Featured Application
E. coli in seawater can be simply and efficiently detected by direct PCR. The sensitivity of detection is significantly improved using magnetic beads based bacterial pre-concentration.
abstract
Foodborne and waterborne E. coli remains a major economic burden worldwide. Assaying seawater for trace levels of E. coli is challenging since it applies time-consuming preparations, expensive instrumentation and complicated procedures. Therefore, there is a continued demand for new analytical technologies that can detect low bacterial concentrations in a more cost- and time-effective manner. In this study, combination of E. coli pre-concentration with a direct polymerase chain reaction (PCR) was shown to enable rapid bacterial detection without enrichment step or DNA extraction/purification. The E1 aptamer that targets E. coli surface epitope grafted onto magnetic beads efficiently concentrated E. coli from water samples containing high concentration of NaCl. When direct PCR was performed on bacteria attached to these aptamer-modified magnetic beads, a limit of 103 CFU/mL was obtained. The overall analysis was performed in less than 3 h. This approach may lead to a future PCR-based biosensor system for online monitoring of enteric bacteria in seawater.
1. Introduction
The ‘One Health’ approach recommended by WHO recognizes the inseparable link between human, animal and environmental health [1]. The most under investigated part of One Health is the environment. A seaside represents a unique ecological system that is highly influenced by industrial development and discharge of wastewater. Fecal contamination of seawater is characterized by an increase in concentration of Escherichia coli and intestinal enterococci. These bacteria are common members of gut microbiome in mammals and birds but are also opportunistic pathogens for human and animals [2]. E. coli and enterococci are used as standard for waterborne pathogens and as fecal indicator bacteria in water quality testing all over the world because these bacteria are easily culturable (EU BDW 2006/7/EC). The regular monitoring of E. coli content in water enables to avoid health risks, especially from exposure in the foreseeable context of short-term pollution on unusual circumstances. In addition, marine recreational waters were shown to be potential source of resistant bacteria because they may contribute to the dissemination of microorganisms carrying antimicrobial resistance genes [3].
To monitor and control microbiological quality of bathing and recreational seawater, the common routine practice is regular water sampling and analysis for E. coli levels on a weekly or monthly basis. European directive 2006/7/EC concerning the management of bathing water quality refers as “excellent quality” coastal water with less than 250 CFU of E. coli in 100 mL, and as a “good quality” water E. coli levels between 250 and 500 CFU/100 mL based upon a 95th-percentile of microbiological values. The number of E. coli colonies should not exceed 500 CFU/100 mL upon a 90th percentile of microbiological values to be considered as “sufficient”. The assessment of the potential risk is dependent on detection methods that quantify bacterial indicator concentrations. The reference methods of analysis are membrane filtration methods, ISO 9308-3 or ISO 9308-1, that enable to concentrate large amount of water samples (minimum capacity is typically 250 mL) before bacteria enumeration. Official detection methods are sensitive and accurate but time consuming as they are based on bacterial culture on agar plates and colony counting. The results take 2–3 days, which does not meet the demand of rapid seawater testing because an outdated information is forwarded to the public in some cases. Rapid test ISO 9308-1:2000 is faster than the reference tests, since it provides results within 24 h. The rapid test includes 4–5 h resuscitation period after water filtration to enhance the recovery of viable bacteria, and bacterial enumeration after culturing. The specificity of the test can be compromised by background bacterial flora that can be also concentrated by the filtration and then grow on the non-selective agar plates. Another limitation of the official and rapid methods is that in seawater under multi-stress situation, bacteria may enter ‘viable but nonculturable’ (VBNC) status. Although VBNC bacterial cells do not divide and, thus, do not form colonies on nutritional agars that normally support their growth, they were shown to be metabolically active in seawater retaining their pathogenicity [4]. Using this survival strategy, E. coli may rest undetected in seawater by routinely used methods but resuscitate and cause infections.
