Ensemble Discrete Wavelet Transform and Gray-Level Co-Occurrence Matrix for Microcalcification Cluster Classification in Digital Mammography
Abstract
1. Introduction
2. Materials And Methods
2.1. Experimental Data
2.2. Texture Analysis
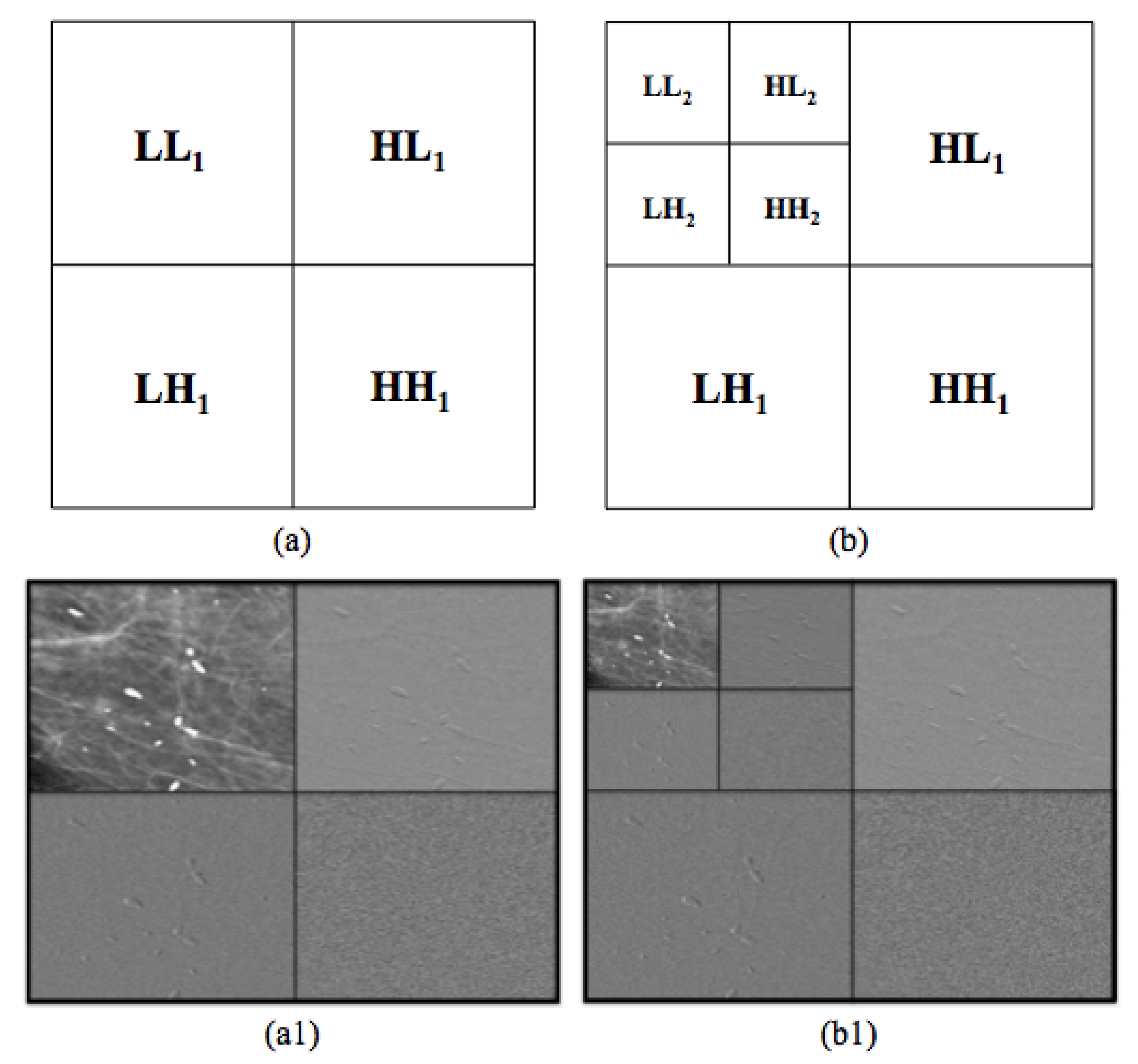
2.2.1. Multi-Scale Wavelet Decomposition
2.2.2. Gray-Level Co-Occurrence Matrix
2.3. Classification Model
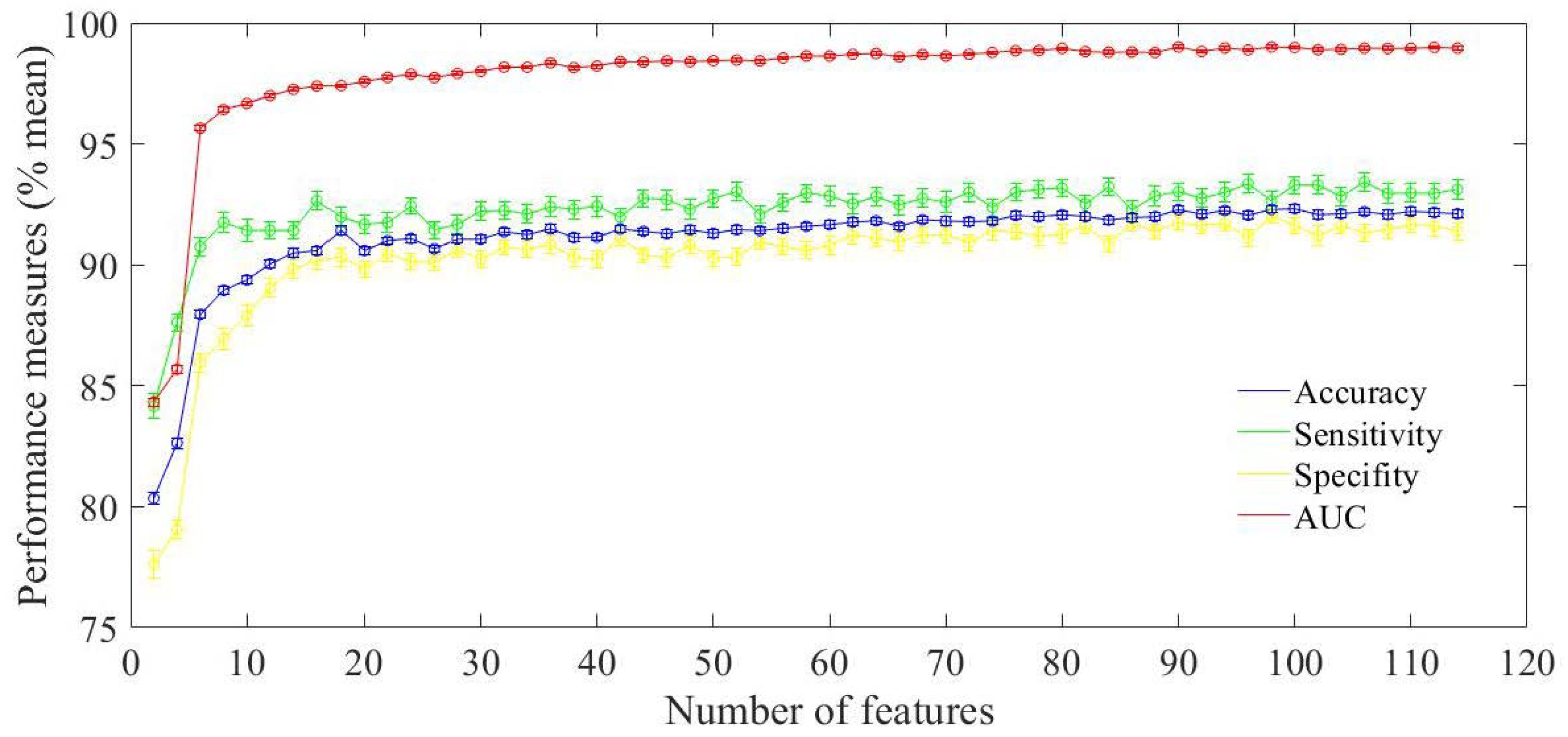
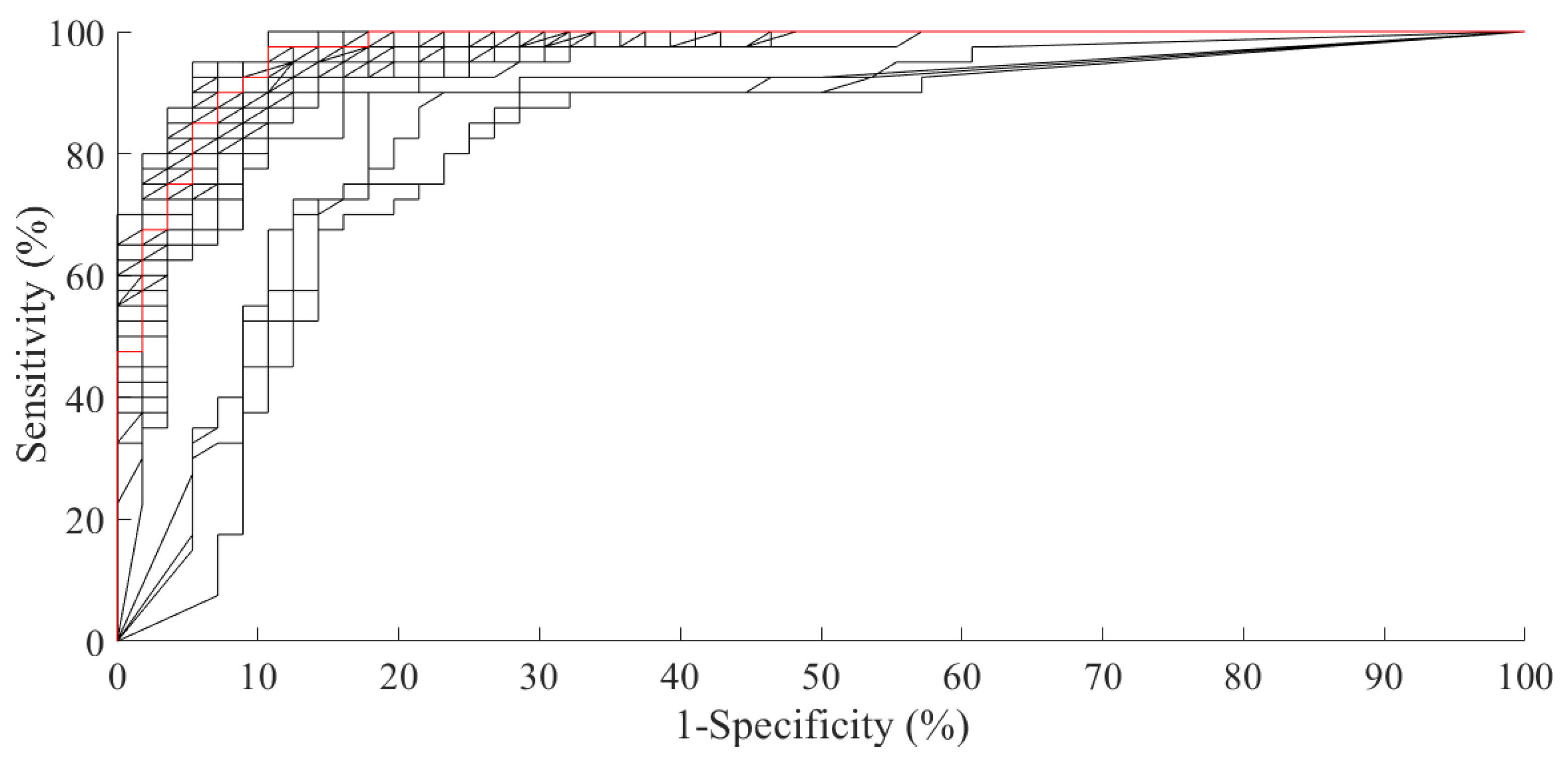
3. Performance Evaluation
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AUC | Area Under the Curve |
| BCDR | Breast Cancer Digital Repository |
| BCDR-DM | Breast Cancer Digital Repository-Digital Mammography |
| CAD | Computer Automated Detection |
| CADx | Computer Automated Diagnosis |
| CNN | Convolutional Neural Network |
| dir1 | right to left direction in GLCM |
| dir2 | left to right direction in GLCM |
| FFDM | Full-Field Digital Mammography |
| GLCM | Gray-Level Co-occurrence Matrix |
| GLCM-SF | Gray-Level Co-occurrence Matrix Statistical Features |
| HAAR-SF | Haar Statistical Features |
| HE | High-Energy |
| HH | High-High |
| HL | High-Low |
| LE | Low-Energy |
| LH | Low-High |
| LL | Low-Low |
| LDA | Linear Discriminant Analysis |
| MC | >Microcalcification |
| RF | Random Forest |
| ROC | Receiver Operating Characteristic |
| ROI | Region Of Interest |
| SVM | Support Vector Machine |
References
- Puliti, D.; Duffy, S.W.; Miccinesi, G.; De Koning, H.; Lynge, E.; Zappa, M.; Paci, E. Overdiagnosis in mammographic screening for breast cancer in Europe: A literature review. J. Med. Screen. 2012, 19, 42–56. [Google Scholar] [CrossRef] [PubMed]
