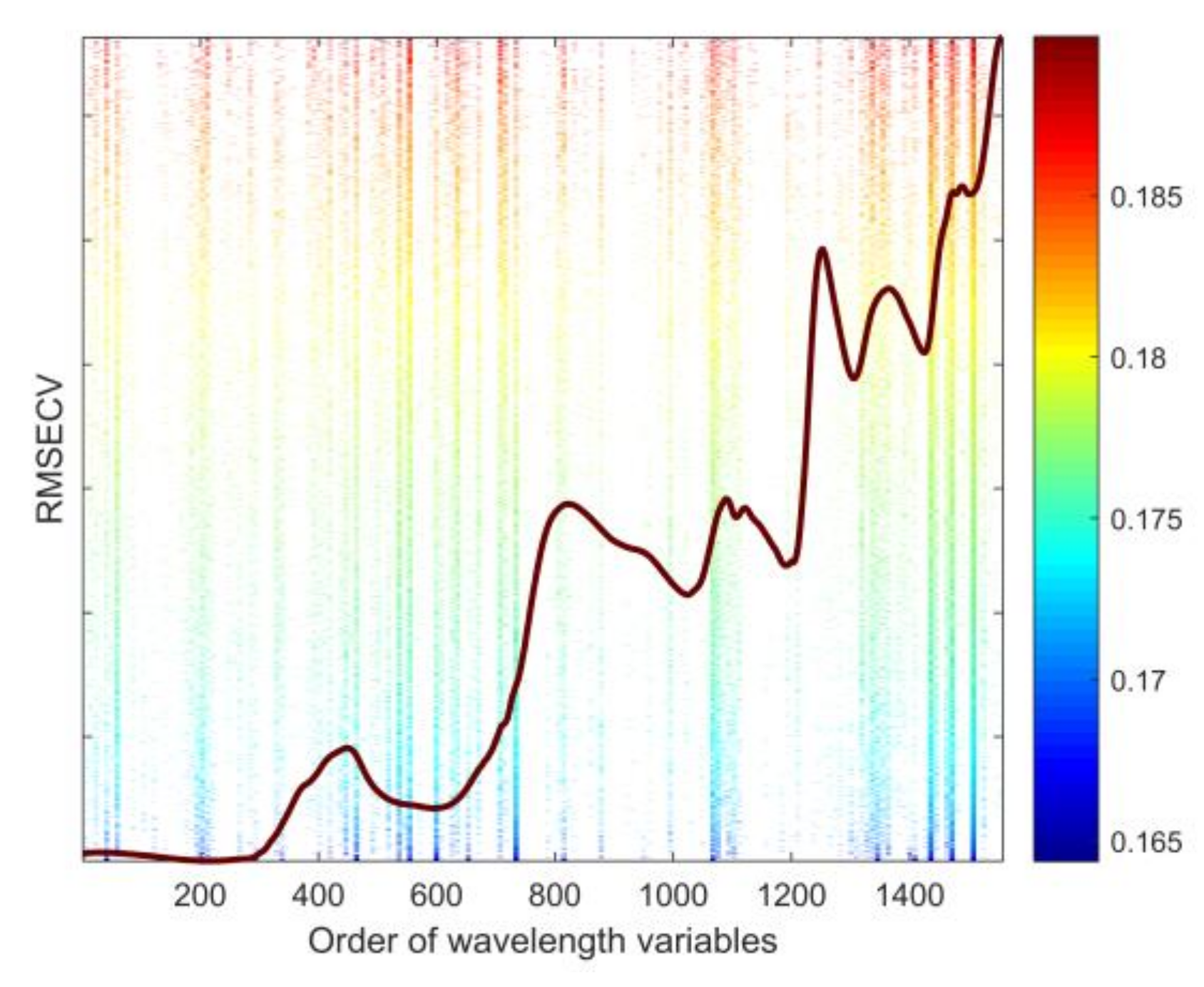
| Cultivar | Training Set | Testing Set | Total |
|---|---|---|---|
| H8 | 250 | 123 | 373 |
| H168 | 250 | 123 | 373 |
| Total | 500 | 246 | 746 |
| Modeling Algorithms | Pre-Processing Methods | Calibration Accuracy | Cross-Validation Accuracy | Prediction Accuracy | Parameters |
|---|---|---|---|---|---|
| KNN | Raw | 88.0% | 88.8% | 90.24% | K = 5 |
| Smoothing | 88.0% | 88.8% | 90.24% | K = 5 | |
| Smoothing + MSC | 99.0% | 99.2% | 97.56% | K = 3 | |
| a S-G 1st | 98.4% | 98.6% | 97.56% | K = 7 | |
| SIMCA | Raw | 99.2% | 99.4% | 98.37% | b PCs1 = 10, c PCs2 = 9 |
| Smoothing | 99.4% | 99.4% | 98.78% | PCs1 = 9, PCs2 = 7 | |
| Smoothing + MSC | 99.4% | 99.2% | 98.78% | PCs1 = 8, PCs2 = 5 | |
| S-G 1st | 96.2% | 96.0% | 95.53% | PCs1 = 6, PCs2 = 9 | |
| PLS-DA | Raw | 99.4% | 99.4% | 97.97% | LVs = 5 |
| Smoothing | 99.4% | 99.4% | 98.37% | LVs = 5 | |
| Smoothing + MSC | 99.6% | 99.4% | 99.19% | LVs = 5 | |
| S-G 1st | 99.4% | 99.2% | 98.78% | LVs = 3 | |
| SVM-DA | Raw | 100% | 99.8% | 99.19% | Cost = 106, gamma = 10−4 |
| Smoothing | 100% | 99.8% | 99.19% | Cost = 106, gamma = 10−4 | |
| Smoothing + MSC | 100% | 100% | 99.59% | Cost = 106, gamma = 10−4 | |
| S-G 1st | 99.6% | 99.2% | 98.78% | Cost = 103, gamma = 10−2 |
| Modeling Algorithms | Calibration Accuracy | Cross-Validation Accuracy | Prediction Accuracy | Parameters |