The public expects to receive adequate and timely information on results of microbiological quality of bathing water. For this, an efficient and accurate control system of bathing water is one of the priorities of coastal touristic area. In some cases, noncompliance to water quality standard would not come to a regulatory agency’s attention until the next scheduled monitoring. When monitoring is infrequent, it is often not known whether the noncompliance is due to a pollution incident that has occurred shortly prior to the monitoring or an incident that has occurred days or weeks before [5]. A new technology, which would provide result in a few hours, is needed to enable the public to be informed effectively and to improve measures of health protection through “One Health” approach.
In this work, we tested the feasibility to detect E. coli in seawater by a direct PCR method after original pre-concentration step. Because pathogens are typically low concentrated in seawaters, direct monitoring of waterborne E. coli is costly, technically challenging, and in some cases not feasible. High concentration of NaCl in seawater may interfere with commonly used molecular detection technics based on PCR or ELISA. That is because proteins (antibodies or enzymes, like DNA polymerase) may be inhibited by NaCl at high concentration. To overcome problems of low pathogen and high salt concentration in seawater, we combine the aptamer coated magnetic beads pre-concentration modality with the direct PCR detection of E. coli. A specific DNA aptamer selected by bacterial cell- systematic evolution of ligands by exponential enrichment (SELEX) that binds whole E. coli cells with high affinity and specificity [6] was chosen for the development of the inexpensive and rapid pre-concentration step. The analytical performance of the direct PCR on E. coli whole cells carried by magnetic beads is evaluated using a procedure without any DNA extraction or purification.
2. Materials and Methods
2.1. Reagents
All reagents were of analytical grade and used as purchased. Luria-Bertani (LB) broth was purchased from Becton Dickinson (DB, Le Pont de Claix, France) and used as 25 g/L water solution (10 g/L tryptone, 5 g/L yeast extract, 5 g/L NaCl). Peptone Bacto (PB) from DB (Le Pont de Claix France) was used as 1 g/L water solution. NaCl (99%) and Tween 20 were purchased from Merck (Fontenay sous Bois, France). Dynabeads M-280 streptavidin magnetic particles were purchased from Invitrogen (Saint Aubin, France). Promega Wizard Genomic DNA purification kit was used for extraction of bacterial genomic DNA. Colorant Midori green advance was purchased from Dutcher (Brumath, France), and 1 Kb Plus DNA ladder from New England Biolabs (Beverly, MA, USA). Phosphate-buffered saline (PBS) 10× was purchased from Lch Chime (Les Aires, France).
2.2. Bacterial Strain and Growth Condition.
Permanent stock of E. coli K-12 TG1strain was maintained at −80 °C in LB broth supplemented with 15% glycerol. Bacterial cells were routinely grown in LB broth or on LB agar (Difco) medium at 37 °C. An overnight liquid culture was washed and used to inoculate 25 mL various culture media in a 250 mL Erlenmeyer flask. Flasks were incubated with shaking (200 rpm) at 37 °C. The Wizard genomic DNA purification kit (Promega, Charbonnieres, France) was used for DNA extraction. The optical density of bacterial solutions was measured at 600 nm using Vis Spectrophotometer Genesys (Theromo Fisher).
2.3. Drop Plate Method for Enumerating Bacteria
Bacterial solutions were serially diluted by placing 100 µL of the suspension into a dilution tube containing 900 µL of BP. This tube was vortexed, and 100 µL was removed and placed into a second dilution tube containing 900 µL of BP. This process was repeated six times. Then, 10µL of each dilution was plated in triplicate on LB agar and incubated for 18 h at 37 °C. Colony counts from triplicate plates were converted to CFU/mL.
2.4. Aptamer
In this study, we used previously selected ssDNA E1aptamer that specifically binds to E. coli [6]. The aptamer was custom-synthesized by Eurogenotec (Angers, France) with biotin tag: 5′-biotin GCA ATG GTA CGG TAC TTC CAC TTA GGT CGA GGT TAG TTT GTC TTG CTG GCG CAT CCA CTG AGC GCA AAA GTG CAC GCT ACT TTG CTA A-3′. Before use, E1 aptamer was dissolved into a concentration of 20 μM in distilled water.