- Elmore, J.G.; Armstrong, K.; Lehman, C.D.; Fletcher, S.W. Screening for breast cancer. JAMA 2005, 293, 1245–1256. [Google Scholar] [CrossRef] [PubMed]
- Khehra, B.S.; Pharwaha, A.P.S. Least-squares support vector machine for characterization of clusters of microcalcifications. World Acad. Sci. Eng. Technol. Int. J. Comput. Inf. Sci. Eng. 2013, 7, 932–941. [Google Scholar]
- Jalalian, A.; Mashohor, S.B.; Mahmud, H.R.; Saripan, M.I.B.; Ramli, A.R.B.; Karasfi, B. Computer-aided detection/diagnosis of breast cancer in mammography and ultrasound: A review. Clin. Imaging 2013, 37, 420–426. [Google Scholar] [CrossRef]
- Beura, S.; Majhi, B.; Dash, R. Mammogram classification using two dimensional discrete wavelet transform and gray-level co-occurrence matrix for detection of breast cancer. Neurocomputing 2015, 154, 1–14. [Google Scholar] [CrossRef]
- Buciu, I.; Gacsadi, A. Directional features for automatic tumor classification of mammogram images. Biomed. Signal Process. Control 2011, 6, 370–378. [Google Scholar] [CrossRef]
- Fanizzi, A.; Tangaro, S.; Bellotti, R.; Basile, T.; Bottigli, U.; Losurdo, L.; Massafra, R.; Tamborra, P.; Didonna, V.; La Forgia, D. Automatised detection of microcalcification in mammography. Phys. Med. Eur. J. Med. Phys. 2016, 32, 217. [Google Scholar] [CrossRef]
- Bozkurt, S.; Gimenez, F.; Burnside, E.S.; Gulkesen, K.H.; Rubin, D.L. Using automatically extracted information from mammography reports for decision-support. J. Biomed. Inform. 2016, 62, 224–231. [Google Scholar] [CrossRef]
- Verma, B. Novel network architecture and learning algorithm for the classification of mass abnormalities in digitized mammograms. Artif. Intell. Med. 2008, 42, 67–79. [Google Scholar] [CrossRef]
