Abstract
The concept of liquid biopsy has emerged as a novel approach for cancer screening, which is based on the analysis of circulating cancer biomarkers in body fluids. Among the various circulating cancer biomarkers, including Food and Drug Administration (FDA)-approved circulating tumor cells (CTC) and circulating tumor DNA (ctDNA), exosomes have attracted tremendous attention due to their ability to diagnose cancer in its early stages with high efficiency. Recently, surface-enhanced Raman spectroscopy (SERS) has been applied for the detection of cancer exosomes due to its high sensitivity, specificity, and multiplexing capability. In this article, we review recent progress in the development of SERS-based technologies for in vitro identification of circulating cancer exosomes. The accent is made on the detection strategies and interpretation of the SERS data. The problems of detecting cancer-derived exosomes from patient samples and future perspectives of SERS-based diagnostics are also discussed.
1. Introduction
The accurate detection and diagnosis of cancer in the very early stages of its progression is highly important to increase the probability of achieving a successful treatment. Recently, liquid biopsy has emerged as a minimally invasive approach that can potentially be used in medicine for early diagnosis and monitoring of patient’s health. This approach is based on the reliable detection of tumor markers circulating in biological fluids using various techniques. Recently, identification and quantification of circulating tumor cells (CTC), circulating tumor DNA (ctDNA), and exosomes have attracted particular attention for early cancer detection. CTC is one of FDA-approved liquid biopsy methods with relatively high specificity that allows performing in vitro examination by cell culturing as well as phenotypic and genotypic analysis. On the other hand, CTC’s complex heterogeneity, low abundance (1–10 cells/10 mL of peripheral blood of cancer patients [1]), and difficulty to develop an enrichment technology in order to achieve a high limit of detection (LOD) present a challenge for CTC to be a single, universal, and reliable approach for cancer diagnostics in its early stages [2]. Quantification and analysis of ctDNA present in cancer patient’s blood is another Food and Drug Administration (FDA)-approved liquid biopsy method that can become a supplement to conventional biopsies due to its high sensitivity and easy access to genomic information of the patient. Access to such information allows the selection of the most appropriate treatment and to monitor its effectiveness in real-time. In addition, it was shown that ctDNA levels often correlate with tumor burden [3]. However, ctDNA has a short half-life in blood (~2 h) [4] and can interfere with cell-free DNA (cfDNA) that is not related to the tumor [5]. Similar to CTC, current FDA-approved ctDNA assays have low LOD at ~25 copies/mL [6] making its detection and ability to obtain a detailed molecular profile a difficult task. Although detection of CTC and ctDNA can become highly effective approaches for cancer diagnosis, the mentioned challenges impact the feasibility of their implementation for diagnosis especially at the initial stage of cancer. In this respect, analysis of exosomes circulating in the patient’s body may be an innovative and promising method for the diagnosis of cancer and solving the most important problems of sensitivity, specificity, and reliability of currently available diagnostic methods.
Exosomes are stable, membrane-bound nano-vesicles with a diameter of 20–150 nm [7,8] and density ranging 1.15–1.19 g/mL [9], that are secreted into circulation by virtually all cell types including epithelial cells, mesenchymal cells, lymphocytes, and cancer cells [10,11]. Exosomes are transported into the extracellular space as a result of fusion of late multivesicular bodies (MVBs) with the plasma membrane [12,13]. As a result, exosomes entering the extracellular space can be found in any body fluid, including blood (reported concentration ranges 107–1012 exosomes/mL of plasma or serum [14,15,16,17,18]), urine, and saliva [19]. They contain cargo such as nucleic acids (including microRNAs and other non-coding RNAs) and surface proteins inherited from the mother cell [20,21,22,23,24]. The amount and type of such membrane proteins and nucleic acids contained in exosomes are now the main focus of research and discussion [25], as exosomes play an important role in short- and long-range intercellular communication [26,27,28,29]. However, delivery of a distinct type of cargo by tumor-derived exosomes to a normal cell can induce changes and cause the recipient cell to become malignant, causing cancer progression [30]. Finding tumor-derived exosomes in a patient’s sample can allow for diagnosis of cancers in their earliest stage when symptoms are not yet present and dramatically increase the success rate of treatment.
To date, numerous methods have been developed to isolate, detect, and characterize exosome-like vesicles. The most common protocol that is considered to be a gold standard for isolation of exosomes is differential centrifugation, which includes three main steps: low speed centrifugation during a short time period; high speed centrifugation with extended time period to remove cell debris and microvesicles; and ultra-high speed centrifugation, or ultracentrifugation (UC), at forces of 100,000 g or greater to precipitate exosomes [31,32]. An additional centrifugation step, using sucrose or iodixanol to form a density gradient, can be coupled with differential centrifugation to further purify the sample from contaminants [33]. Size-exclusion chromatography [34], immuno-affinity capture [35], filtration [36,37], polymer-based precipitation [38], field-flow fractionation [39], and different combinations of these methods [36,40] have been introduced in recent years to reduce the cost and time needed for exosome isolation and achieve high yield and purity of exosome samples. Microfluidic technology is yet another new and effective technique for the non-invasive separation of exosome-like vesicles preserving the structure and composition of intact exosomes [41,42] with the ability to process and characterize exosomes by using immunolabeled entities simultaneously [43,44,45]. At present, the enzyme-linked immunosorbent assay (ELISA) is the gold standard for exosome quantification using their membrane proteins, such as CD63, CD9, and CD81 [46,47,48]. Although ELISA provides rapid analysis of exosomal membrane proteins with relatively low cost, it is difficult to capture all subpopulations of exosomes containing the protein of interest and perform fast analysis of multiple samples.
To enhance efficiency in the quantification and multiplex detection of exosomes, optical methods have been developed. While nanoparticle tracking analysis (NTA) [49,50], dynamic light scattering (DLS) [51], and flow cytometry (FC) [52,53] can be used to obtain the size distributions and concentrations of exosomes in the fluid samples, fluorescence microscopy is used for in vitro imaging and tracking of exosomes labeled by specific fluorophores [54,55]. Surface plasmon resonance (SPR) provides quantitative information about exosomes attached to the surface of the SPR sensor with an exosome specific antibody [56,57].
One of the most powerful approaches to detect biomolecules with high sensitivity, specificity, and multiplexing capability is surface-enhanced Raman scattering (SERS) [58,59,60]. SERS is also widely used to interrogate conformational changes in molecules under various external conditions [61,62,63]. It is a non-invasive method, which operates in a small volume and is suitable for real-time measurements with minimum sample preparation [64,65,66]. SERS is based on strong amplification of Raman scattering intensity from molecules adsorbed on noble metal (Ag, Au) nanostructures or located in the vicinity of the nanostructure surface [67,68,69]. Specifically, huge SERS enhancement (more than 109–1011) can be achieved when molecules are located within an interparticle junction (“plasmonic hot spot”) or around sharp tips [70,71,72]. There are two generally accepted mechanisms that contribute to SERS enhancement: (i) The electromagnetic mechanism, when local electromagnetic field is increased due to the resonance with the surface plasmons [73]; (ii) and chemical enhancement, which attributes to the charge transfer between a metal nanoparticle and molecule [74]. In addition, a molecule can be stimulated to an excited state by surface plasmons, this mechanism is assigned to molecular resonance or surface-enhanced resonance Raman scattering (SERRS) [75]. In order to obtain extremely high SERS enhancement, tremendous efforts for fabricating different metallic nanostructures to generate the intense electromagnetic field have been made [76,77,78,79], which results in highly effective and reproducible SERS sensors [80,81,82,83]. Biomolecules can be detected by using the SERS method in a label-free manner—directly interacting with SERS substrates, which results in a fingerprint SERS spectrum unique to the biomolecule [68,84]. Alternatively, antibody/aptamer-coated SERS substrates or SERS tags can be used to capture biomolecules from complex biofluids such as blood and serum with high specificity. The SERS signals are then generated by a Raman reporter encapsulated in the SERS tags or the spectral shifts are monitored as a result of the structural deformation of the bond in the event of antibody—antigen binding [85,86,87]. These strategies have been implemented for the detection of relevant biomolecules like proteins, DNA, RNA, and cancer markers, as well as macromolecular structures such as bacterial cells and viruses [88,89,90,91,92,93]. In this respect, SERS is becoming increasingly popular for the detection and analysis of exosome-like vesicles using the above-mentioned approaches. In this review, we summarize the latest achievements and ongoing research in the identification and analysis of exosome-like vesicles using the SERS method. We introduce different types of SERS substrates used for the detection of exosomes, which in most cases determine the detection efficiency and interpretation of SERS data. In addition, we discuss the methodology for detection of exosomes and interpretation of SERS results in terms of sensitivity, specificity, and performance.