2.5. Magnetic Beads Functionalization
Prior to immobilization to magnetic beads, the E1 aptamer was denatured by heating at 95 °C for 5 min; then cooled at 4 °C for 15 min, and finally renatured at 25 °C for 5 min. Streptavidin magnetic beads were prepared according to the manufacturer’s instructions, as previously described [7]. Initially, 200 µL of magnetic beads were gently washed with 0.5 mM ethylenediaminetetraacetic acid (EDTA), 1 M NaCl, 5 mM Tris-HCl buffer, pH 7, and concentrated using a magnetic stand DynaMag2, (Thermo Fisher Scientific, Lissieu, France). The washed beads were incubated with 2.5 µM E1 aptamer in 0.5 mM EDTA, 1 M NaCl, 5 mM Tris-HCl buffer, pH 7, in a bench-top rotator mixer at room temperature for 30 min. The aptamer-bead conjugates were concentrated using a magnetic stand and washed with 1 mL of 0.05% Tween20, PBS (PBST) for four times before storing at 4 °C.
2.6. Bacterial Capturing by Apta-Magnetic Beads
Varying concentrations of E. coli cells (from 102 CFU/mL to 108 CFU/mL, in BP + 0.4M NaCl, 1 mL volume) were each incubated with the aptamer-magnetic beads (10 μg/mL) under shaking for 30 min at room temperature. The complex of E. coli-aptamer-magnetic bead was washed three times in 1 mL PBST using a magnetic stand, to remove non-bound bacteria. In the final step, beads were concentrated using the magnetic stand, and dissolved in 50 µL PBS. A 20 µL of this bead suspension was used for direct PCR.
2.7. Optical Microscopy
Bacterial cells and magnetic beads were observed with an AxioObserver.Z1 Zeiss inverted optical microscope equipped with a Zeiss AxioCam MRm digital camera. Images were processed using the ZEN software package.
2.8. PCR Tests
All PCRs ware performed using specific primers targeting the yalO gene of E. coli as previously published [8]. Classical PCRs were performed on 50 μL solutions containing DreamTaq DNA polymerase (2× DreamTaq Green PCR Master Mix, Thermo Scientific), 0.02 mM forward YalO primer (5′-TGATTTCCGTGCGTCTGAATG-3′) and 0.02 mM reverse YalO primer (5′-ATGCTGCCGTAGCGTGTTTC-3′) and various concentration of template DNA (genomic DNA extracted from E. coli). Following template volumes were used for PCR reaction: 0.39 μL for DNA sourced from BP, 0.55 μL for DNA sourced from BP + 0.4 M NaCl, and 0.38 μL for DNA sourced from LB and LB + 0.4 M NaCl solutions. The concentration and quality of the DNA were determined by a spectrophotometer NanoDrop 2000c (Thermo Fisher Scientific, Lissieu, France) and Qubit Fluorometric quantification (Thermo Fisher Scientific, Lissieu, France). The DNA preparations were stored at −20 °C until use. The total volume was adjusted with RNase-free water.
Direct PCR was performed with functionalized magnetic beads carrying E. coli cells. Before detection, 7.5 µM biotinylated E1 aptamer conjugates with 0.5 mg of magnetic beads were mixed with bacterial culture and washed four times with PBST. Direct PCRs were performed on 50 µL total volume containing 20 µL of magnetic beads carrying bacteria cells, 25 µL of DreamTag Green PCR Master Mix 2×, 0.02 mM reversed and forward yalO primers (1 µL of each primer at 1 mM, and 3 µL distillated water). PCR was performed under the following conditions: initial denaturation at 95 °C for 3 min, followed by 35 cycles of denaturation at 95 °C for 30 s, primer annealing at 58 °C for 30 s, primer extension at 72 °C for 30 s, and a final extension at 72 °C for 10 min. In every assay, a buffer control, to which no DNA template was added, was used as a negative control.
