Intelligent Microarray Data Analysis through Non-negative Matrix Factorization to Study Human Multiple Myeloma Cell Lines
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture
2.3. Western Blotting
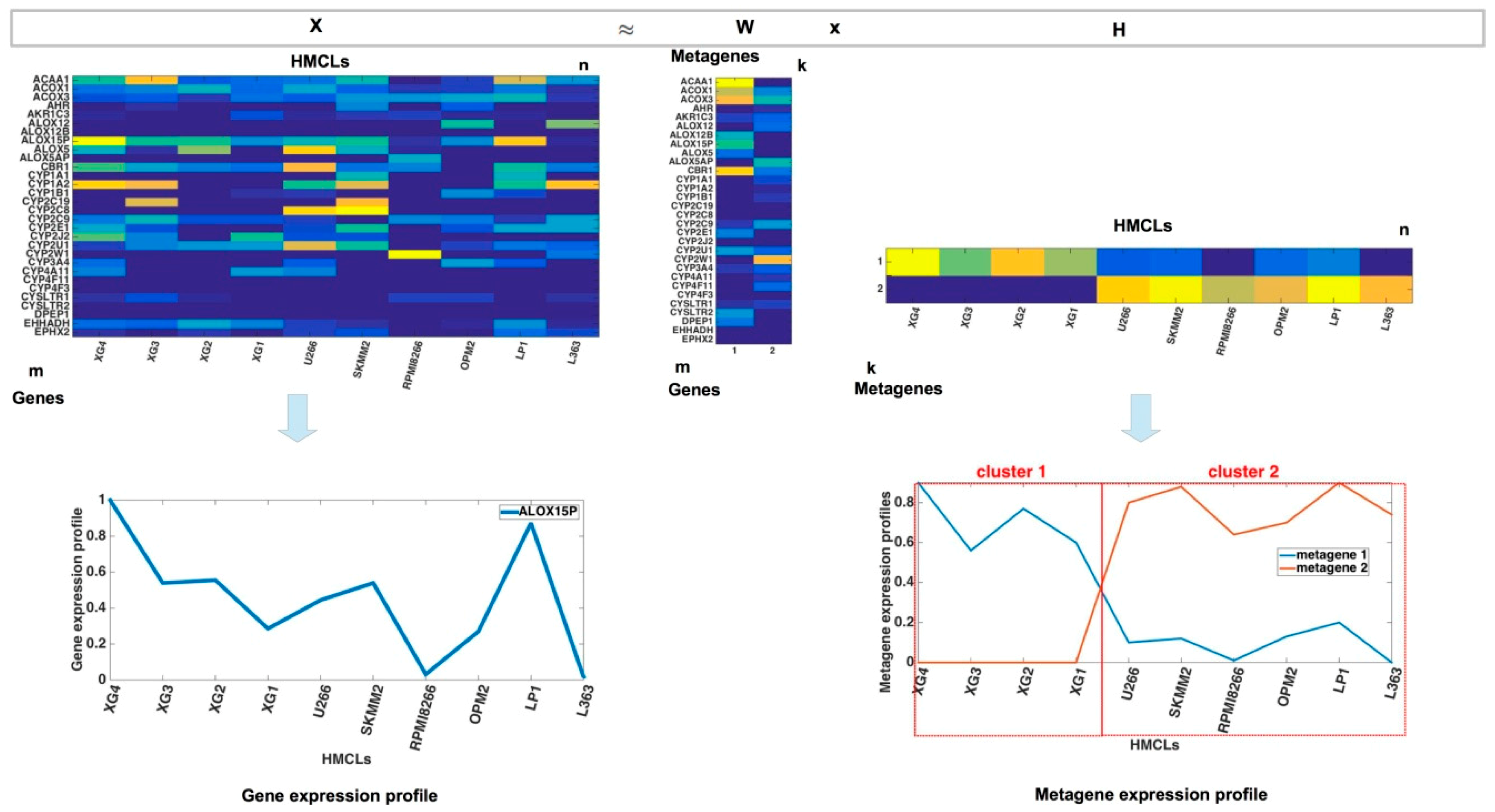
2.4. Non-Negative Matrix Factorization
3. Results and Discussion
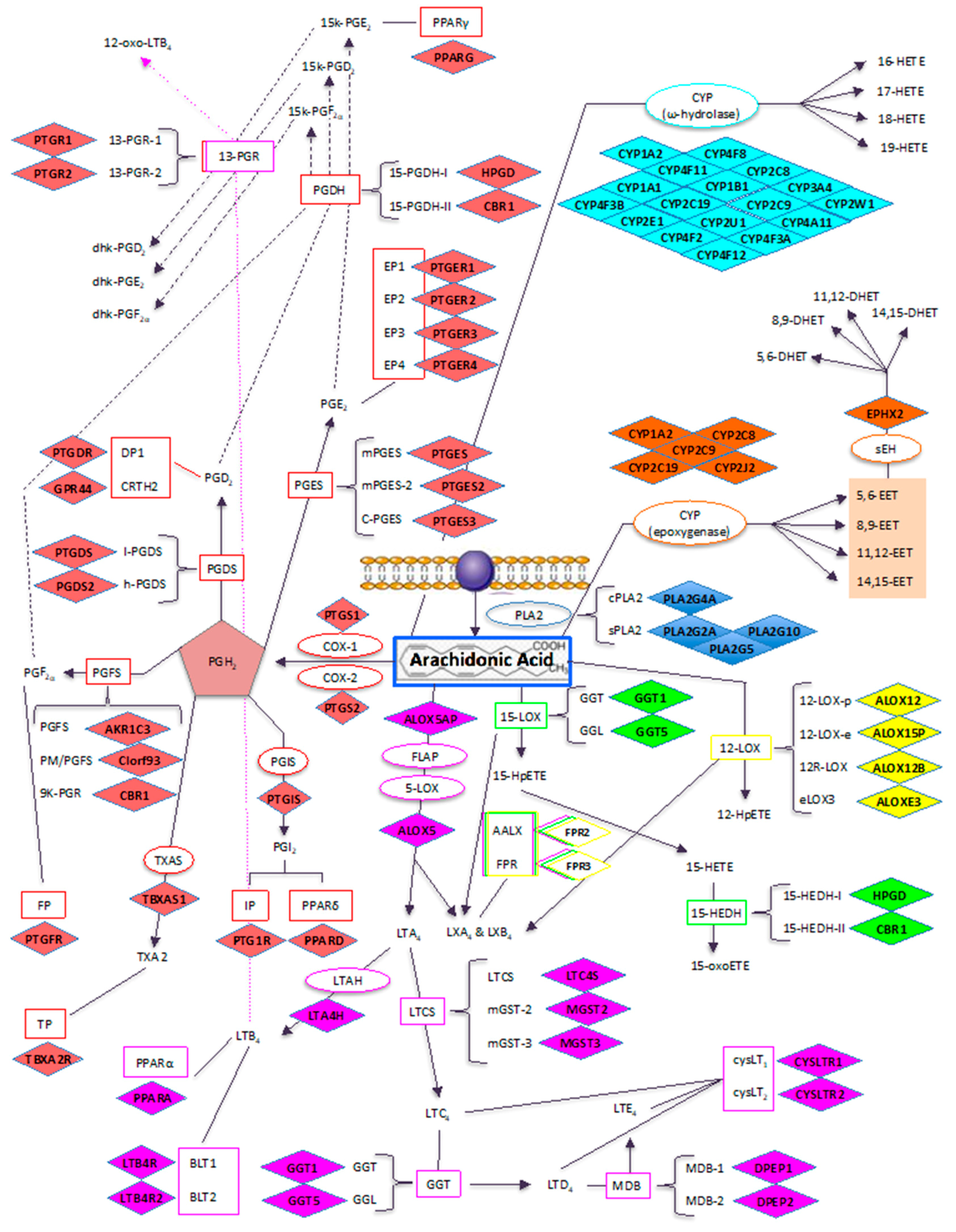
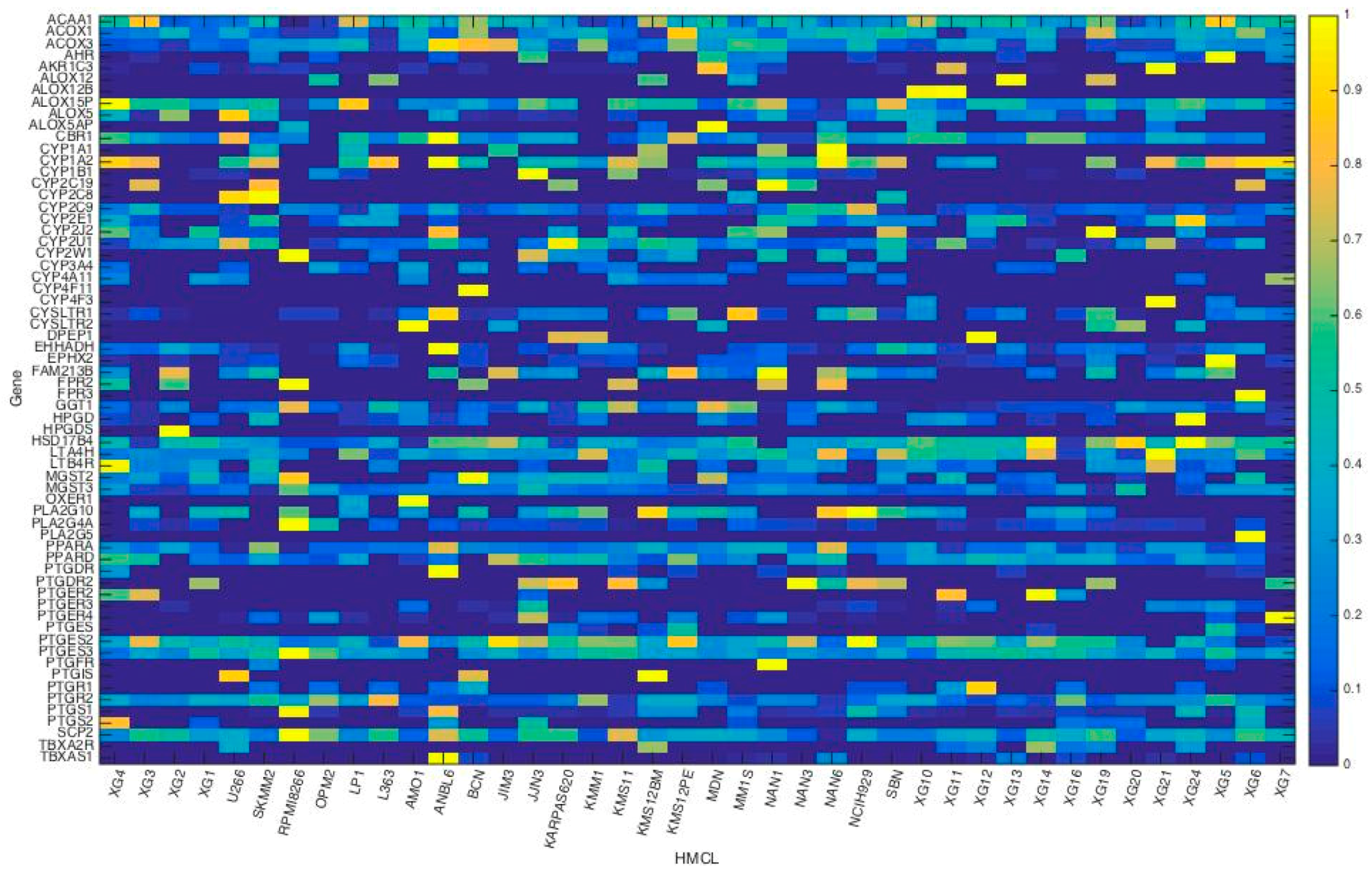
3.1. Microarray Data
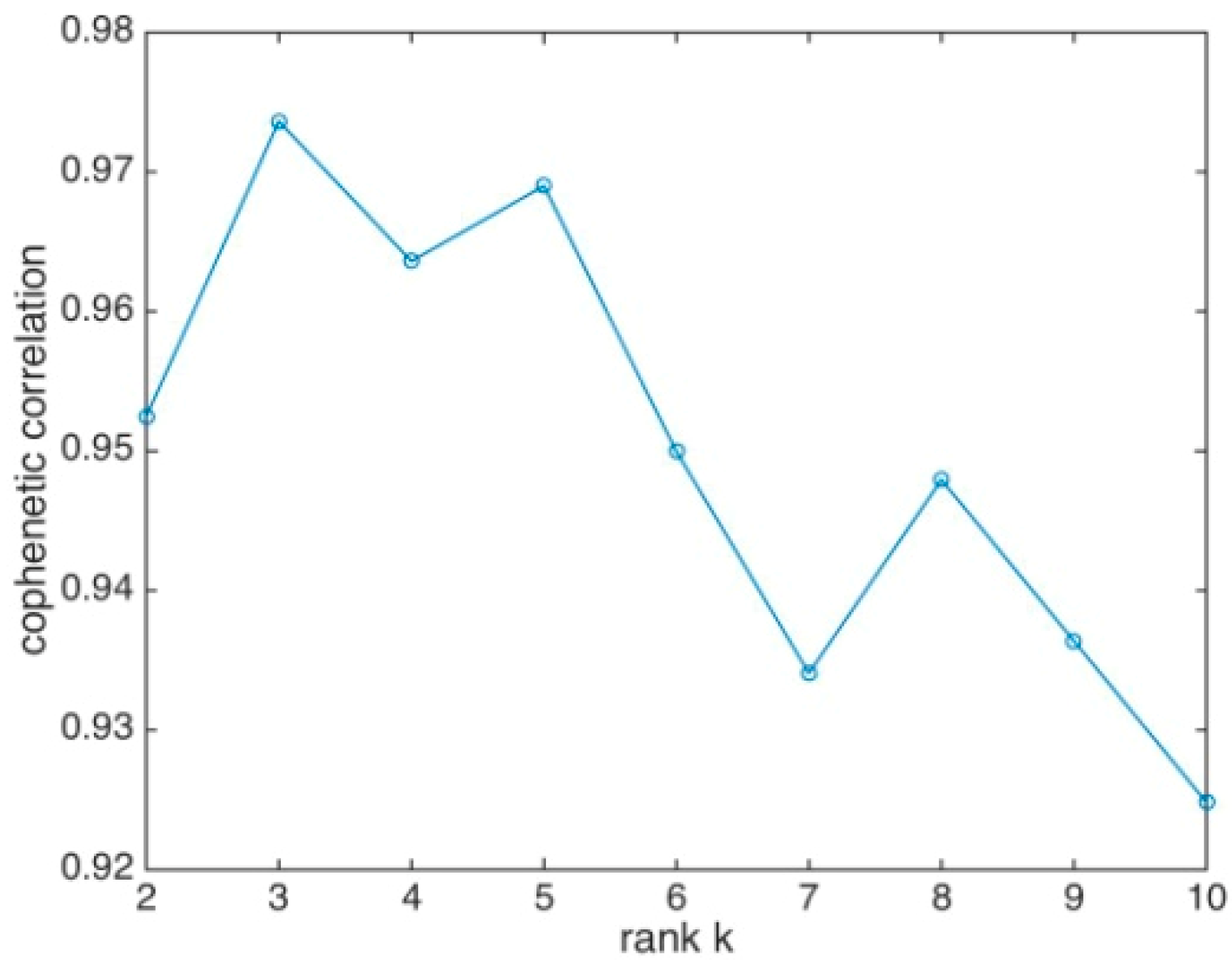
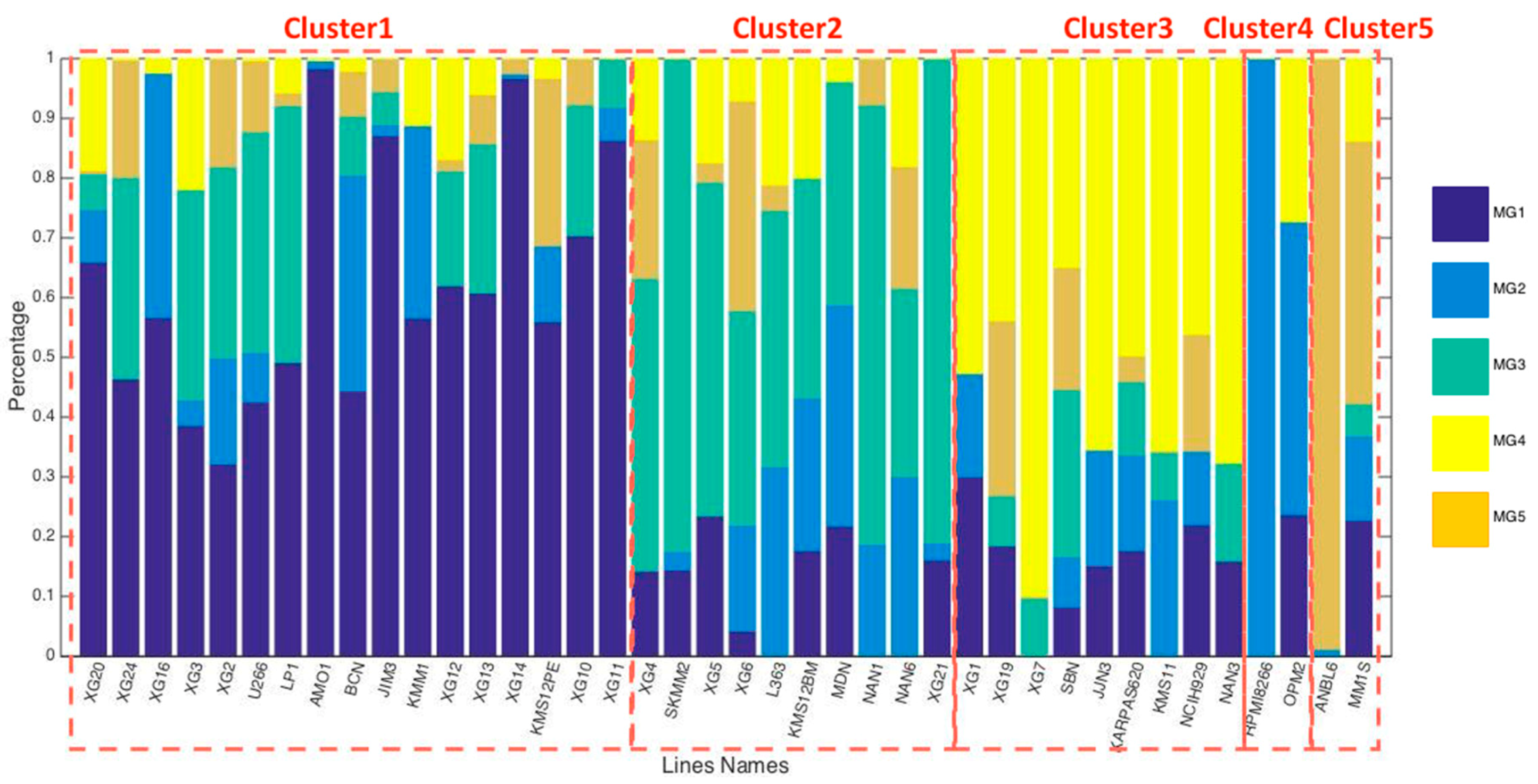
3.2. HMCL Clustering Results
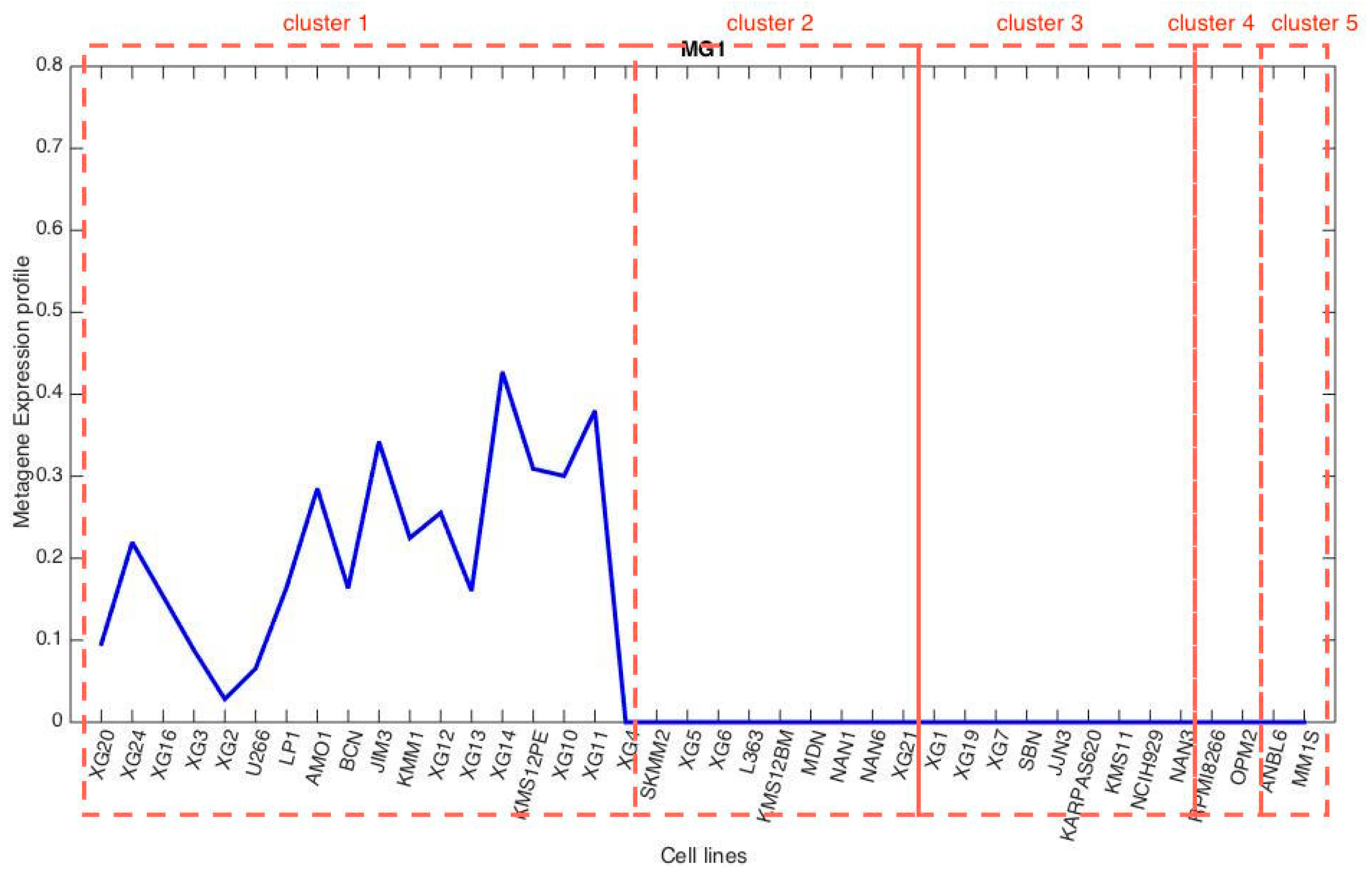
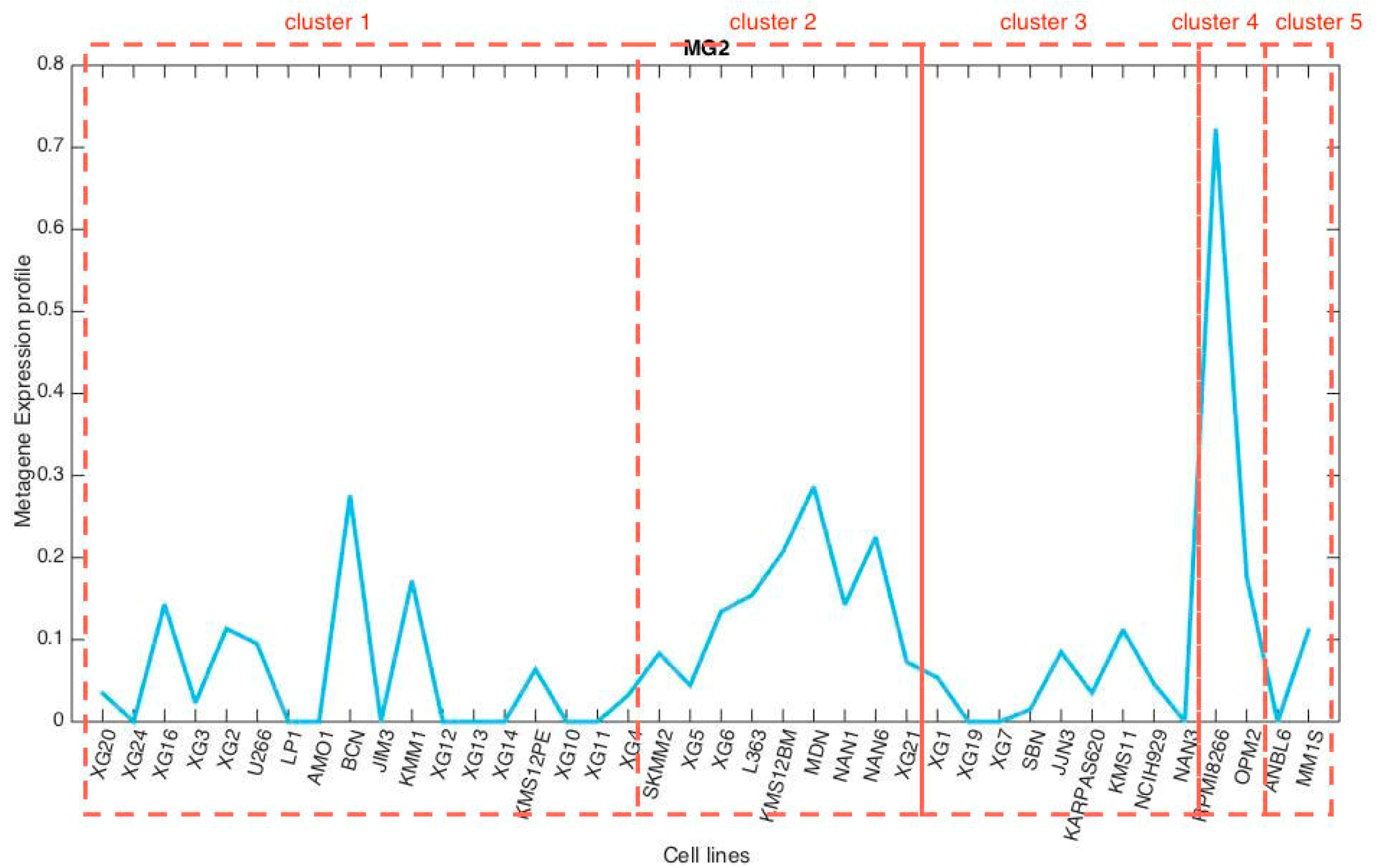
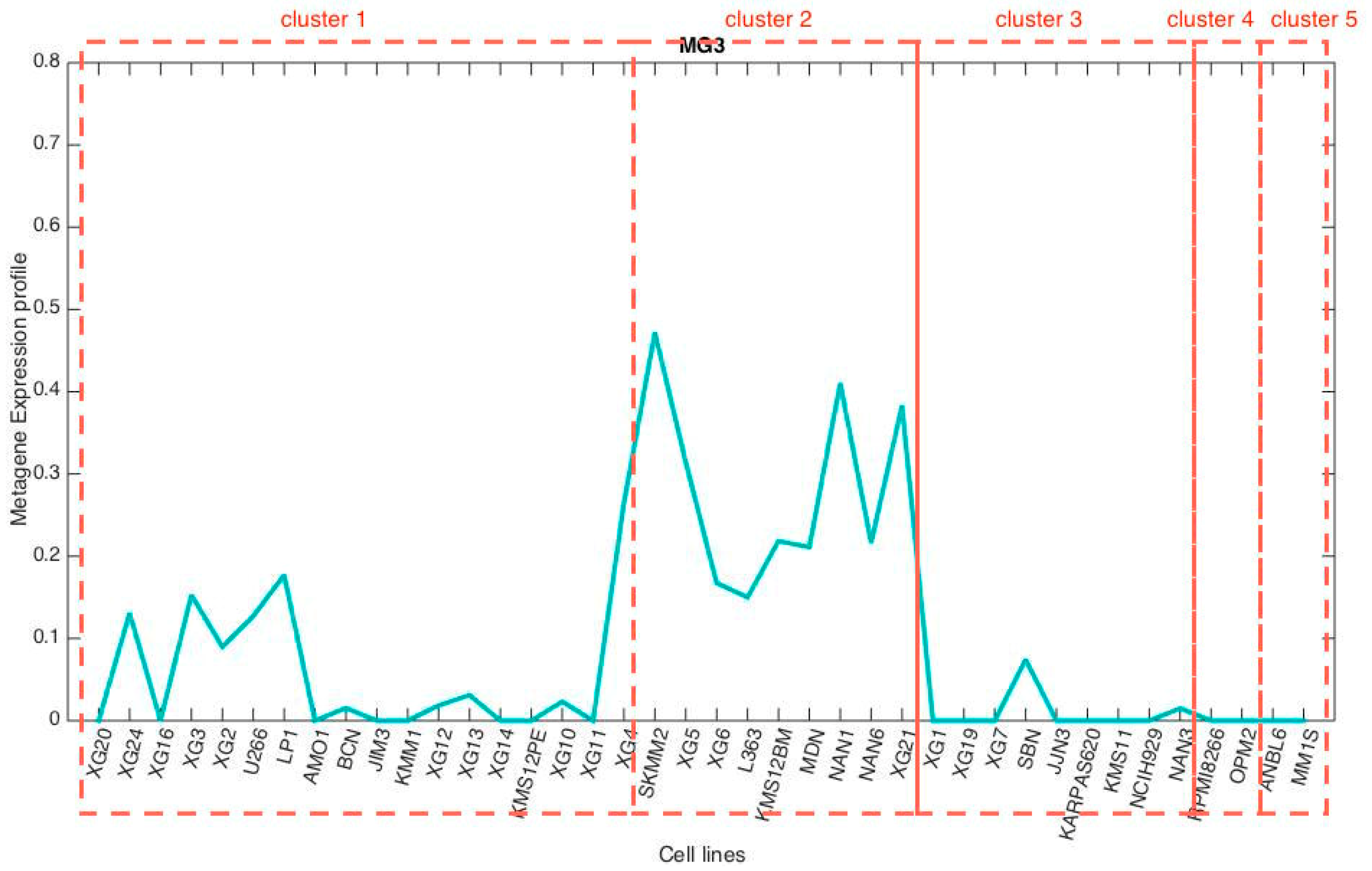
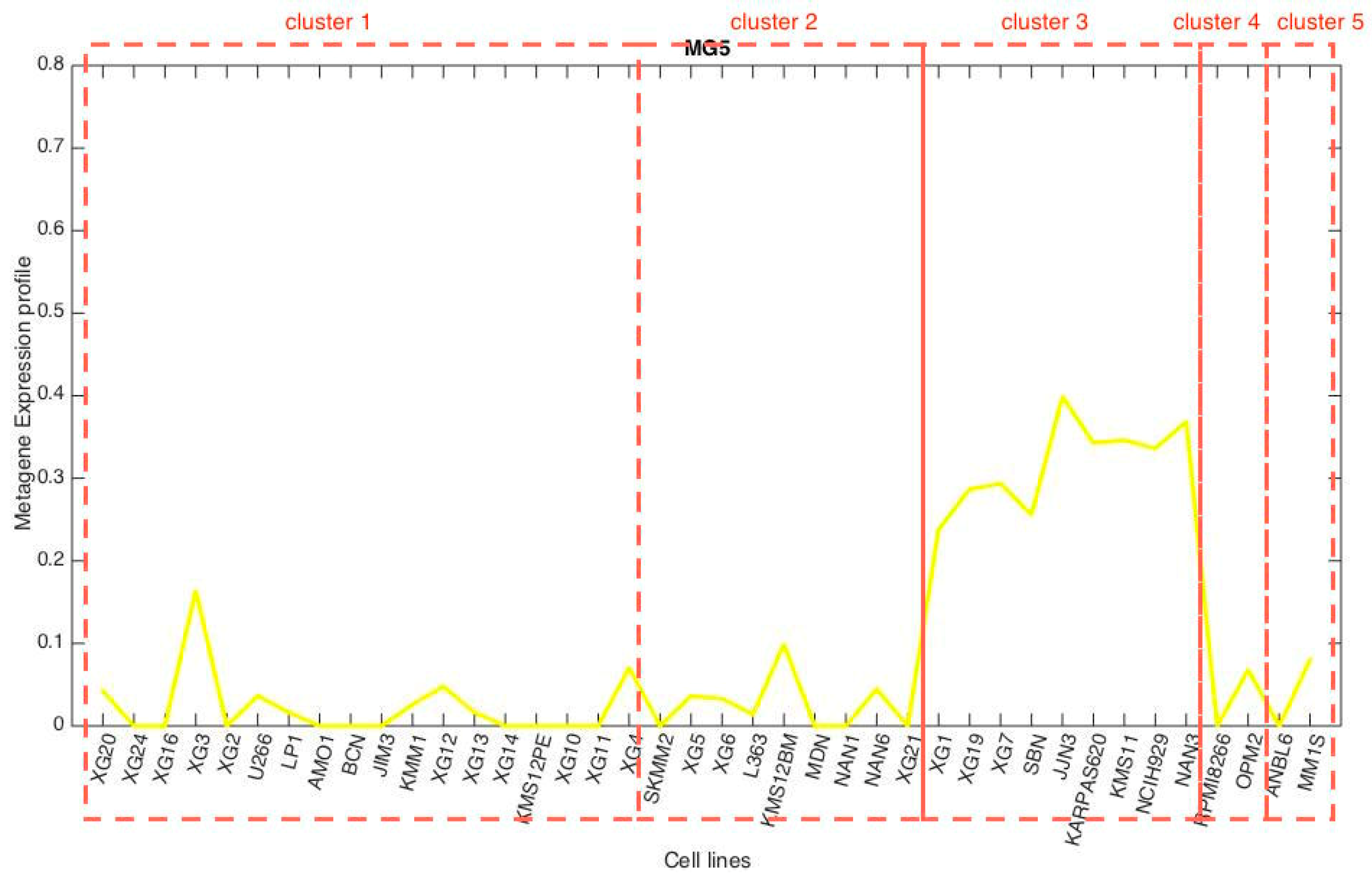
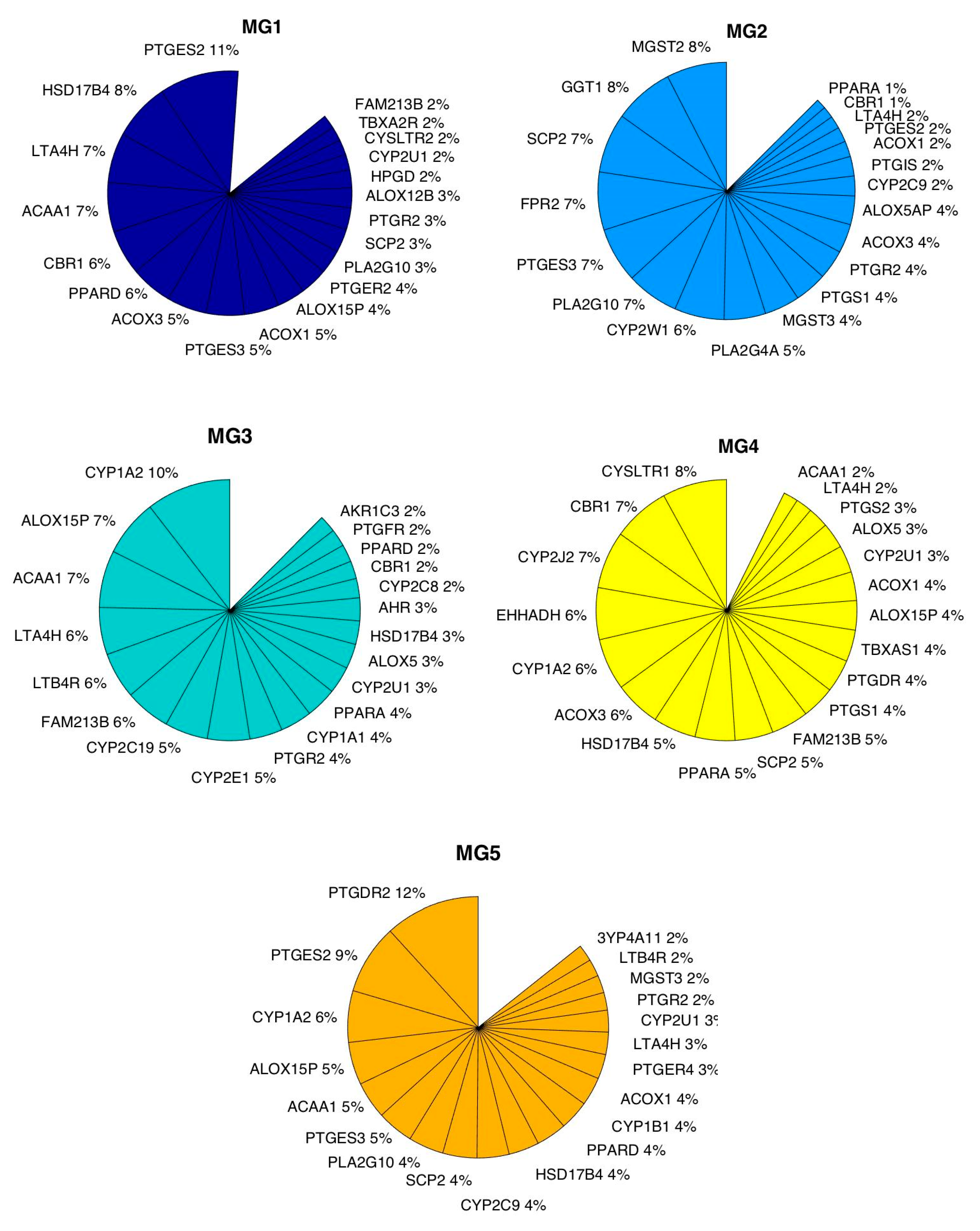
3.3. Metagene Analysis
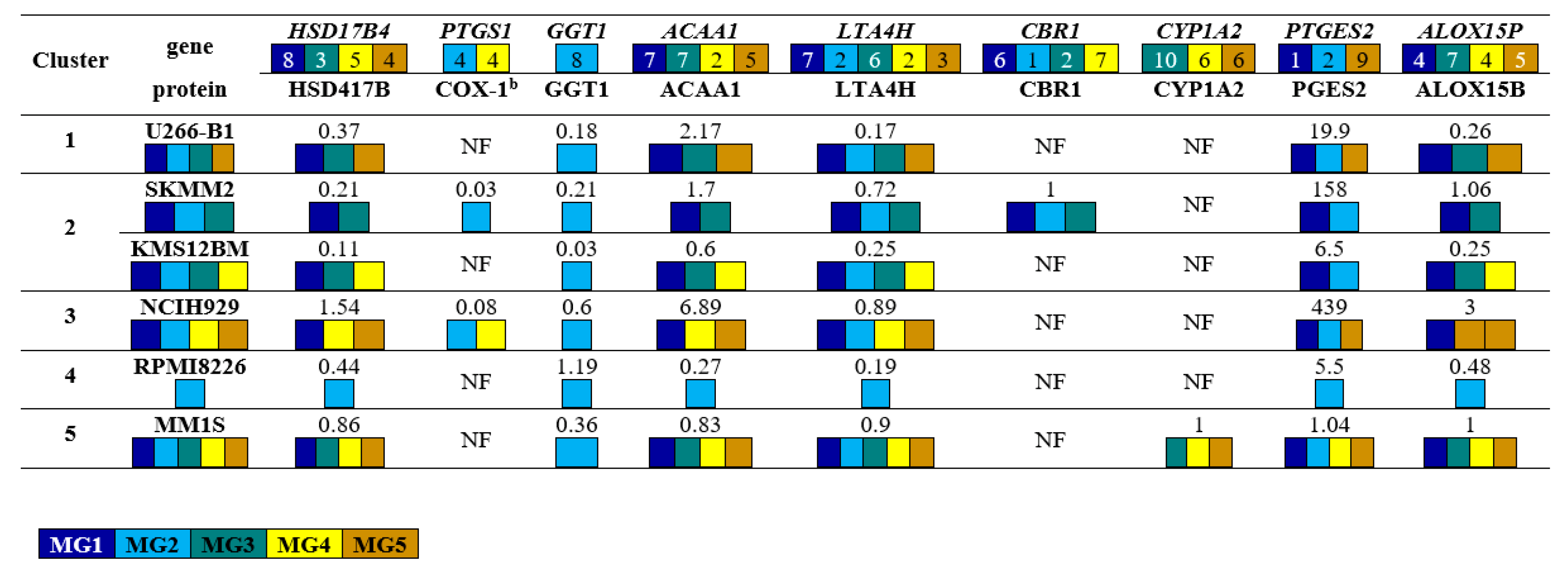
3.4. Proofs of Concepts: Western Blotting Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A

References
- Berthold, M.; Hand, D.J. Intelligent Data Analysis: An Introduction; Springer: New York, NY, USA; Secaucus, NJ, USA, 2007. [Google Scholar]
- Berthold, M.R.; Borgelt, C.; Hoppner, F.; Klawonn, F. Guide to Intelligent Data Analysis: How to Intelligently Make Sense of Real Data, 1st ed.; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Nieto, J.J.; Torres, A.; Georgiou, D.N.; Karakasidis, T.E. Fuzzy polynucleotide spaces and metrics. Bull. Math. Biol. 2006, 68, 703–725. [Google Scholar] [CrossRef] [PubMed]
- Casalino, G.; Del Buono, N.; Mencar, C. Non-Negative Matrix Factorizations for Intelligent Data Analysis. In Non-Negative Matrix Factorization Techniques, Signals and Communication Technology; Naik, G.R., Ed.; Springer: Berlin/Heidelberg, Germany, 2016; ISBN 978-3-662-48330-5. [Google Scholar]