2. The Label-Free Approach
The label-free approach is one of major strategies for the detection of biomolecules using the SERS method. Since in the label-free approach a molecule of interest directly interacts with a SERS sensor, the SERS signal comes from the reporter molecule. This provides reliable spectral information about the molecular structure of the analyte without uncertainties due to the labeling of the sensors used in indirect strategy. Using the label-free approach makes it possible to identify the analyte in a complex mixture without a separation step or functionalization of a sensor. In this section, the latest achievements in the identification and analysis of exosome-like vesicles using non-functionalized SERS substrates are discussed. For the convenience of the readers, we have summarized these in Table 1.

Table 1.
Surface-enhanced Raman spectroscopy (SERS) performance in detecting exosomes using label-free approaches.
Stremersch et al. showed a novel approach to identifying single exosome-like vesicles (ELVs) derived from red blood cells (RBC) and B16F10 melanoma cancer cells using the SERS method. The diagnostic approach is based on the electrostatic adsorption of cationic gold nanoparticles (AuNP) onto the anionic surface of ELVs, resulting in AuNP coating around ELVs. To obtain a colloidally stable suspension of AuNP coated ELVs, the optimal AuNP numbers were determined to be 600 AuNP per B16F10 vesicle and 1200 AuNP per RBC vesicle, respectively. SERS spectra of individual ELVs with a high density of AuNP were then acquired along with performing spectral analyses of the obtained Raman spectra by applying two statistical models. Furthermore, the authors were able to discriminate between AuNP coated B16F10 cancerous and RBC-derived ELVs upon mixture at two different ratios, which simulates the in vivo situation. The method appears promising for detecting cancerous ELVs in complex mixtures containing different types of vesicles simultaneously. Although there were some variations in the SERS spectra of individual ELVs, the developed approach is very simple and can be integrated with standard methods, for example, microfluidics enabling automated and rapid SERS diagnostics [94].
The label-free analysis of exosomes derived from the SKOV3 cell line was performed using SERS-active 3D nanobowls by trapping the exosomes into multiple and individual SERS-active 3D nanobowls [95]. Nanobowl-based SERS substrates, illustrated in Figure 1a, provide more reproducible SERS intensities when compared to the nanoparticle-based approaches due to a more uniform “hot-spots” density. Very few exosomes can be trapped into a single nanobowl, which enables the analysis of few particles. As the exosomes stay inside the nanobowls and are intact due to a solution inside the bowls, the SERS signals were exclusively originated from the solution kit used to isolate the exosome. The enhancement is greatest within 5 nm from the nanobowl surface, which corresponds to the lipid membrane thickness and thus allows SERS signals only from the exosome surface. The changes in SERS spectra were observed due to the release of exosomal contents on the bowl surface as a result of exosome damage during drying of the samples. Principal component analysis (PCA) of time-dependent SERS measurements and SERS spectra obtained from ultracentrifugation purified exosomes confirmed the interaction of exosome cargo with the bowl surface.
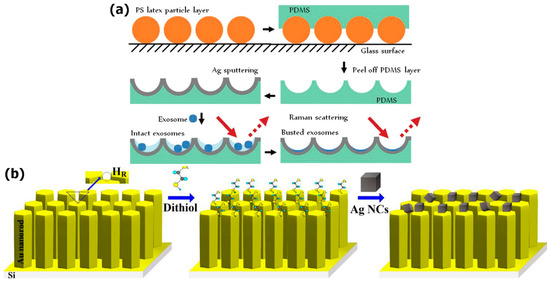
Figure 1.
Surface-enhanced Raman spectroscopy (SERS) substrates fabrication: (a) A nanobowl-like substrate; Reproduced with permission from [95], The Royal Society of Chemistry, 2015; (b) a bimetallic substrate designed as an AuNR array surface with self-assembled AgNC. Reproduced with permission from [96], Elsevier, 2017.
The biochemical composition of normal and lung cancer exosomes was examined by utilizing a hot-spot originating between well-aligned gold nanorods (AuNR) and silver nanocubes (AgNC) as demonstrated in Figure 1b [96]. The optimized SERS substrates have shown the possibility of detecting the exosomes at concentrations 104–105 times lower than the normal blood amount. The peaks of proteins, nucleic acids, and lipids were dominated in the SERS spectra of normal lung exosomes, whereas the contributions from proteins were observed in the SERS spectra of lung cancer exosomes. The spectral differences were also found between the exosomes derived from fibroblast and epithelium cell lines. The time-dependent measurements revealed intense SERS signals from cancer-derived exosomes to that of the SERS signals of exosomes from the normal cell line.
The effective discrimination between exosomes derived from healthy and colorectal cancer cell lines was achieved by utilizing super-hydrophobic silicon micropillars coated with silver nanograins, as shown in Figure 2 [97]. The exosomes were concentrated in the SERS active area due to the reduced friction coefficient, which results in the greatest signal enhancement of Raman exosome signatures compared to other areas on the SERS substrate. The spectral analysis exhibited differences in the biochemical composition of healthy and tumor exosomes. It was found that the exosomes derived from the CCD841-CoN cell line showed higher SERS intensity peaks related to vibrations of lipids, whereas the peaks assigned to proteins and ribonucleic acid bases were dominated in the SERS spectra of exosomes purified from the HCT116 cell line.
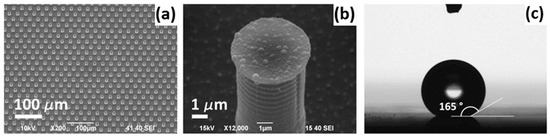
Figure 2.
A super-hydrophobic substrate: (a) A scanning electron microscope (SEM) picture of the substrate made as a periodic hexagonal pattern of cylindrical micropillars; (b) a SEM picture of the single silicon pillar with randomly distributed silver nanograins; (c) a drop put on the implemented super-hydrophobic surface. Reproduced with permission from [97], Elsevier, 2012.
To distinguish non-small cell lung cancer (NSCLC) and healthy cell lines, Park et al. combined a label-free SERS approach and statistical pattern analysis, conducting the whole SERS spectra analysis by PCA instead of taking into account only the positions and intensities of peaks. For SERS signals they used spherical gold nanoparticles. Extracting meaningful patterns from obtained Raman spectra and analyzing first and second principal components (PCA1 and PCA respectively) the authors demonstrated a good distinction between exosomes from lung cancer cells (H522 and H1299) and normal alveolar cells, as shown in Figure 3. However, when applied to blood samples of real patients, this approach did not show a clear separation of the two diseased and two healthy patients. This finding suggests that a real blood sample contains lots of exosomes of different origins and only few exosomes are from cancer cells, so the classification is not clear, but the tendency can be seen, thus it is a subject for future studies [98].

Figure 3.
Exosome classification by principal component analysis (PCA): (a) Principal component scatter plot; (b) highlighting points related to different exosome types. Reproduced with permission from [98], American Chemical Society, 2017.
A similar approach based on PCA was used to investigate the origin of the peaks of NSCLC-derived exosomes and several exosomal surface protein markers using gold aggregated clusters on a glass substrate [99]. To reveal differences between normal and cancerous exosomes (derived from PC9 and H1299 cells), the 26 peaks were evaluated based on the first principal component loading data. The obtained peaks were selected as unique Raman bands. Also, ratiometric analysis revealed that 13 out of 20 Raman bands are well correlated with cancerous exosomes ration, therefore they were assigned as unique Raman bands of NSCLC exosomes. To investigate a correlation between the Raman bands of CD9, CD81, EGFR, and EpCAM protein markers and unique Raman bands of NSCLC exosomes, an antibody-coated SERS substrate was used before and after the conjugation of antigens. Proteins CD9 and CD81 are common on the surfaces of all exosomes, while EGFR and EpCAM are diagnostic and prognostic markers for NSCLC exosomes. The PC1 loading data and the calculation of Euclidian distance revealed that only the EFGP marker showed high similarity with the unique Raman bands of NSCLC exosomes, while the other protein markers had poor similarity. These results were strongly correlated with immunoblotting data.
Combining SERS with multivariate analysis, the Raman spectral signatures of single exosomes derived from different biological sources were measured on a SERS substrate made of a gold periodic pyramid nanostructure with a graphene monolayer on the top (Figure 4) [100]. The SERS spectra of the exosomes isolated by two different methods were studied. It was found that SERS peaks of individual exosomes isolated by ultracentrifugation/filtration can be observed in the SERS spectra, whereas the peaks of the exosomes derived by a commercially available isolation kit, ExoQuick, were absent. For a test on the detection of specificity, exosomes from fetal bovine serum, human serum, and two human lung cancer cell lines were taken. All samples were distinct in their spectral characteristics, mostly by the relative peak intensities.

Figure 4.
A hybrid substrate made of a gold periodic pyramid nanostructure covered with a single-layer of graphene. Reproduced with permission from [100], American Chemical Society, 2019.