- Rodríguez-Ruiz, A.; Krupinski, E.; Mordang, J.J.; Schilling, K.; Heywang-Köbrunner, S.H.; Sechopoulos, I.; Mann, R.M. Detection of breast cancer with mammography: Effect of an artificial intelligence support system. Radiology 2018, 290, 305–314. [Google Scholar] [CrossRef]
- Fanizzi, A.; Losurdo, L.; Basile, T.M.A.; Bellotti, R.; Bottigli, U.; Delogu, P.; Diacono, D.; Didonna, V.; Fausto, A.; Lombardi, A.; et al. Fully Automated Support System for Diagnosis of Breast Cancer in Contrast-Enhanced Spectral Mammography Images. J. Clin. Med. 2019, 8, 891. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Ferrero, G.V.; Arribas, J.I. A statistical-genetic algorithm to select the most significant features in mammograms. In Proceedings of the International Conference on Computer Analysis of Images and Patterns, Vienna, Austria, 27–29 August 2007; Springer: New York, NY, USA, 2007; pp. 189–196. [Google Scholar]
- Malar, E.; Kandaswamy, A.; Chakravarthy, D.; Dharan, A.G. A novel approach for detection and classification of mammographic microcalcifications using wavelet analysis and extreme learning machine. Comput. Biol. Med. 2012, 42, 898–905. [Google Scholar] [CrossRef]
- Phadke, A.C.; Rege, P.P. Detection and Classification of Microcalcifications Using Discrete Wavelet Transform. Int. J. Emerg. Trends Technol. Comput. Sci. 2013, 2, 130–134. [Google Scholar]
- Losurdo, L.; Fanizzi, A.; Basile, T.M.; Bellotti, R.; Bottigli, U.; Dentamaro, R.; Didonna, V.; Fausto, A.; Massafra, R.; Monaco, A.; et al. A Combined Approach of Multiscale Texture Analysis and Interest Point/Corner Detectors for Microcalcifications Diagnosis. In Proceedings of the International Conference on Bioinformatics and Biomedical Engineering, Granada, Spain, 25–27 April 2018; Springer: New York, NY, USA, 2018; pp. 302–313. [Google Scholar]