- Casalino, G.; Gillis, N. Sequential dimensionality reduction for extracting localized features. Pattern Recognit. 2017, 63, 15–29. [Google Scholar] [CrossRef][Green Version]
- Boccarelli, A.; Esposito, F.; Coluccia, M.; Frassanito, M.A.; Vacca, A.; Del Buono, N. Improving knowledge on the activation of bone marrow fibroblasts in MGUS and MM disease through the automatic extraction of genes via a nonnegative matrix factorization approach on gene expression profiles. J. Transl. Med. 2018, 16, 217–233. [Google Scholar] [CrossRef] [PubMed]
- Esposito, F.; Gillis, N.; Del Buono, N. Orthogonal joint sparse NMF for microarray data analysis. J. Math. Biol. 2019, 79, 223. [Google Scholar] [CrossRef] [PubMed]
- Casalino, G.; Castiello, C.; Del Buono, N.; Mencar, C. A framework for intelligent Twitter data analysis with non-negative matrix factorization. Int. J. Web Inf. Syst. 2018, 14, 334–356. [Google Scholar] [CrossRef]
- Filippone, M.; Camastra, F.; Masulli, F.; Rovetta, S. A survey of kernel and spectral methods for clustering. Pattern Recognit. 2008, 41, 176–190. [Google Scholar] [CrossRef]
- Brunet, J.P.; Tamayo, P.; Golub, T.R.; Mesirov, J.P. Metagenes and molecular pattern discovery using matrix factorization. Proc. Natl. Acad. Sci. USA 2004, 101, 4164–4169. [Google Scholar] [CrossRef]
- Marin, J.J.; Briz, O.; Monte, M.J.; Blazquez, A.G.; Macias, R.I. Genetic variants in genes involved in mechanisms of chemoresistance to anticancer drugs. Curr. Cancer Drug Targets 2012, 12, 402–438. [Google Scholar] [CrossRef]
- Lombardi, L.; Poretti, G.; Mattioli, M.; Fabris, S.; Agnelli, L.; Bicciato, S.; Lambertenghi-Deliliers, G. Molecular characterization of human multiple myeloma cell lines by integrative genomics: Insights into the biology of the disease. Genes Chromosomes Cancer 2007, 46, 226–238. [Google Scholar] [CrossRef]