Twenty μL of the PCR product were separated by electrophoresis on 1.5% agarose gel in 1 mM EDTA, 20 mM acetic acid, 40 mM Tris-base buffer, pH 8.0 (TAE buffer) and stained with midori green. Gels were visualized under UV light using a Fusion Fx gel documentation system (Vilber Lourmat, Marne-la-Vallee, France). The 1 Kb Plus DNA ladder was used as a molecular marker to indicate the size of the amplicons.
3. Results
3.1. Survival of E. coli in Highly Salted Media
In seawater, enteric bacteria encounter a variety of challenges to survive because high salt concentration and osmolarity induce a stress to bacterial cells [9]. Thus, we first compared the survival of E. coli cells in a rich bacterial medium, LB, and LB complemented with 0.4 M NaCl, and its survival in a poor bacterial medium, Peptone Bacto, and Peptone Bacto supplemented with 0.4 mM NaCl. Because bacteria under the stress may become VBNC, two parameters were evaluated: the optical density of the solutions at 600 nm (OD600), which reflects total number of bacteria, and the colony forming ability, which is a viability parameter. Figure 1a shows that when solutions were inoculated to a cell density of approximately 108 CFU/mL, similar survival rates were obtained in both media supplemented or not with NaCl upon 30 h, although some differences were observed during the first 5 h post inoculation. In addition, no drastic morphological modification of E. coli cells was observed in solutions containing high NaCl concentration compared to the corresponding pure medium (Figure 1b). Recently, Zhang et al. [10] showed that there was no significant difference in E. coli cell growth when fresh water was replaced by 60% artificial seawater. Thus, it seems that viability of E. coli cells is no affected by high NaCl concentration (0.4 M) in solutions that normally support bacterial survival.
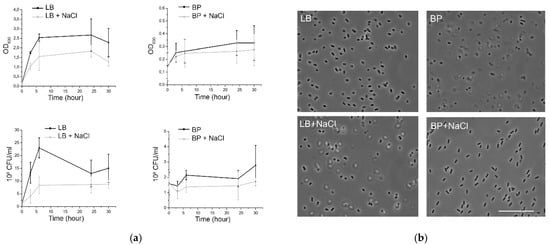
Figure 1.
(a) Survival curves of E. coli at 108 CFU/mL suspended in different solutions. (b) Phase contrast transmission micrographs of E. coli cells incubated for 30 h in different media. Scale bar, 10 µm stains for all images.
3.2. PCR Detection of E. coli Genomic DNA
Second, we wondered whether high NaCl concentration inhibits DNA polymerase and, in that way, decreases the sensitivity of PCR detection of bacteria. For this, genomic DNA were extracted from bacterial cells cultivated in four above media (LB, LB + NaCl; PB, and PB + NaCl) and tested for the efficiency of classical PCR. Previously validated primers that target consensus sequence of the orphan gene yaiO of E. coli yielding 115 bp PCR product [8] were used for PCR amplification. Figure 2 points out that the expected amplicon of 115 bp was detected with similar sensitivity using DNA from bacteria cultivated in pure bacterial media and in corresponding media supplemented with 0.4 M NaCl. This indicates that classical PCR technique can be successfully applied for E. coli detection in seawater.
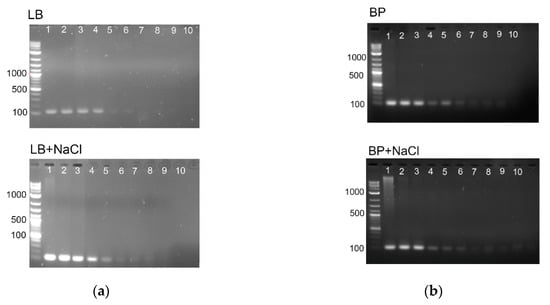
Figure 2.