- Hu, K.; Yang, W.; Gao, X. Microcalcification diagnosis in digital mammography using extreme learning machine based on hidden Markov tree model of dual-tree complex wavelet transform. Expert Syst. Appl. 2017, 86, 135–144. [Google Scholar] [CrossRef]
- Pérez, N.; Silva, A.; Ramos, I. Ensemble features selection method as tool for breast cancer classification. Int. J. Image Min. 2015, 1, 224–244. [Google Scholar] [CrossRef]
- Singh, B.; Kaur, M. An approach for classification of malignant and benign microcalcification clusters. Sādhanā 2018, 43, 39. [Google Scholar] [CrossRef]
- Ren, J. ANN vs. SVM: Which one performs better in classification of MCCs in mammogram imaging. Knowl.-Based Syst. 2012, 26, 144–153. [Google Scholar] [CrossRef]
- Fanizzi, A.; Basile, T.; Losurdo, L.; Amoroso, N.; Bellotti, R.; Bottigli, U.; Dentamaro, R.; Didonna, V.; Fausto, A.; Massafra, R.; et al. Hough transform for clustered microcalcifications detection in full-field digital mammograms. In Proceedings of the Applications of Digital Image Processing XL International Society for Optics and Photonics, San Diego, CA, USA, 19 September 2017; Volume 10396, p. 1039616. [Google Scholar]
- Gonzalez, R.C.; Woods, R.E. Digital Image Processing; Prentice-Hall, Inc.: Upper Saddle River, NJ, USA, 2002. [Google Scholar]
- Mallat, S.G. A theory for multiresolution signal decomposition: The wavelet representation. IEEE Trans. Pattern Anal. Mach. Intell. 1989, 11, 674–693. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973, 6, 610–621. [Google Scholar] [CrossRef]