- Richardson, P.G.; Sonneveld, P.; Schuster, M.W.; Irwin, D.; Stadtmauer, E.A.; Facon, T.; Harousseau, J.L.; Ben-Yehuda, D.; Lonial, S.; San Miguel, J.F.; et al. Safety and efficacy of bortezomib in high-risk and elderly patients with relapsed multiple myeloma. Br. J. Haematol. 2007, 137, 429–435. [Google Scholar] [CrossRef]
- Fonseca, R.; Bergsagel, P.L.; Drach, J.; Shaughnessy, J.; Gutierrez, N.; Stewart, A.K.; Morgan, G.; Van Ness, B.; Chesi, M.; Minvielle, S.; et al. International Myeloma Working Group molecular classification of multiple myeloma: Spotlight review. Leukemia 2009, 23, 2210–2221. [Google Scholar] [CrossRef] [PubMed]
- Vangsted, A.; Klausen, T.W.; Vogel, U. Genetic variations in multiple myeloma II: Association with effect of treatment. Eur. J. Haematol. 2012, 88, 93–117. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Rajkumar, S.V. Many facets of bortezomib resistance/susceptibility. Blood 2008, 112, 2177–2178. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, S.V.; Dimopoulos, M.A.; Palumbo, A.; Blade, J.; Merlini, G.; Mateos, M.V.; Kumar, S.; Hillengass, J.; Kastritis, E.; Richardson, P.; et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014, 15, 538–548. [Google Scholar] [CrossRef]
- Rajkumar, S.V. Myeloma today: Disease definitions and treatment advances. Am. J. Hematol. 2016, 91, 90–100. [Google Scholar] [CrossRef]
- Mitra, A.K.; Harding, T.; Mukherjee, U.K.; Jang, J.S.; Li, Y.; HongZheng, R.; Jen, J.; Sonneveld, P.; Kumar, S.; Kuehl, W.M.; et al. A gene expression signature distinguishes innate response and resistance to proteasome inhibitors in multiple myeloma. Blood Cancer J. 2017, 7, e581. [Google Scholar] [CrossRef]