(a) PCR sensitivity with yaiO primer set obtained with different concentrations of DNA extracted from E. coli cells cultivated in LB (upper panel) and LB + 0.4 mM NaCl (lower panel). Line 1: 200 ng/µL; line 2: 20 ng/µL; line 3: 2 ng/µL; line 4: 20 pg/µL; line 5: 2 pg/µL; line 6: 200 fg/µL; line 7: 20 fg/µL; line 8: 2 fg/µL; line 10: no template control. (b) PCR sensitivity with yaiO primer set obtained with different concentrations of DNA extracted from E. coli cells cultivated in Bacto Pepton medium, BP, (upper panel) and BP + 0.4 mM NaCl (lower panel). Line 1: 200 ng/µL; line 2: 20 ng/µL; line 3: 2 ng/µL; line 4: 200 pg/µL; line 5: 20 pg/µL; line 6: 2 pg/µL; line 7: 200 fg/µL; line 8: 20 fg/µL; line 9: 2 fg/µL; line 10: no template control. DNA ladder (1 bp +) was used in all experiments.
However, although very robust, sensitive and specific classical PCR demands previously extracted and purified genomic DNA. Thus, we also tested whether direct PCR (without performing DNA isolation and purification steps) can be used to evidence E. coli in highly salted media.
3.3. Bacterial Pre-Concentration with Aptamer Decorated Magnetic Beads
The principle of the direct PCR method for E. coli detection in salted media is described in Figure 3. The first step was pre-concentration of bacterial cells with magnetic beads decorated with the specific E1 aptamer that recognizes an epitope on the surface of E. coli vegetative cells. The second step was a direct PCR detection of E. coli attached to beads.
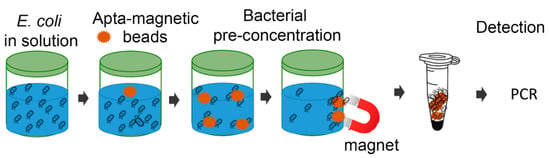
Figure 3.
Schematic representation of the developed integrated apta-magnetic direct PCR method.
The feasibility of E. coli detection by an integrated aptamer-magnetic direct PCR method was studied on bacterial cells cultured in 0.4 M NaCl, BP medium. This supplemented medium was considered as artificial seawater because most natural oligotrophic seawaters contain about 1 mg of dissolved organic carbon in addition to the high salt concentration [11].
Streptavidin-biotin conjugated chemistry was used to decorate magnetic beads with the E1 aptamers. Bright monodisperse metallic beads of a spherical shape were observed before bacterial immobilization (Figure 4a). The technique did not enable to visualize aptamer molecules on the beads surface because of nanometric sizes of biotinylated aptamer molecules. When admixed to a PB + 0.4 M NaCl culture containing E. coli, functionalized beads captured efficiently bacterial cells. Figure 4b shows that functionalized beads after 30 min incubation with bacteria were covered with bacterial cells, even after multiple washings.

Figure 4.
(a) Phase contrast transmission micrographs of magnetic beads functionalized with the E1 aptamer. Scale bar, 10µm. (b) Phase contrast study of E. coli-bead complex after capturing. Scale bar, 10µm stains for both images.
The efficiency of capturing was calculated by enumerating bacteria in supernatant after magnetic concentration using a drop plate method remained. It was estimated to 90% for 104 CFU/mL of E. coli in BP + NaCl.
3.4. Direct PCR Detection of E. coli after Pre-Concentration
Figure 5a shows that direct PCR obtained without the pre-PCR DNA extraction and purification steps can reveal the presence of E. coli in artificial seawater but only at high bacterial concentration of 108 CFU/mL. However, when bacterial cells were pre-concentrated with magnetic beads, the limit of detection was decreased to 103 CFU/mL (Figure 5b).