A structured polycarbonate substrate from regular recordable disks (CD-R and DVD-R) coated with silver was used as a cost-effective platform for label-free SERS exosome analysis [101]. The substrate fabrication process is shown in Figure 5. The Raman fingerprints of hemoglobin and exosomes and the impact of the silver thickness, the polycarbonate nanostructure, and the excitation wavelength on Raman enhancement were investigated. The results demonstrated the feasibility of the proposed approach for the analysis of complex biomolecular targets. Another potential application is a Raman scanner based on optical disk drives.
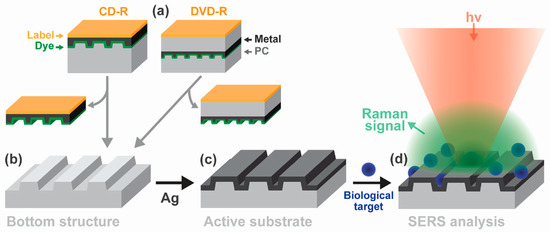
Figure 5.
(a–c) Disk-based SERS-active substrate fabrication; (d) sample analysis. Reproduced with permission from [101], Elsevier, 2017.
Kerr et al. implemented Raman and SERS spectroscopy to study the compositional differences between exosomes derived from ovarian carcinoma cells, cultivated at normal (21%) and reduced (1%) oxygen conditions [102]. Using a trained multivariate algorithm based on PCA and DFA the authors showed biomolecular differences between exosomes derived from normoxic and hypoxic tumor cells. The Raman spectra of parent ovarian cells showed a biomolecular composition similar to that of their corresponding exosomes. Distinct peak differences are demonstrated in Figure 6. Sensitivity and specificity of 100% were achieved by applying multivariate statistical algorithms to Raman microspectrometry, whereas poorer results were obtained when using the SERS method due to damage of the samples caused by the laser.
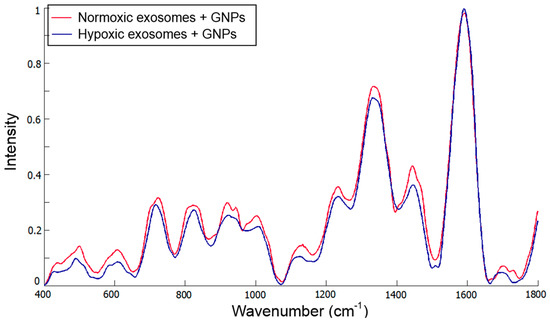
Figure 6.
Mean spectra from normoxic and hypoxic exosomes. Reproduced with permission from [102].
It is worth noting that in all cited articles of this section the authors performing the SERS analysis of cancer-derived exosomes demonstrated exosome spectra only for specific types of cell lines, without extension to all exosomes. Therefore, it is particularly difficult to determine common exosome peaks from SERS spectra. The difference between exosomal cargo was provided either by statistical analysis of a spectrum data set or by recognizing specific peaks of tumor-derived exosomes, which don’t overlap with the peaks of exosomes derived from healthy cell lines. Thus, examination of the general and specific unique peaks of exosomes derived from different types of both cancerous and healthy cells is an urgent need for reliable and efficient SERS diagnosis.
3. Immunolabeling Approach
Although the label-free SERS detection approach represents the simplest method for the identification of biomolecules, there are limitations that can reduce its efficiency, especially when measuring molecules in a complex medium such as blood or serum. In particular, the probability for direct contact between the target molecule and the SERS substrate can be reduced due to absorption of other molecules in the sample. In addition, data analysis can be complicated due to contributions of SERS signals in the SERS spectrum from co-adsorbed molecules. In this respect, the indirect detection approach using antibody-coated SERS substrates and SERS tags increases the degree of specificity and provides specifically strong SERS output signals (Table 2). In this section we review immunolabeling SERS assays that have recently been applied for extraction and identification of exosomes.

Table 2.
SERS performance in detecting exosomes by applying immunolabeling approaches.
A sandwich-type immunocomplex containing/based on antibody-labeled magnetic beads and AuNR has been shown to be an effective approach for the detection of tumor-derived exosomes [103]. Two types of antibodies, anti-CD63 and anti-HER2, were used to recognize two corresponding proteins on the surface of exosomes. The sandwich-type immunocomplex was formed by mixing of AuNR, targeted exosomes, and magnetic beads, as shown in Figure 7. The immunocomplex was then precipitated by а magnet and SERS signals detected in the pellet using 5,5’-Dithiobis(2-nitrobenzoic acid)-labeled AuNR. By using this approach, the authors examined the exosomes derived from human breast cancer cells (SKBR3) and compared them to exosomes isolated from normal human embryonic lung fibroblasts (MRC5). The authors tested the different number of exosomes and found that the proposed approach provides a limit of detection of approximately 1200 exosomes. In addition, the presented method is relatively fast with an operation period/analysis time of 2 h and can be used without the purification of exosomes—for example, by using the cell culture media.
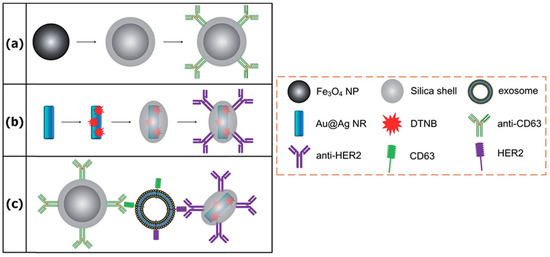
Figure 7.
A scheme of the sandwich-type immunocomplex approach: (a) Magnetic nanobead fabrication; (b) SERS nanoprobe fabrication; (c) sandwich-type structure forming. Reproduced with permission from [103], The Royal Society of Chemistry, 2016.
The selective targeting of exosomes derived from ovarian cancer SKOV3 cells by using an α3ß1 integrin-specific peptide conjugated to silver nanoparticles (AgNP) for SERS-based detection and analysis was demonstrated by Chan’s group [104]. The peculiarity of this approach is demonstrated in Figure 8. The LXY30 peptide binds first to the α3β1 integrin on the SKOV3 exosomes followed by the addition of AgNP enabling formation of the exosome–Ag complex via thiol binding. The successful binding of the exosomes to AgNP was verified by the appearance of several distinct SERS peaks that can be assigned to SKOV3 exosomes. The absence of these peaks in the SERS spectra for AgNP-LXY30-Jurkat-derived exosomes confirmed the efficiency/specificity of the proposed method.
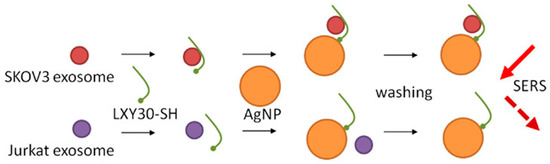
Figure 8.
Selective exosome binding to metallic nanoparticles and SERS detection. Reproduced with permission from [104], John Wiley and Sons, Inc. Copyright © 2000 by John Wiley Sons, Inc, 2017.
Highly sensitive detection of tumor-derived exosomes using the immunocomplex between SERS-active gold nanostars (AuNS) and magnetic beads was reported in a prior study [105]. SERS nanoprobes comprising of 4-mercaptobenzoic acid (4-MBA) as a Raman reporter were functionalized with two cholesterol-modified DNA single strands to form the complex between cholesterol moieties and the exosomal membrane (Figure 9). At the same time, biotin-labeled anti-CD9 was immobilized onto the surfaces of streptavidin-linked magnetic beads for specific capture of CD9-binding exosomes. In the presence of the exosomes, the immunocomplex between magnetic beads, tumor-derived exosomes, and SERS nanoprobes was formed providing strong SERS signals whereas in the absence of the exosomes just poor SERS signals were acquired. The exosomes isolated from liver cancer HepG2 cells were successfully detected with a sensitivity of 27 particles per μL and analysis time of 50 min. The potential of the immunocomplex for detection of clinical samples was also demonstrated.
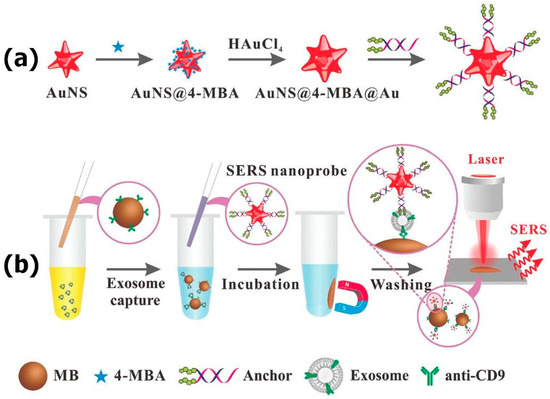
Figure 9.
Mechanisms of (a) AuNS@4-MBA@Au SERS nanoprobe fabrication; (b) exosome detection. Reproduced with permission from [105], The Royal Society of Chemistry, 2018.