- Williams, T.J.; Peck, M.J. Role of prostaglandin-mediated vasodilatation in inflammation. Nature 1977, 270, 530–532. [Google Scholar] [CrossRef]
- Pai, R.; Soreghan, B.; Szabo, I.L.; Pavelca, M.; Baatar, D.; Tarnawski, A.S. Prostaglandin E2 transactivates EGF receptor: A novel mechanism for promoting colon cancer growth and gastrointestinal hypertrophy. Nat. Med. 2002, 8, 289–293. [Google Scholar] [CrossRef]
- Salcedo, R.; Zhang, X.; Young, H.A.; Michael, N.; Wasserman, K.; Ma, W.H. Angiogenic effects of prostaglandin E2 are mediated by up-regulation of CXCR4 on human microvascular endothelial cells. Blood 2003, 102, 1966–1977. [Google Scholar] [CrossRef]
- Perrone, M.G.; Scilimati, A.; Simone, L.; Vitale, P. Selective COX-1 inhibition: A therapeutic target to be reconsidered. Curr. Med. Chem. 2010, 17, 3769–3805. [Google Scholar] [CrossRef]
- Palumbo, A.; Cavo, M.; Bringhen, S.; Zamagni, E.; Romano, A.; Patriarca, F. Aspirin, Warfarin, or Enoxaparin thromboprophylaxis in patients with multiple myeloma treated with Thalidomide: A Phase III, Open-Label, Randomized Trial. J. Clin. Oncol. 2011, 29, 986–993. [Google Scholar] [CrossRef] [PubMed]
- Baz, R.; Li, L.; Kottke-Marchant, K.; Srkalovic, G.; McGowan, B.; Yiannaki, E. The Role of Aspirin in the Prevention of Thrombotic Complications of Thalidomide and Anthracycline-Based Chemotherapy for Multiple Myeloma. Mayo Clin. Proc. 2005, 80, 1568–1574. [Google Scholar] [CrossRef] [PubMed]