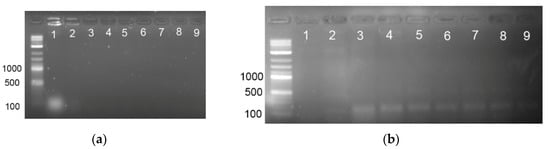
Figure 5.
(a) Direct PCR detection of E. coli with yaiO primer set using 20 µL of BP + NaCl medium containing different concentrations of E. coli. Line 1: 108 CFU/mL; line 2: 107 CFU/ml; line 3: 106 CFU/mL; line 4: 105 CFU/mL; line 5: 104 CFU/mL; line 6: 103 CFU/mL; line 7: 102 CFU/mL; line 8: 101 CFU/mL; line 9: no template control. (b) Direct PCR detection of E. coli with yaiO primer set using 20 µL of magnet pre-concentrated bacterial cells from 1 mL of BP + NaCl medium containing different concentrations of E. coli. Line 1: no template control; line 2: no template control performed with magnetic beads carrying E1 aptamer; line 3: 108 CFU/mL; line 4: 107 CFU/mL; line 5: 106 CFU/mL; line 6: 105 CFU/mL; line 7: 104 CFU/mL; line 8: 103 CFU/mL; line 9: 102 CFU/mL. DNA ladder (1 bp +) was used in all experiments.
4. Discussion
Microbiological quality of seawater is a public health problem that can have serious repercussions in the economy and tourism. The prevention of waterborne diseases relies on microbiological detection methods. Recently, several new analytic methods have been proposed to replace standard procedures that take a few days to provide results. For instance, optical biosensors that use filters and camera enable to detect bacteria in water by fluorescence or hemiluminescence [12,13,14,15]. In the literature, electrochemical sensors are proposed that detect pathogens thanks to the current or potential variations that occur upon pathogen or its biomarker interaction with an electrode [16,17,18,19,20,21]. Among various detection methods, the most robust are those based on ELISA immunological tests and on PCR-based molecular tests [22]. Both methods may provide results within 24 h. However, their sensitivity is not sufficient for on-field applications, as an enrichment step is needed prior to detection. The enrichment step is particularly essential for detection of waterborne bacteria that are typically highly diluted. Moreover, PCR-based methods (like qPCR, digital PCR, isothermal methods) are performed on extracted and purified bacterial DNA. Purification of DNA enables to avoid possible inhibitors of the downstream amplification. Multi-steps that include bacterial concentration by filtration, pathogen recovery/enrichment, and DNA extraction/purification significantly increase the cost and time of water analysis.
Detection of E. coli is of great importance to the quality control of food and water. The combination of pathogen magnetic pre-concentration and direct PCR strategy proposed in this study enables detection of E. coli without long multi-step DNA extraction. In addition, amplification of DNA sequences by PCR allows detection of VBNC cells that pose problem in conventional culture methods.
In our procedure, the enrichment step (that takes from few hours to a few days) was replaced by the magnetic pre-concentration that was performed in less than 1 h. The pre-concentration increased the limit of detection by five orders of magnitude (from 108 CFU/mL to 103 CFU/mL of E. coli). Regarding the low concentration of contaminating bacteria in potable, recreational, and bathing waters, we believe that this procedure based on aptamer-coated magnetic beads may successfully replace membrane filtration to concentrate bacteria. It can be performed on-site, as no electricity source is needed nor a specific equipment for sample preparation. A simple magnet enables to collect beads carrying bacterial cells for analysis.
As the present work was a proof of principle, a lab PCR was used to verify the feasibility of the system. Portable PCR have already been presented in [14,18,23,24,25], and the aim of future work is to miniaturize the system for direct PCR detection in order to obtain a transportable system for in-field measurements.
Author Contributions
Conceptualization, J.V.; methodology, J.V.; validation, J.V., D.K. and N.R., formal analysis, Z.K., F.R. and J.V.; investigation, Z.K., D.K., and J.V.; resources, A.V., J.V. and N.R.; data curation, J.V.; writing—original draft preparation, J.V.; writing—review and editing, all authors; supervision, J.V., A.V., and N.R.; funding acquisition, A.V., N.R. and J.V.