Wang et al. developed a SERS method for the identification of several sorts of exosomes simultaneously using magnetic beads as a captured substrate and gold nanoparticles decorated with specific aptamers as a SERS probe (Figure 10). SERS probes were also comprised of three types of Raman reporters providing spectral separation of the SERS signals. HER2 aptamers bind to the HER2 protein, which is a biomarker for SKBR3 breast cancer cells, in the same way PSMA and CEA aptamers recognize LNCaP prostate and T84 colorectal cancer cells, respectively. Gold nanoparticles functionalized with DTNB Raman reporter (strong peak at 1326 cm−1) and HER2 aptamer, 2NAT Raman reporter (one of strong peaks at 1378 cm−1) and PSMA aptamer, and MMC Raman reporter (strong peak at 1170 cm−1) and CEA aptamer allowed simultaneous observation of several sorts of cancerous exosomes in the assay spectrum without spectroscopic overlapping. Upon forming of apta-immunocomplex between magnetic beads, exosomes, and SERS probes, a magnet is applied to collect the pellet followed by SERS measurements of the supernatant. The limit of detection (LOD) of three types of exosomes was found to be 32 SKBR3, 73 T84, and 203 LNCaP exosomes per microliter, respectively. The addition of one exosome sort to the mixture of SERS probes and magnetic beads decreased the SERS intensity of the corresponding probe. The same trend was observed upon the addition of two or three types of exosomes to the mixture. This approach was successfully tested for detection of exosomes derived from blood samples of three patients suffering from/diagnosed with breast, colorectal, and prostate cancers [106].
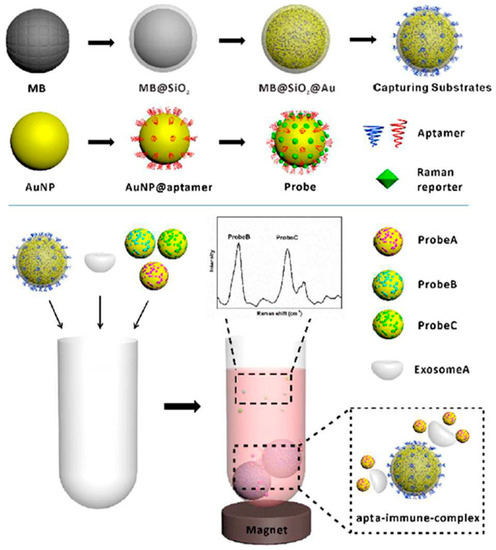
Figure 10.
A scheme of multiple exosome detectionReproduced with permission from [106], The Royal Society of Chemistry, 2018.
Kwizera et al. used a 3D-printed array template and AuNR to implement a simple, cheap, highly efficient, and miniaturized array device. It captures exosomes in a target-specific manner on the antibody array template situated on standard glass microscopy slides coated with silver. Figure 11a demonstrates the measurement sequence: First, the antibody array preparation; then, labeling the captured exosomes by AuNR; and finally, exosome detection. SERS nanotags represent AuNR with the QSY21 Raman reporter incorporated in the cetyltrimethylammonium bromide (CTAB) bilayer. AuNRs attach to exosome lipid membranes due to their opposite charges. Their interaction with the exosome lipid membrane is illustrated in Figure 11b,c. Protein profiling (CD9, CD44, CD 63, EGFR, CD81, EpCAM, HER2, IGF1R) of cancerous and non-cancerous exosomes (derived from breast cancer MM231, MM468, SKBR3, and normal MCF12A cells) demonstrated that exosomes from different cell lines are related to different surface marker profiles. Proteins CD9, CD63, CD81 are present in all considered cell lines. Within breast cancer patients’ serum assays, CD44 was not able to distinguish probes from cancer patients and controls, while EpCAM and HER2 succeeded in this task. Thus, the device provides a quantitative detection of the target proteins and this process has no significant nonspecific interference. The productivity is 80 purified samples per 2 h. The developed method is meant for initially isolated exosomes, but in future it is proposed to allow direct exosome detection in plasma [107].
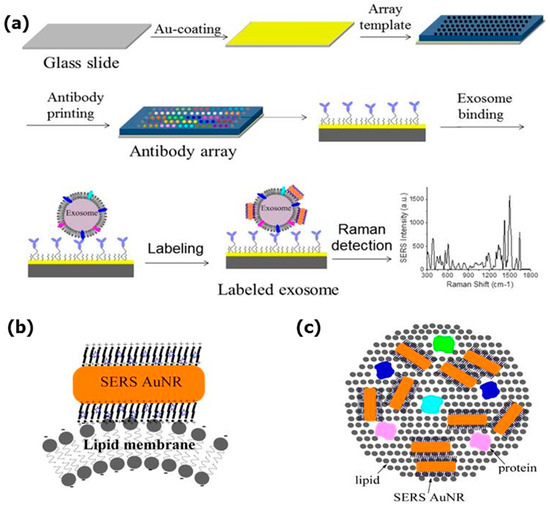
Figure 11.
A miniaturized antibody array-based device for exosome analysis in a target-specific manner: (a) A schematic design of the Raman exosome assay using a 3D-printed array template. AuNR binding to the exosome’s lipid membrane: (b) Side view; (c) top view. Reproduced with permission from [107], Ivyspring International Publisher Pty Ltd. Licensed under CC BY-NC 4.0., 2018.
A high-sensitivity approach for early diagnosis, classification, and metastasis monitoring of pancreatic cancer was developed by Li et al. [108] by using a sandwich structure formed by a polydopamine (PDA)-modified immunocapture substrate, exosomes, and polydopamine-encapsulated antibody-reporter-Ag(shell)-Au(core) multilayer (PEARL) nanotags, as illustrated in Figure 12. Due to a self-polymerizing porous hydrophilic PDA layer, specific antibodies are simultaneously encapsulated and captured by the surface. PDA encapsulation provides higher antibody capture efficiency and exosome detection sensitivity compared with other antibody capture methods. A PEARL tag consists of AuNP as a core, a silver shell, 4-aminobenzenethiol (pATP) as a Raman reporter, BSA, PDA, and antibodies. The intensity of the pATP Raman peak at 1072 cm−1 was chosen as a quantitative signal, because it is one of the strongest peaks in immunoassays’ SERS spectra and is not influenced by any impure surrounding peaks. An optimal low PDA layer thickness in PEARL provides stability and high Raman intensity of the nanotag. Comparing the intensities of anti-CD9, anti-CD63, anti-GPC1, and anti-MIF groups for exosomes derived from cancerous and healthy pancreatic cells revealed that anti-MIF and anti-GPC1 platforms can distinguish between them. The developed immunoassay provides an extra-low detection limit of one exosome in a 2 μL sample. Validation of the immunosensor in diluted clinical serum samples from pancreatic cancer patients proved its capability to discriminate between cancer patients and healthy controls as well as between different cancer stages (the latter can be provided merely by anti-MIF).
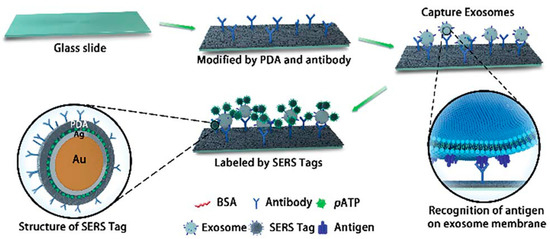
Figure 12.
A scheme of the “chip–exosome–PEARL tag” sandwich-structured sensor. Reproduced with permission from [108], The Royal Society of Chemistry, 2018.
A microfluidic biochip, which is potentially appropriate for exosome analysis, was implemented by Xiong et al. [89]. They used SERS for a rapid parallel analysis of several cancer protein biomarkers in a microfluidic chip based on magnetic nanochains (Magchains), and cellular and subcellular analysis is considered among its potential applications. The bioconjugated Magchains are driven by an external magnetic field and act as both nanoscale stir bars for rapid liquid mixing and capturing agents for bioseparation. The developed magnetic nanochain integrated microfluidic biochip (MiChip) has a simple planar design shown in Figure 13a,b, also it can be easily transformed to a multichannel array for a multiple parallel analysis (Figure 13c). The Magchains are made as superparamagnetic Fe3O4 nanoparticles coated with polydopamine that is self-polymerized under weak basic conditions, providing strong adhesion and forming nanochains, which are then antibody(Ab-I)-functionalized (Figure 13d). The Raman nanotags are AuNR with different Raman reporter molecules, coated with thiolated PEG for surface passivation and functionalized by detection antibodies (Ab-II). The assay process is the following: A mixture of Magchains and SERS probes is injected via the first inlet while a liquid specimen is injected via the second one. Then, they are both transferred and confined in the mixing chamber by two pneumatic valves and their mixing is operated by a spinning magnetic field generated by electromagnetic coils. Hereby sandwich-type immunocomplexes are formed between Magchains, targets of interest, and SERS probes. Finally, valve 2 opens, and the Magchains are magnetically driven to the detection chamber, where Raman spectroscopic detection is provided. By this route, the authors simultaneously detected cancer protein biomarkers for prostate, colorectal, and hepatocellular cancers—prostate-specific antigen (PSA), carcinoembryonic antigen (CEA), and α-fetoprotein (AFP). They also examined clinical serum samples and successfully distinguished patients with these types of cancer.