- Zonder, J.A.; Barlogie, B.; Durie, B.G.; McCoy, J.; Crowley, J.; Hussein, M.A. Thrombotic complications in patients with newly diagnosed multiple myeloma treated with lenalidomide and dexamethasone: Benefit of aspirin prophylaxis. Blood 2006, 108, 403–404. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2015. CA Cancer J. Clin. 2015, 65, 5–29. [Google Scholar] [CrossRef] [PubMed]
- Cuzick, J.; Thorat, M.A.; Bosetti, C.; Brown, P.H.; Burn, J.; Cook, N.R.; Ford, L.G.; Jacobs, E.J.; Jankowski, J.A.; La Vecchia, C.; et al. Estimates of benefits and harms of prophylactic use of aspirin in the general population. Ann. Oncol. 2015, 26, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Perrone, M.G.; Malerba, P.; Uddin, M.J.; Vitale, P.; Panella, A.; Crews, B.C.; Daniel, C.K.; Ghebreselasie, K.; Nickels, M.; Tantawy, M.N.; et al. PET radiotracer [18F]-P6 selectively targeting COX-1 as a novel biomarker in ovarian cancer: Preliminary investigation Eur. J. Med. Chem. 2014, 80, 562–568. [Google Scholar]
- Vitale, P.; Perna, F.M.; Perrone, M.G.; Scilimati, A. Screening on the use of Kluyveromyces marxianus CBS 6556 growing cells as enantioselective biocatalyst for ketones reduction. Tetrahedron Asymmetry 2011, 22, 1985–1993. [Google Scholar] [CrossRef]
- Catalano, A.; Carocci, A.; Corbo, F.; Franchini, C.; Muraglia, M.; Scilimati, A.; De Bellis, M.; De Luca, A.; Camerino Conte, D.; Sinicropi, M.S.; et al. Constrained analogues of tocainide as potent skeletal muscle sodium channel blockers towards the development of antimyotonic agents. Eur. J. Med. Chem. 2008, 43, 2535–2540. [Google Scholar] [CrossRef]
- Di Nunno, L.; Vitale, P.; Scilimati, A.; Simone, L.; Capitelli, F. Stereoselective dimerization of 3-arylisoxazoles to cage-shaped bis-beta-lactams syn 2,6-diaryl-3,7-diazatricyclo[4.2.0.02,5]- octan-4,8-diones induced by hindered lithium amides. Tetrahedron 2007, 63, 12388–12395. [Google Scholar] [CrossRef]