Funding
This research benefited from INRA-MICALIS facilities. In part it was funded by the European Union and Greek national funds through the Operational Program Competitiveness, Entrepreneurship and Innovation, under the call RESEARCH-CREATE-INNOVATE (project code: T1EDK-04615) of A.K., and by the University Paris-Saclay through the Programme d’Investissement d’Avenir, AAP Prematuration Idex (funding agreement 2019-13001227) of N.R. and J.V.
Acknowledgments
We thank Elena Bidnenko (INRA, France) for valuable advices and Marisa Manzano (University of Udine, Italy) for critical reading of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- One Health. Available online: https://www.who.int/features/qa/one-health/en/ (accessed on 09 December 2019).
- Bäumler, A.J.; Sperandio, V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 2016, 535, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Da Costa Andrade, V.; Zampieri, B.D.B.; Ballesteros, E.R.; Pinto, A.B.; De Oliveira, A.J.F.C. Densities and antimicrobial resistance of Escherichia coli isolated from marine waters and beach sands. Environ. Monit. Assess. 2015, 187, 342. [Google Scholar] [CrossRef] [PubMed]
- Pommepuy, M.; Butin, M.; Derrien, A.; Gourmelon, M.; Colwell, R.; Cormier, M. Retention of enteropathogenicity by viable but nonculturable Escherichia coli exposed to seawater and sunlight. Appl. Environ. Microbiol. 1996, 62, 4621–4626. [Google Scholar] [PubMed]
- Sokolova, E.; Åström, J.; Pettersson, T.J.; Bergstedt, O.; Hermansson, M. Estimation of pathogen concentrations in a drinking water source using hydrodynamic modelling and microbial source tracking. J. Water Health 2012, 10, 358–370. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Song, M.Y.; Jurng, J.; Kim, B.C. Isolation and characterization of DNA aptamers against Escherichia coli using a bacterial cell–systematic evolution of ligands by exponential enrichment approach. Anal. Biochem. 2013, 436, 22–28. [Google Scholar] [CrossRef]
- Vidic, J.; Noiray, M.; Bagchi, A.; Slama-Schwok, A. Identification of a Novel Complex between the Nucleoprotein and PA (1–27) of Influenza A Virus Polymerase. Biochemistry 2016, 55, 4259–4262. [Google Scholar] [CrossRef]
- Molina, F.; López-Acedo, E.; Tabla, R.; Roa, I.; Gómez, A.; Rebollo, J.E. Improved detection of Escherichia coli and coliform bacteria by multiplex PCR. BMC Biotechnol. 2015, 15, 48. [Google Scholar] [CrossRef]
- Rozen, Y.; Belkin, S. Survival of enteric bacteria in seawater. FEMS Microbiol. Rev. 2001, 25, 513–529. [Google Scholar] [CrossRef]
- Zhang, W.; Zhu, J.; Zhu, X.; Song, M.; Zhang, T.; Xin, F.; Dong, W.; Ma, J.; Jiang, M. Expression of global regulator IrrE for improved succinate production under high salt stress by Escherichia coli. Bioresour. Technol. 2018, 254, 151–156. [Google Scholar] [CrossRef]
- Hansell, D.A.; Carlson, C.A.; Repeta, D.J.; Schlitzer, R. Dissolved organic matter in the ocean: A controversy stimulates new insights. Oceanography 2009, 22, 202–211. [Google Scholar] [CrossRef]