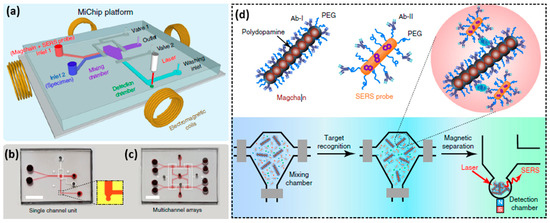
Figure 13.
A magnetic nanochain integrated microfluidic biochip: (a) A design of the platform; (b) a top view for a single channel platform; and (c) a multichannel array; (d) structures of a magnetic nanochain, a SERS probe, and detection algorithm. Reproduced with permission from [89], Licensed under CC BY-NC 4.0, 2018.
Although the immunolabeling strategy for exosome detection using the SERS method has demonstrated high specificity to particular markers that are present on cancer-derived exosomes and more stable signal output in case of SERS nanotags, it still suffers from some shortcomings. For example, this approach is laborious, time-consuming, and relatively expensive. In addition, it usually requires tedious pretreatment and washing procedures. One of the possible solutions of these limitations can be: (i) The use of magnetic beads to reduce washing steps and the enrichment of the targets by an external magnetic field; (ii) more application-specific and robust capture ligands, for instance aptamer-based receptors.
4. Conclusions
In this work we reviewed the latest results when applying the SERS method for isolation, identification, and analysis of exosome-like vesicles. The literature search has shown that these SERS analysis methods can be divided into two: label-free and immunolabeling approaches. While both strategies suffer from some limitations such as spectral overlapping, complex data analysis in the label-free approach, and the difficulties of sensor functionalization with receptor molecules for immunolabeling assay, they were successfully applied for in vitro SERS identification of exosomes isolated from the corresponding cell lines or blood samples from patients. In particular, the label-free SERS regime along with a trained multivariate analysis has shown potential in discriminating between the exosomes isolated from healthy and cancerous cell lines. The possibility to collect the biochemical composition of intact exosomes is another valuable advantage of label-free SERS detection. At the same time, the immunolabeling approach has demonstrated higher specificity in detecting exosomes compared to data obtained in the label-free regime. In addition, antibody functionalized SERS nanotags demonstrated multiplexing capabilities, which are extremely important to probe exosomes in complex media without a purification step or in a sample with different populations of exosomes.
However, despite recent success in the identification of exosome-like vesicles by the SERS method, several problems need to be solved to enable highly effective and rapid detection of tumor-derived exosomes in real patient samples. One of the most difficult problems in exosome detection in clinical applications is to specifically detect tumor-derived exosomes in the samples containing exosomes that are isolated from normal cells. The heterogeneity of disease-specific exosomes and exosomal cargo that eventually influence the accuracy of the SERS analysis is another considerable challenge. The possible solutions to overcome the biological and technical challenges can be: (i) The right selection of the established exosomal markers; (ii) measuring a sufficiently large population of exosomes to differentiate between common exosomal peaks and specific ones; (iii) the development of normative exosomal pools of healthy heterogeneous populations of exosomes; (iv) controlling the spatial localization of SERS sensors on the exosome samples to obtain enhancements from components of interest.
The combination of optical and spectroscopic components, and novel assay design into an integrated system for selective isolation of various exosome subpopulations in heterogeneous samples can substantially enhance the potential of the SERS method for use in practical applications. The integrated optofluidic chip designed for multiwavelength fluorescence and Raman spectroscopy is a good example to be used in the analysis of liquid samples [109]. An important step towards the fabrication of an on-chip SERS platform combining the sensing area, filters, and spectrometer on a single chip was demonstrated in prior research [110]. Furthermore, the reduction of both the size and cost of the Raman spectrometers will enable the development of ultrasensitive, user-friendly, and low-cost miniature integrated systems for on-site SERS diagnosis.
Author Contributions
All of the authors contribute equally to this work.
Funding
This work was supported by Skoltech Translational Research and Innovation Program (for projects 2018/2019).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Alix-Panabières, C.; Pantel, K. Challenges in circulating tumour cell research. Nat. Rev. Cancer 2014, 14, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Sun, Y. Liquid Biopsy: Advances, Limitations and Clinical Applications. JSM Biotechnol. Bioeng. 2017, 4, 404–408. [Google Scholar]
- Schøler, L.V.; Reinert, T.; Ørntoft, M.B.W.; Kassentoft, C.G.; Arnadøttir, S.S.; Vang, S.; Nordentoft, I.; Knudsen, M.; Lamy, P.; Andreasen, D.; et al. Clinical implications of monitoring circulating Tumor DNA in patients with colorectal cancer. Clin. Cancer Res. 2017, 23, 5437–5445. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Su, L.; Qian, C. Circulating tumor DNA: A promising biomarker in the liquid biopsy of cancer. Oncotarget 2015, 48832–48841. [Google Scholar] [CrossRef]
- Johann, D.J.; Steliga, M.; Shin, I.J.; Yoon, D.; Arnaoutakis, K.; Hutchins, L.; Liu, M.; Liem, J.; Walker, K.; Pereira, A.; et al. Liquid biopsy and its role in an advanced clinical trial for lung cancer. Exp. Biol. Med. 2018, 243, 262–271. [Google Scholar] [CrossRef]
- Gale, D.; Lawson, A.R.J.; Howarth, K.; Madi, M.; Durham, B.; Smalley, S.; Calaway, J.; Blais, S.; Jones, G.; Clark, J.; et al. Development of a highly sensitive liquid biopsy platform to detect clinically-relevant cancer mutations at low allele fractions in cellfree DNA. PLoS ONE 2018. [Google Scholar] [CrossRef]
- Martins, T.S.; Catita, J.; Rosa, I.M.; Da Cruz e Silva, O.A.B.; Henriques, A.G. Exosome isolation from distinct biofluids using precipitation and column-based approaches. PLoS ONE 2018, 13, e0198820. [Google Scholar] [CrossRef]
- Chernyshev, V.S.; Rachamadugu, R.; Tseng, Y.H.; Belnap, D.M.; Jia, Y.; Branch, K.J.; Butterfield, A.E.; Pease, L.F.; Bernard, P.S.; Skliar, M. Size and shape characterization of hydrated and desiccated exosomes. Anal. Bioanal. Chem. 2015, 407, 3285–3301. [Google Scholar] [CrossRef]
- Alexander, R.P.; Chiou, N.-T.; Ansel, K.M. Improved exosome isolation by sucrose gradient fractionation of ultracentrifuged crude exosome pellets. Protoc. Exch. 2016. [Google Scholar] [CrossRef]
- Trams, E.G.; Lauter, C.J.; Norman Salem, J.; Heine, U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. BBA Biomembr. 1981, 645, 63–70. [Google Scholar] [CrossRef]