- Vitale, P.; Scilimati, A. Functional 3-Arylisoxazoles and 3-Aryl-2-isoxazolines from reaction of aryl nitrile oxides and enolates: Synthesis and reactivity. Synthesis 2013, 45, 2940–2948. [Google Scholar] [CrossRef]
- Perrone, M.G.; Santandrea, E.; Bleve, L.; Vitale, P.; Colabufo, N.A.; Jockers, R.; Milazzo, F.M.; Sciarroni, A.F.; Scilimati, A. Stereospecific synthesis and bio-activity of novel beta3-adrenoceptor agonists and inverse agonists. Bioorg. Med. Chem. 2008, 16, 473–2488. [Google Scholar] [CrossRef] [PubMed]
- Perrone, M.G.; Santandrea, E.; Scilimati, A.; Tortorella, V.; Capitelli, F.; Bertolasi, V. Baker’s yeast-mediated reduction of ethyl 2-(4-chlorophenoxy)-3-oxoalkanoates suitable intermediates for potential PPARalpha ligands. Tetrahedron Asymmetry 2004, 15, 3501–3510. [Google Scholar] [CrossRef]
- Perrone, M.G.; Santandrea, E.; Dell’Uomo, N.; Giannessi, F.; Milazzo, F.M.; Sciarroni, A.F.; Scilimati, A.; Tortorella, V. Synthesis and Biological Evaluation of New Clofibrate Analogues as Potential PPARalpha Agonists. Eur. J. Med. Chem. 2005, 40, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Smith, W.L. A Cyclooxygenase-2-dependent Prostaglandin E2 Biosynthetic System in the Golgi Apparatus. J. Biol. Chem. 2015, 290, 5606–5620. [Google Scholar] [CrossRef]
- Pati, M.L.; Vitale, P.; Ferorelli, S.; Iaselli, M.; Miciaccia, M.; Boccarelli, A.; Di Mauro, G.D.; Fortuna, C.G.; Souza Domingos, T.F.; Rodrigues Pereira da Silva, L.C.; et al. Translational impact of novel widely pharmacological characterized mofezolac-derived COX-1 inhibitors combined with bortezomib on human multiple myeloma cell lines viability. Eur. J. Med. Chem. 2019, 164, 59–76. [Google Scholar] [CrossRef]