- Heery, B.; Briciu-Burghina, C.; Zhang, D.; Duffy, G.; Brabazon, D.; O’Connor, N.; Regan, F. ColiSense, today’s sample today: A rapid on-site detection of β-D-Glucuronidase activity in surface water as a surrogate for E. coli. Talanta 2016, 148, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Tawil, N.; Sacher, E.; Mandeville, R.; Meunier, M. Surface plasmon resonance detection of E. coli and methicillin-resistant S. aureus using bacteriophages. Biosens. Bioelectron. 2012, 37, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Vidic, J.; Manzano, M.; Chang, C.-M.; Jaffrezic-Renault, N. Advanced biosensors for detection of pathogens related to livestock and poultry. Vet. Res. 2017, 48, 11. [Google Scholar] [CrossRef]
- Zhu, P.; Shelton, D.R.; Karns, J.S.; Sundaram, A.; Li, S.; Amstutz, P.; Tang, C.-M. Detection of water-borne E. coli O157 using the integrating waveguide biosensor. Biosens. Bioelectron. 2005, 21, 678–683. [Google Scholar] [CrossRef]
- Manzano, M.; Viezzi, S.; Mazerat, S.; Marks, R.S.; Vidic, J. Rapid and label-free electrochemical DNA biosensor for detecting hepatitis A virus. Biosens. Bioelectron. 2018, 100, 89–95. [Google Scholar] [CrossRef]
- Zhao, Y.-W.; Wang, H.-X.; Jia, G.-C.; Li, Z. Application of aptamer-based biosensor for rapid detection of pathogenic Escherichia coli. Sensors 2018, 18, 2518. [Google Scholar] [CrossRef]
- Vidic, J.; Vizzini, P.; Manzano, M.; Kavanaugh, D.; Ramarao, N.; Zivkovic, M.; Radonic, V.; Knezevic, N.; Giouroudi, I.; Gadjanski, I. Point-of-need DNA testing for detection of foodborne pathogenic bacteria. Sensors 2019, 19, 1100. [Google Scholar] [CrossRef]
- Vizzini, P.; Braidot, M.; Vidic, J.; Manzano, M. Electrochemical and Optical Biosensors for the Detection of Campylobacter and Listeria: An Update Look. Micromachines 2019, 10, 500. [Google Scholar] [CrossRef]
- Rocha, A.M.; Yuan, Q.; Close, D.M.; O’Dell, K.B.; Fortney, J.L.; Wu, J.; Hazen, T.C. Rapid detection of microbial cell abundance in aquatic systems. Biosens. Bioelectron. 2016, 85, 915–923. [Google Scholar] [CrossRef]
- Miodek, A.; Sauriat-Dorizon, H.; Chevalier, C.; Delmas, B.; Vidic, J.; Korri-Youssoufi, H. Direct electrochemical detection of PB1-F2 protein of influenza A virus in infected cells. Biosens. Bioelectron. 2014, 59, 6–13. [Google Scholar] [CrossRef]
- Nurliyana, M.; Sahdan, M.; Wibowo, K.; Muslihati, A.; Saim, H.; Ahmad, S.; Sari, Y.; Mansor, Z. The Detection Method of Escherichia Coli in Water Resources: A Review. J. Phys. Conf. Ser. 2018, 995, 012065. [Google Scholar] [CrossRef]
- Lepej, S.Z.; Poljak, M. Portable molecular diagnostic instruments in microbiology: Current status. Clin. Microbiol. Infect. 2019. [Google Scholar] [CrossRef]
- Andrews, D.; Chetty, Y.; Cooper, B.S.; Virk, M.; Glass, S.K.; Kelly, P.A.; Sudhanva, M.; Jeyaratnam, D. Multiplex PCR point of care testing versus routine, laboratory-based testing in the treatment of adults with respiratory tract infections: A quasi-randomised study assessing impact on length of stay and antimicrobial use. BMC Infect. Dis. 2017, 17, 671. [Google Scholar] [CrossRef] [PubMed]
- Schreckenberger, P.C.; McAdam, A.J. Point-counterpoint: Large multiplex PCR panels should be first-line tests for detection of respiratory and intestinal pathogens. J. Clin. Microbiol. 2015, 53, 3110–3115. [Google Scholar] [CrossRef] [PubMed][Green Version]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).