- Ramos-Pollán, R.; Guevara-López, M.A.; Suárez-Ortega, C.; Díaz-Herrero, G.; Franco-Valiente, J.M.; Rubio-del Solar, M.; González-de Posada, N.; Vaz, M.A.P.; Loureiro, J.; Ramos, I. Discovering mammography-based machine learning classifiers for breast cancer diagnosis. J. Med. Syst. 2012, 36, 2259–2269. [Google Scholar] [CrossRef]
- Pathak, B.; Barooah, D. Texture analysis based on the gray-level co-occurrence matrix considering possible orientations. Int. J. Adv. Res. Electr. Electron. Instrum. Eng. 2013, 2, 4206–4212. [Google Scholar]
- Mallat, S. A Wavelet Tour of Signal Processing; Academic Press: New York, NY, USA, 1999. [Google Scholar]
- Mohanaiah, P.; Sathyanarayana, P.; GuruKumar, L. Image texture feature extraction using GLCM approach. Int. J. Sci. Res. Publ. 2013, 3, 1. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Saeys, Y.; Inza, I.; Larrañaga, P. A review of feature selection techniques in bioinformatics. Bioinformatics 2007, 23, 2507–2517. [Google Scholar] [CrossRef] [PubMed]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Youden, W. Index for rating diagnostic tests. Cancer 1950, 3, 32–35. [Google Scholar] [CrossRef]
- Alam, N.; RE Denton, E.; Zwiggelaar, R. Classification of microcalcification clusters in digital mammograms using a stack generalization based classifier. J. Imaging 2019, 5, 76. [Google Scholar] [CrossRef]
- Hepsağ, P.U.; Özel, S.A.; Yazıcı, A. Using deep learning for mammography classification. In Proceedings of the 2017 IEEE International Conference on Computer Science and Engineering (UBMK), Antalya, Turkey, 5–8 October 2017; pp. 418–423. [Google Scholar]