- Lee, D.D.; Seung, H. Learning the parts of objects by nonnegative matrix factorization. Nature 1999, 401, 788–791. [Google Scholar] [CrossRef]
- Boutsidis, C.; Gallopoulos, E. SVD-based initialization: A head start for nonnegative matrix factorization. Pattern Recognit. 2008, 41, 1350–1362. [Google Scholar] [CrossRef]
- Moreaux, J.; Klein, B.; Bataille, R.; Descamps, G.; Maïga, S.; Hose, D. A high-risk signature for patients with multiple myeloma established from the molecular classification of human myeloma cell lines. Haematologica 2011, 96, 574–582. [Google Scholar] [CrossRef]
- Buczynski, M.W.; Dumlao, D.S.; Dennis, E.A. An integrated omics analysis of eicosanoid biology. J. Lipid Res. 2009, 50, 1505. [Google Scholar] [CrossRef]
- Liu, W.; Yuan, K.; Ye, D. Reducing microarray data via nonnegative matrix factorization for visualization and clustering analysis. J. Biomed. Inform. 2008, 41, 602–606. [Google Scholar] [CrossRef]
- Park, J.; Bae, E.K.; Lee, C.; Choi, J.H.; Jung, W.J.; Ahn, K.S.; Yoon, S.S. Establishment and characterization of bortezomib-resistant U266 cell line: Constitutive activation of NF-κB-mediated cell signals and/or alterations of ubiquitylation-related genes reduce bortezomib-induced apoptosis. BMB Rep. 2014, 47, 274–279. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, D.; Singh, A.V.; Ciccarelli, B.; Richardson, P.G.; Palladino, M.A.; Anderson, K.C. Combination of proteasome inhibitors bortezomib and NPI-0052 trigger in vivo synergistic cytotoxicity in multiple myeloma. Blood 2008, 111, 1654–1664. [Google Scholar] [CrossRef] [PubMed]