|---|---|---|---|---|
| KNN | 88.4% | 89.2% | 89.84% | K = 7 |
| SIMCA | 99.8% | 99.8% | 98.78% | PCs1 = 8, PCs2 = 3 |
| PLS-DA | 99.6% | 99.8% | 99.19% | LVs = 8 |
| SVM-DA | 99.8% | 99.6% | 98.78% | cost = 106, gamma = 10−3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, G.; Lü, E.; Wang, N.; Lu, H.; Wang, F.; Zeng, F. Cultivar Classification of Single Sweet Corn Seed Using Fourier Transform Near-Infrared Spectroscopy Combined with Discriminant Analysis. Appl. Sci. 2019, 9, 1530. https://doi.org/10.3390/app9081530
Qiu G, Lü E, Wang N, Lu H, Wang F, Zeng F. Cultivar Classification of Single Sweet Corn Seed Using Fourier Transform Near-Infrared Spectroscopy Combined with Discriminant Analysis. Applied Sciences. 2019; 9(8):1530. https://doi.org/10.3390/app9081530
Chicago/Turabian StyleQiu, Guangjun, Enli Lü, Ning Wang, Huazhong Lu, Feiren Wang, and Fanguo Zeng. 2019. "Cultivar Classification of Single Sweet Corn Seed Using Fourier Transform Near-Infrared Spectroscopy Combined with Discriminant Analysis" Applied Sciences 9, no. 8: 1530. https://doi.org/10.3390/app9081530
APA StyleQiu, G., Lü, E., Wang, N., Lu, H., Wang, F., & Zeng, F. (2019). Cultivar Classification of Single Sweet Corn Seed Using Fourier Transform Near-Infrared Spectroscopy Combined with Discriminant Analysis. Applied Sciences, 9(8), 1530. https://doi.org/10.3390/app9081530