- Cai, H.; Huang, Q.; Rong, W.; Song, Y.; Li, J.; Wang, J.; Chen, J.; Li, L. Breast Microcalcification Diagnosis Using Deep Convolutional Neural Network from Digital Mammograms. Comput. Math. Methods Med. 2019, 2019, 2717454. [Google Scholar] [CrossRef]
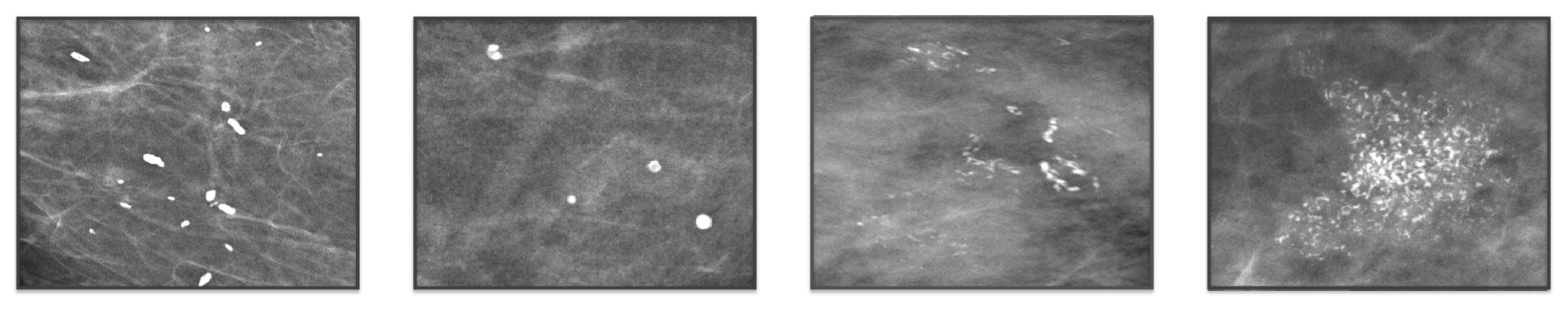

| Benign/Malignant | |
|---|---|
| Significant Features | Frequency (%) |
| Relative Smoothness Haar LL1 | 100 |
| Variance Haar LL1 | 100 |
| Relative Smoothness Haar LL2 | 100 |
| Variance Haar LL2 | 100 |
| Sum Variance GLCM HH1 dir2 | |
| Sum Variance GLCM HH1 dir1 | |
| Sum Average GLCM HH1 dir2 | |
| Autocorrelation GLCM HH1 dir2 | |
| Sum Average GLCM HH1 dir1 | |
| Mean Haar HH1 | |
| Autocorrelation GLCM HH1 dir1 | |
| Difference Entropy GLCM HH1 dir1 | |
| Sum Entropy GLCM LH1 dir2 | |
| Sum Entropy GLCM HH1 dir2 | |
| Entropy GLCM HH1 dir1 | |
| Kurtosis Haar HH2 | |
| Difference Entropy GLCM HH1 dir2 | |
| Entropy GLCM HH1 dir2 | |
| Entropy GLCM HH2 dir1 | |
| Mean Haar LL2 | |
| Kurtosis Haar HL2 | |
| Methods | DB | No. of ROIs | Features | Classifier | Acc (%) | AUC (%) |
|---|---|---|---|---|---|---|
| Ren et al. (2012) [19] | DDSM | 150 | statistical, shape and structural | SVM | - | 94 |
| Khehra et al. (2013) [3] | DDSM | 380 | statistical, shape, textural | LS-SVM | 89 | - |
| Perez et al. (2015) [17] | BCDR | 76 | shape and textural | NB | - | 79 |
| Hu et al. (2017) [16] | DDSM | 150 | textural | Extreme Learning Machine | - | 92 |
| Hepsağ et al. (2017) [33] | BCDR | 134 | shape and textural | CNN | 84 | - |
| Singh et al. (2018) [18] | DDSM | 428 | shape and textural | SVM | 94 | 93 |
| Alam et al. (2019) [32] | DDSM | 280 | shape, textural, topological | Ensemble learning | 85 | 82 |
| Cai et al. (2019) [34] | Private database | 749 | shape and textural | CNN | 87 | 94 |