- Ling, S.C.; Lau, E.K.; Al-Shabeeb, A.; Nikolic, A.; Catalano, A.; Iland, H.; Horvath, N.; Ho, P.J.; Harrison, S.; Fleming, S.; et al. Response of myeloma to the proteasome inhibitor bortezomib is correlated with the unfolded protein response regulator XBP-1. Haematologica 2012, 97, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Maïga, S.; Gomez-Bougie, P.; Bonnaud, S.; Gratas, C.; Moreau, P.; Le Gouill, S.; Pellat-Deceunynck, C.; Amiot, M. Paradoxical effect of lenalidomide on cytokine/growth factor profiles in multiple myeloma. Br. J. Cancer 2013, 108, 1801–1806. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Casalino, G.; Coluccia, M.; Pati, M.L.; Pannunzio, A.; Vacca, A.; Scilimati, A.; Perrone, M.G. Intelligent Microarray Data Analysis through Non-negative Matrix Factorization to Study Human Multiple Myeloma Cell Lines. Appl. Sci. 2019, 9, 5552. https://doi.org/10.3390/app9245552
Casalino G, Coluccia M, Pati ML, Pannunzio A, Vacca A, Scilimati A, Perrone MG. Intelligent Microarray Data Analysis through Non-negative Matrix Factorization to Study Human Multiple Myeloma Cell Lines. Applied Sciences. 2019; 9(24):5552. https://doi.org/10.3390/app9245552
Chicago/Turabian StyleCasalino, Gabriella, Mauro Coluccia, Maria L. Pati, Alessandra Pannunzio, Angelo Vacca, Antonio Scilimati, and Maria G. Perrone. 2019. "Intelligent Microarray Data Analysis through Non-negative Matrix Factorization to Study Human Multiple Myeloma Cell Lines" Applied Sciences 9, no. 24: 5552. https://doi.org/10.3390/app9245552
APA StyleCasalino, G., Coluccia, M., Pati, M. L., Pannunzio, A., Vacca, A., Scilimati, A., & Perrone, M. G. (2019). Intelligent Microarray Data Analysis through Non-negative Matrix Factorization to Study Human Multiple Myeloma Cell Lines. Applied Sciences, 9(24), 5552. https://doi.org/10.3390/app9245552