- Palma, J.; Yaddanapudi, S.C.; Pigati, L.; Havens, M.A.; Jeong, S.; Weiner, G.A.; Weimer, K.M.E.; Stern, B.; Hastings, M.L.; Duelli, D.M. MicroRNAs are exported from malignant cells in customized particles. Nucleic Acids Res. 2012, 40, 9125–9138. [Google Scholar] [CrossRef]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Fernando, M.R.; Jiang, C.; Krzyzanowski, G.D.; Ryan, W.L. New evidence that a large proportion of human blood plasma cell-free DNA is localized in exosomes. PLoS ONE 2017, 12. [Google Scholar] [CrossRef]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom. 2013, 14. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Kano, M.; Akutsu, Y.; Hanari, N.; Hoshino, I.; Murakami, K.; Usui, A.; Suito, H.; Takahashi, M.; Otsuka, R.; et al. Quantification of plasma exosome is a potential prognostic marker for esophageal squamous cell carcinoma. Oncol. Rep. 2016, 36, 2535–2543. [Google Scholar] [CrossRef]
- Tamkovich, S.N.; Bakakina, Y.S.; Tutanov, O.S.; Somov, A.K.; Kirushina, N.A.; Dubovskaya, L.V.; Volotovski, I.D.; Laktionov, P.P. Proteome analysis of circulating exosomes in health and breast cancer. Russ. J. Bioorganic Chem. 2017, 43, 126–134. [Google Scholar] [CrossRef]
- Li, M.; Zeringer, E.; Barta, T.; Schageman, J.; Cheng, A.; Vlassov, A.V. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130502. [Google Scholar] [CrossRef]
- de la Torre Gomez, C.; Goreham, R.V.; Bech Serra, J.J.; Nann, T.; Kussmann, M. “Exosomics”-A review of biophysics, biology and biochemistry of exosomes with a focus on human breast milk. Front. Genet. 2018, 9. [Google Scholar] [CrossRef]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Simons, M.; Raposo, G. Exosomes-vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009, 21, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Kharaziha, P.; Ceder, S.; Li, Q.; Panaretakis, T. Tumor cell-derived exosomes: A message in a bottle. Biochim. Biophys. Acta Rev. Cancer 2012, 1826, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta Gen. Subj. 2012, 1820, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Chevillet, J.R.; Kang, Q.; Ruf, I.K.; Briggs, H.A.; Vojtech, L.N.; Hughes, S.M.; Cheng, H.H.; Arroyo, J.D.; Meredith, E.K.; Gallichotte, E.N.; et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. USA 2014, 111, 14888–14893. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, S.; Weber, J.; Baxter, D.; Galas, D.J. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010, 38, 7248–7259. [Google Scholar] [CrossRef]
- Ge, R.; Tan, E.; Sharghi-Namini, S.; Asada, H.H. Exosomes in cancer microenvironment and beyond: Have we overlooked these extracellular messengers? Cancer Microenviron. 2012, 5, 323–332. [Google Scholar] [CrossRef]
- Pucci, F.; Pittet, M.J. Molecular pathways: Tumor-derived microvesicles and their interactions with immune cells In vivo. Clin. Cancer Res. 2013, 19, 2598–2604. [Google Scholar] [CrossRef]
- Regev-Rudzki, N.; Wilson, D.W.; Carvalho, T.G.; Sisquella, X.; Coleman, B.M.; Rug, M.; Bursac, D.; Angrisano, F.; Gee, M.; Hill, A.F.; et al. Cell-cell communication between malaria-infected red blood cells via exosome-like vesicles. Cell 2013, 153, 1120–1133. [Google Scholar] [CrossRef]
- Whiteside, T.L. Tumor-Derived Exosomes and Their Role in Cancer Progression. In Advances in Clinical Chemistry; Elsevier Science: Amsterdam, The Netherlands, 2016; ISBN 9780128046890. [Google Scholar]
- Witwer, K.W.; Buzás, E.I.; Bemis, L.T.; Bora, A.; Lässer, C.; Lötvall, J.; Nolte-’t Hoen, E.N.; Piper, M.G.; Sivaraman, S.; Skog, J.; et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef]
- Taylor, D.D.; Shah, S. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods 2015, 87, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Cantin, R.; Diou, J.; Bélanger, D.; Tremblay, A.M.; Gilbert, C. Discrimination between exosomes and HIV-1: Purification of both vesicles from cell-free supernatants. J. Immunol. Methods 2008, 338, 21–30. [Google Scholar] [CrossRef]
- Baranyai, T.; Herczeg, K.; Onódi, Z.; Voszka, I.; Módos, K.; Marton, N.; Nagy, G.; Mäger, I.; Wood, M.J.; El Andaloussi, S.; et al. Isolation of exosomes from blood plasma: Qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods. PLoS ONE 2015, 10, e0145686. [Google Scholar] [CrossRef] [PubMed]
- Tauro, B.J.; Greening, D.W.; Mathias, R.A.; Ji, H.; Mathivanan, S.; Scott, A.M.; Simpson, R.J. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 2012, 56, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Nordin, J.Z.; Lee, Y.; Vader, P.; Mäger, I.; Johansson, H.J.; Heusermann, W.; Wiklander, O.P.B.; Hällbrink, M.; Seow, Y.; Bultema, J.J.; et al. Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 879–883. [Google Scholar] [CrossRef]
- Merchant, M.L.; Powell, D.W.; Wilkey, D.W.; Cummins, T.D.; Deegens, J.K.; Rood, I.M.; McAfee, K.J.; Fleischer, C.; Klein, E.; Klein, J.B. Microfiltration isolation of human urinary exosomes for characterization by MS. Proteom. Clin. Appl. 2010, 4, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Rekker, K.; Saare, M.; Roost, A.M.; Kubo, A.L.; Zarovni, N.; Chiesi, A.; Salumets, A.; Peters, M. Comparison of serum exosome isolation methods for microRNA profiling. Clin. Biochem. 2014, 47, 135–138. [Google Scholar] [CrossRef]
- Petersen, K.E.; Shiri, F.; White, T.; Bardi, G.T.; Sant, H.; Gale, B.K.; Hood, J.L. Exosome Isolation: Cyclical Electrical Field Flow Fractionation in Low-Ionic-Strength Fluids. Anal. Chem. 2018, 90, 12783–12790. [Google Scholar] [CrossRef]
- Yamada, T.; Inoshima, Y.; Matsuda, T.; Ishiguro, N. Comparison of Methods for Isolating Exosomes from Bovine Milk. J. Vet. Med. Sci. 2012, 74, 1523–1525. [Google Scholar] [CrossRef]
- Wu, M.; Ouyang, Y.; Wang, Z.; Zhang, R.; Huang, P.-H.; Chen, C.; Li, H.; Li, P.; Quinn, D.; Dao, M.; et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc. Natl. Acad. Sci. USA 2017, 114, 10584–10589. [Google Scholar] [CrossRef]
- Liu, C.; Guo, J.; Tian, F.; Yang, N.; Yan, F.; Ding, Y.; Wei, J.; Hu, G.; Nie, G.; Sun, J. Field-Free Isolation of Exosomes from Extracellular Vesicles by Microfluidic Viscoelastic Flows. ACS Nano 2017, 11, 6968–6976. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Yang, Y.; Zeng, Y.; He, M. A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab Chip 2016, 16, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Kanwar, S.S.; Dunlay, C.J.; Simeone, D.M.; Nagrath, S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip 2014, 14, 1891–1900. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Crow, J.; Roth, M.; Zeng, Y.; Godwin, A.K. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip Miniat. Chem. Biol. 2014, 14, 3773–3780. [Google Scholar] [CrossRef]
- Logozzi, M.; De Milito, A.; Lugini, L.; Borghi, M.; Calabrò, L.; Spada, M.; Perdicchio, M.; Marino, M.L.; Federici, C.; Iessi, E.; et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS ONE 2009, 4, e5219. [Google Scholar] [CrossRef]
- Khan, S.; Bennit, H.F.; Turay, D.; Perez, M.; Mirshahidi, S.; Yuan, Y.; Wall, N.R. Early diagnostic value of survivin and its alternative splice variants in breast cancer. BMC Cancer 2014, 14, 176. [Google Scholar] [CrossRef]
- Lamparski, H.G.; Metha-Damani, A.; Yao, J.Y.; Patel, S.; Hsu, D.H.; Ruegg, C.; Le Pecq, J.B. Production and characterization of clinical grade exosomes derived from dendritic cells. J. Immunol. Methods 2002, 270, 211–226. [Google Scholar] [CrossRef]