| Proposed approach | BCDR | 96 | textural | RF | 92 | 97 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fanizzi, A.; Basile, T.M.; Losurdo, L.; Bellotti, R.; Bottigli, U.; Campobasso, F.; Didonna, V.; Fausto, A.; Massafra, R.; Tagliafico, A.; et al. Ensemble Discrete Wavelet Transform and Gray-Level Co-Occurrence Matrix for Microcalcification Cluster Classification in Digital Mammography. Appl. Sci. 2019, 9, 5388. https://doi.org/10.3390/app9245388
Fanizzi A, Basile TM, Losurdo L, Bellotti R, Bottigli U, Campobasso F, Didonna V, Fausto A, Massafra R, Tagliafico A, et al. Ensemble Discrete Wavelet Transform and Gray-Level Co-Occurrence Matrix for Microcalcification Cluster Classification in Digital Mammography. Applied Sciences. 2019; 9(24):5388. https://doi.org/10.3390/app9245388
Chicago/Turabian StyleFanizzi, Annarita, Teresa Maria Basile, Liliana Losurdo, Roberto Bellotti, Ubaldo Bottigli, Francesco Campobasso, Vittorio Didonna, Alfonso Fausto, Raffaella Massafra, Alberto Tagliafico, and et al. 2019. "Ensemble Discrete Wavelet Transform and Gray-Level Co-Occurrence Matrix for Microcalcification Cluster Classification in Digital Mammography" Applied Sciences 9, no. 24: 5388. https://doi.org/10.3390/app9245388
APA StyleFanizzi, A., Basile, T. M., Losurdo, L., Bellotti, R., Bottigli, U., Campobasso, F., Didonna, V., Fausto, A., Massafra, R., Tagliafico, A., Tamborra, P., Tangaro, S., Lorusso, V., & La Forgia, D. (2019). Ensemble Discrete Wavelet Transform and Gray-Level Co-Occurrence Matrix for Microcalcification Cluster Classification in Digital Mammography. Applied Sciences, 9(24), 5388. https://doi.org/10.3390/app9245388