- Dragovic, R.A.; Gardiner, C.; Brooks, A.S.; Tannetta, D.S.; Ferguson, D.J.P.; Hole, P.; Carr, B.; Redman, C.W.G.; Harris, A.L.; Dobson, P.J.; et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomed. Nanotechnol. Biol. Med. 2011, 7, 780–788. [Google Scholar] [CrossRef]
- Gardiner, C.; Ferreira, Y.J.; Dragovic, R.A.; Redman, C.W.G.; Sargent, I.L. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef]
- Akers, J.C.; Ramakrishnan, V.; Nolan, J.P.; Duggan, E.; Fu, C.C.; Hochberg, F.H.; Chen, C.C.; Carter, B.S. Comparative analysis of technologies for quantifying extracellular vesicles (EVs) in clinical cerebrospinal fluids (CSF). PLoS ONE 2016, 11, e0149866. [Google Scholar] [CrossRef]
- Nolan, J.P. Flow cytometry of extracellular vesicles: Potential, pitfalls, and prospects. Curr. Protoc. Cytom. 2015, 73, 13.14.1–13.14.16. [Google Scholar]
- Van Der Pol, E.; Van Gemert, M.J.C.; Sturk, A.; Nieuwland, R.; Van Leeuwen, T.G. Single vs. swarm detection of microparticles and exosomes by flow cytometry. J. Thromb. Haemost. 2012, 10, 919–930. [Google Scholar] [CrossRef]
- Tian, T.; Zhu, Y.L.; Hu, F.H.; Wang, Y.Y.; Huang, N.P.; Xiao, Z.D. Dynamics of exosome internalization and trafficking. J. Cell. Physiol. 2013, 228, 1487–1495. [Google Scholar] [CrossRef]
- Suetsugu, A.; Honma, K.; Saji, S.; Moriwaki, H.; Ochiya, T.; Hoffman, R.M. Imaging exosome transfer from breast cancer cells to stroma at metastatic sites in orthotopic nude-mouse models. Adv. Drug Deliv. Rev. 2013, 65, 383–390. [Google Scholar] [CrossRef]
- Im, H.; Shao, H.; Park, Y.I.; Peterson, V.M.; Castro, C.M.; Weissleder, R.; Lee, H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014, 32, 490–495. [Google Scholar] [CrossRef]
- Sina, A.A.I.; Vaidyanathan, R.; Dey, S.; Carrascosa, L.G.; Shiddiky, M.J.A.; Trau, M. Real time and label free profiling of clinically relevant exosomes. Sci. Rep. 2016, 6, 30460. [Google Scholar] [CrossRef]
- Alvarez-Puebla, R.; Liz-Marzán, L.M.; García De Abajo, F.J. Light concentration at the nanometer scale. J. Phys. Chem. Lett. 2010, 1, 2428–2434. [Google Scholar] [CrossRef]
- Kahraman, M.; Mullen, E.R.; Korkmaz, A.; Wachsmann-Hogiu, S. Fundamentals and applications of SERS-based bioanalytical sensing. Nanophotonics 2017, 6, 831–852. [Google Scholar] [CrossRef]
- Wang, X.; Qian, X.; Beitler, J.J.; Chen, Z.G.; Khuri, F.R.; Lewis, M.M.; Shin, H.J.C.; Nie, S.; Shin, D.M. Detection of circulating tumor cells in human peripheral blood using surface-enhanced raman scattering nanoparticles. Cancer Res. 2011, 71, 1526–1532. [Google Scholar] [CrossRef]
- Cui, Q.; Yashchenok, A.; Zhang, L.; Li, L.; Masic, A.; Wienskol, G.; Möhwald, H.; Bargheer, M. Fabrication of bifunctional gold/gelatin hybrid nanocomposites and their application. ACS Appl. Mater. Interfaces 2014, 6, 1999–2002. [Google Scholar] [CrossRef]
- Han, X.; Wang, H.; Ou, X.; Zhang, X. Silicon nanowire-based surface-enhanced Raman spectroscopy endoscope for intracellular pH detection. ACS Appl. Mater. Interfaces 2013, 5, 5811–5814. [Google Scholar] [CrossRef]
- Kong, K.V.; Dinish, U.S.; Lau, W.K.O.; Olivo, M. Sensitive SERS-pH sensing in biological media using metal carbonyl functionalized planar substrates. Biosens. Bioelectron. 2014, 54, 135–140. [Google Scholar] [CrossRef]
- Yashchenok, A.; Masic, A.; Gorin, D.; Shim, B.S.; Kotov, N.A.; Fratzl, P.; Möhwald, H.; Skirtach, A. Nanoengineered colloidal probes for raman-based detection of biomolecules inside living cells. Small 2013, 9, 351–356. [Google Scholar] [CrossRef]
- Radziuk, D.; Moehwald, H. Highly effective hot spots for SERS signatures of live fibroblasts. Nanoscale 2014, 6, 6115–6126. [Google Scholar] [CrossRef]
- Wuytens, P.C.; Subramanian, A.Z.; De Vos, W.H.; Skirtach, A.G.; Baets, R. Gold nanodome-patterned microchips for intracellular surface-enhanced Raman spectroscopy. Analyst 2015, 40, 8080–8087. [Google Scholar] [CrossRef]
- Harper, M.M.; Dougan, J.A.; Shand, N.C.; Grahama, D.; Faulds, K. Detection of SERS active labelled DNA based on surface affinity to silver nanoparticles. Analyst 2012, 137, 2063–2068. [Google Scholar] [CrossRef]
- Xu, L.J.; Zong, C.; Zheng, X.S.; Hu, P.; Feng, J.M.; Ren, B. Label-free detection of native proteins by surface-enhanced Raman spectroscopy using iodide-modified nanoparticles. Anal. Chem. 2014, 86, 2238–2245. [Google Scholar] [CrossRef]
- Tian, L.; Fei, M.; Tadepalli, S.; Morrissey, J.J.; Kharasch, E.D.; Singamaneni, S. Bio-Enabled Gold Superstructures with Built-In and Accessible Electromagnetic Hotspots. Adv. Healthc. Mater. 2015, 4, 1502–1509. [Google Scholar] [CrossRef]
- Emory, S.; Nie, S.M. Near-Field Surface-Enhanced Raman Spectroscopy on Single Silver Nanoparticles. Anal. Chem. 1997, 69, 2631–2635. [Google Scholar] [CrossRef]
- Etchegoin, P.G.; Le Ru, E.C. A perspective on single molecule SERS: Current status and future challenges. Phys. Chem. Chem. Phys. 2008, 10, 6079–6089. [Google Scholar] [CrossRef]
- Cui, Q.; Yashchenok, A.; Li, L.; Möhwald, H.; Bargheer, M. Mechanistic study on reduction reaction of nitro compounds catalyzed by gold nanoparticles using in situ SERS monitoring. Colloids Surf. A Physicochem. Eng. Aspects 2015, 470, 108–113. [Google Scholar] [CrossRef]
- Jeanmaire, D.L.; Van Duyne, R.P. Surface raman spectroelectrochemistry: Part, I. Heterocyclic, aromatic, and aliphatic amines adsorbed on the anodized silver electrode. J. Electroanal. Chem. Interf. Electr. 1977, 84, 1–20. [Google Scholar] [CrossRef]
- Otto, A. Surface enhanced Raman scattering (SERS), What do we know? Appl. Surf. Sci. 1980, 6, 309–355. [Google Scholar] [CrossRef]
- Yamamoto, Y.S.; Ishikawa, M.; Ozaki, Y.; Itoh, T. Fundamental studies on enhancement and blinking mechanism of surface-enhanced Raman scattering (SERS) and basic applications of SERS biological sensing. Front. Phys. 2014, 9, 31–46. [Google Scholar] [CrossRef]
- Haynes, C.L.; Van Duyne, R.P. Nanosphere lithography: A versatile nanofabrication tool for studies of size-dependent nanoparticle optics. J. Phys. Chem. B 2001, 105, 5599–5611. [Google Scholar] [CrossRef]
- Merk, V.; Kneipp, J.; Leosson, K. Gap size reduction and increased SERS enhancement in lithographically patterned nanoparticle arrays by templated growth. Adv. Opt. Mater. 2013, 1, 313–318. [Google Scholar] [CrossRef]
- Keating, M.; Song, S.; Wei, G.; Graham, D.; Chen, Y.; Placido, F. Ordered silver and copper nanorod arrays for enhanced Raman scattering created via guided oblique angle deposition on polymer. J. Phys. Chem. C 2014, 118, 4878–4884. [Google Scholar] [CrossRef]
- Chen, B.S.; Meng, G.W.; Zhou, F.; Huang, Q.; Zhu, C.H.; Hu, X.Y.; Kong, M.G. Ordered arrays of Au-nanobowls loaded with Ag-nanoparticles as effective SERS substrates for rapid detection of PCBs. Nanotechnology 2014, 25, 145605. [Google Scholar] [CrossRef]
- Pazos-Perez, N.; Ni, W.; Schweikart, A.; Alvarez-Puebla, R.A.; Fery, A.; Liz-Marzan, L.M. Highly uniform SERS substrates formed by wrinkle-confined drying of gold colloids. Chem. Sci. 2010, 1, 174–178. [Google Scholar] [CrossRef]
- Wu, K.Y.; Rindzevicius, T.; Schmidt, M.S.; Mogensen, K.B.; Hakonen, A.; Boisen, A. Wafer-scale leaning silver nanopillars for molecular detection at ultra-low concentrations. J. Phys. Chem. C 2015, 119, 2053–2062. [Google Scholar] [CrossRef]
- Rycenga, M.; Xia, X.; Moran, C.H.; Zhou, F.; Qin, D.; Li, Z.-Y.; Xia, Y. Generation of hot spots with silver nanocubes for single-molecule detection by surface-enhanced Raman scattering. Angew. Chem. Int. Ed. 2011, 50, 5473–5477. [Google Scholar] [CrossRef]
- Matteini, P.; Cottat, M.; Tavanti, F.; Panfilova, E.; Scuderi, M.; Nicotra, G.; Menziani, M.C.; Khlebtsov, N.; de Angelis, M.; Pini, R. Site-Selective Surface-Enhanced Raman Detection of Proteins. ACS Nano 2017, 11, 918–926. [Google Scholar] [CrossRef]
- Severyukhina, A.N.; Parakhonskiy, B.V.; Prikhozhdenko, E.S.; Gorin, D.A.; Sukhorukov, G.B.; Möhwald, H.; Yashchenok, A.M. Nanoplasmonic Chitosan Nanofibers as Effective SERS Substrate for Detection of Small Molecules. ACS Appl. Mater. Interfaces 2015, 7, 15466–15473. [Google Scholar] [CrossRef]
- Qian, X.; Peng, X.H.; Ansari, D.O.; Yin-Goen, Q.; Chen, G.Z.; Shin, D.M.; Yang, L.; Young, A.N.; Wang, M.D.; Nie, S. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat. Biotechnol. 2008, 26, 83–90. [Google Scholar] [CrossRef]
- Bohndiek, S.E.; Wagadarikar, A.; Zavaleta, C.L.; Van de Sompel, D.; Garai, E.; Jokerst, J.V.; Yazdanfar, S.; Gambhir, S.S. A small animal Raman instrument for rapid, wide-area, spectroscopic imaging. Proc. Natl. Acad. Sci. USA 2013, 110, 12408–12413. [Google Scholar] [CrossRef]
- Panikkanvalappil, S.R.; MacKey, M.A.; El-Sayed, M.A. Probing the unique dehydration-induced structural modifications in cancer cell DNA using surface enhanced Raman spectroscopy. J. Am. Chem. Soc. 2013, 135, 4815–4821. [Google Scholar] [CrossRef]
- Prikhozhdenko, E.S.; Atkin, V.S.; Parakhonskiy, B.V.; Rybkin, I.A.; Lapanje, A.; Sukhorukov, G.B.; Gorin, D.A.; Yashchenok, A.M. New post-processing method of preparing nanofibrous SERS substrates with a high density of silver nanoparticles. RSC Adv. 2016, 6, 84505–84511. [Google Scholar] [CrossRef]
- Xiong, Q.; Lim, C.Y.; Ren, J.; Zhou, J.; Pu, K.; Chan-Park, M.B.; Mao, H.; Lam, Y.C.; Duan, H. Magnetic nanochain integrated microfluidic biochips. Nat. Commun. 2018, 9, 1743. [Google Scholar] [CrossRef]
- Lin, D.; Gong, T.; Qiu, S.; Wu, Q.; Tseng, C.-Y.; Kong, K.V.; Chen, G.; Chen, R. A dual signal amplification nanosensor based on SERS technology for detection of tumor-related DNA. Chem. Commun. 2019, 55, 1548–1551. [Google Scholar] [CrossRef]
- Wang, Y.; Salehi, M.; Schütz, M.; Schlücker, S. Femtogram detection of cytokines in a direct dot-blot assay using SERS microspectroscopy and hydrophilically stabilized Au-Ag nanoshells. Chem. Commun. 2014, 50, 2711–2714. [Google Scholar] [CrossRef]
- Chao, J.; Cao, W.; Su, S.; Weng, L.; Song, S.; Fan, C.; Wang, L. Nanostructure-based surface-enhanced Raman scattering biosensors for nucleic acids and proteins. J. Mater. Chem. B 2016, 4, 1757–1769. [Google Scholar] [CrossRef]
- Vo-Dinh, T.; Allain, L.R.; Stokes, D.L. Cancer gene detection using surface-enhanced Raman scattering (SERS). J. Raman Spectrosc. 2002, 33, 511–516. [Google Scholar] [CrossRef]
- Stremersch, S.; Marro, M.; Pinchasik, B.-E.; Baatsen, P.; Hendrix, A.; De Smedt, S.C.; Loza-Alvarez, P.; Skirtach, A.G.; Raemdonck, K.; Braeckmans, K. Identification of Individual Exosome-Like Vesicles by Surface Enhanced Raman Spectroscopy. Small 2016, 12, 3292–3301. [Google Scholar] [CrossRef]
- Lee, C.; Carney, R.P.; Hazari, S.; Smith, Z.J.; Knudson, A.; Robertson, C.S.; Lam, K.S.; Wachsmann-Hogiu, S. 3D plasmonic nanobowl platform for the study of exosomes in solution. Nanoscale 2015, 7, 9290–9297. [Google Scholar] [CrossRef]
- Sivashanmugan, K.; Huang, W.L.; Lin, C.H.; Der Liao, J.; Lin, C.C.; Su, W.C.; Wen, T.C. Bimetallic nanoplasmonic gap-mode SERS substrate for lung normal and cancer-derived exosomes detection. J. Taiwan Inst. Chem. Eng. 2017, 80, 149–155. [Google Scholar] [CrossRef]
- Tirinato, L.; Gentile, F.; Di Mascolo, D.; Coluccio, M.L.; Das, G.; Liberale, C.; Pullano, S.A.; Perozziello, G.; Francardi, M.; Accardo, A.; et al. SERS analysis on exosomes using super-hydrophobic surfaces. Microelectron. Eng. 2012, 97, 337–340. [Google Scholar] [CrossRef]
- Park, J.; Hwang, M.; Choi, B.; Jeong, H.; Jung, J.H.; Kim, H.K.; Hong, S.; Park, J.H.; Choi, Y. Exosome Classification by Pattern Analysis of Surface-Enhanced Raman Spectroscopy Data for Lung Cancer Diagnosis. Anal. Chem. 2017, 89, 6695–6701. [Google Scholar] [CrossRef]
- Shin, H.; Jeong, H.; Park, J.; Hong, S.; Choi, Y. Correlation between Cancerous Exosomes and Protein Markers Based on Surface-Enhanced Raman Spectroscopy (SERS) and Principal Component Analysis (PCA). ACS Sens. 2018, 3, 2637–2643. [Google Scholar] [CrossRef]
- Yan, Z.; Dutta, S.; Liu, Z.; Yu, X.; Mesgarzadeh, N.; Ji, F.; Bitan, G.; Xie, Y.-H. A Label-Free Platform for Identification of Exosomes from Different Sources. ACS Sens. 2019, 4, 488–497. [Google Scholar] [CrossRef]
- Avella-Oliver, M.; Puchades, R.; Wachsmann-Hogiu, S.; Maquieira, A. Label-free SERS analysis of proteins and exosomes with large-scale substrates from recordable compact disks. Sens. Actuators B Chem. 2017, 252, 657–662. [Google Scholar] [CrossRef]
- Kerr, L.T.; Gubbins, L.; Weiner Gorzel, K.; Sharma, S.; Kell, M.; McCann, A.; Hennelly, B.M. Raman spectroscopy and SERS analysis of ovarian tumour derived exosomes (TEXs): A preliminary study. Proc. SPIE 2014, 9129, 9. [Google Scholar]
- Zong, S.; Wang, L.; Chen, C.; Lu, J.; Zhu, D.; Zhang, Y.; Wang, Z.; Cui, Y. Facile detection of tumor-derived exosomes using magnetic nanobeads and SERS nanoprobes. Anal. Methods 2016, 8, 5001–5008. [Google Scholar] [CrossRef]
- Lee, C.; Carney, R.; Lam, K.; Chan, J.W. SERS analysis of selectively captured exosomes using an integrin-specific peptide ligand. J. Raman Spectrosc. 2017, 48, 1771–1776. [Google Scholar] [CrossRef]
- Tian, Y.F.; Ning, C.F.; He, F.; Yin, B.C.; Ye, B.C. Highly sensitive detection of exosomes by SERS using gold nanostar@Raman reporter@nanoshell structures modified with a bivalent cholesterol-labeled DNA anchor. Analyst 2018, 143, 4915–4922. [Google Scholar]
- Wang, Z.; Zong, S.; Wang, Y.; Li, N.; Li, L.; Lu, J.; Wang, Z.; Chen, B.; Cui, Y. Screening and multiple detection of cancer exosomes using an SERS-based method. Nanoscale 2018, 10, 9053–9062. [Google Scholar] [CrossRef]
- Kwizera, E.A.; O’Connor, R.; Vinduska, V.; Williams, M.; Butch, E.R.; Snyder, S.E.; Chen, X.; Huang, X. Molecular Detection and Analysis of Exosomes Using Surface-Enhanced Raman Scattering Gold Nanorods and a Miniaturized Device. Theranostics 2018, 8, 2722–2738. [Google Scholar] [CrossRef]
- Li, T.-D.; Zhang, R.; Chen, H.; Huang, Z.-P.; Ye, X.; Wang, H.; Deng, A.-M.; Kong, J.-L. An ultrasensitive polydopamine bi-functionalized SERS immunoassay for exosome-based diagnosis and classification of pancreatic cancer. Chem. Sci. 2018, 9, 5372–5382. [Google Scholar] [CrossRef]
- Persichetti, G.; Grimaldi, I.A.; Testa, G.; Bernini, R. Multifunctional optofluidic lab-on-chip platform for Raman and fluorescence spectroscopic microfluidic analysis. Lab Chip 2017, 17, 2631–2639. [Google Scholar] [CrossRef]
- Wuytens, P.C.; Skirtach, A.G.; Baets, R. On-chip surface-enhanced Raman spectroscopy using nanosphere-lithography patterned antennas on silicon nitride waveguides. Opt. Express 2017, 25, 12926–12934. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).