Yeast Chemogenetic Screening as a Tool to Unravel the Antifungal Mode of Action of Two Selected Selenocyanates
Abstract
1. Introduction
2. Materials and Methods
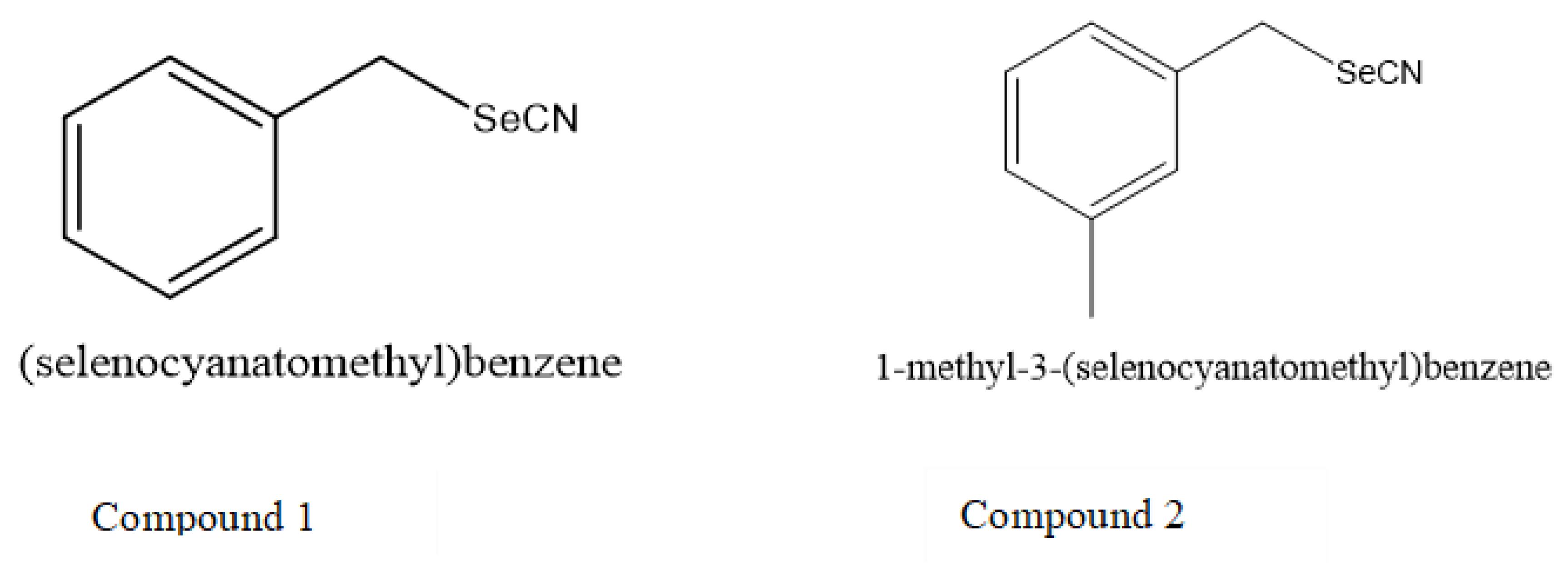
2.1. General Procedure for the Synthesis of Selenocyanates
2.2. Growth Media and Strains
2.2.1. YPD (Yeast Peptone Dextrose) Growth Medium
2.2.2. CSM (Complete Supplement Medium) Growth Medium
2.2.3. Yeast Strains
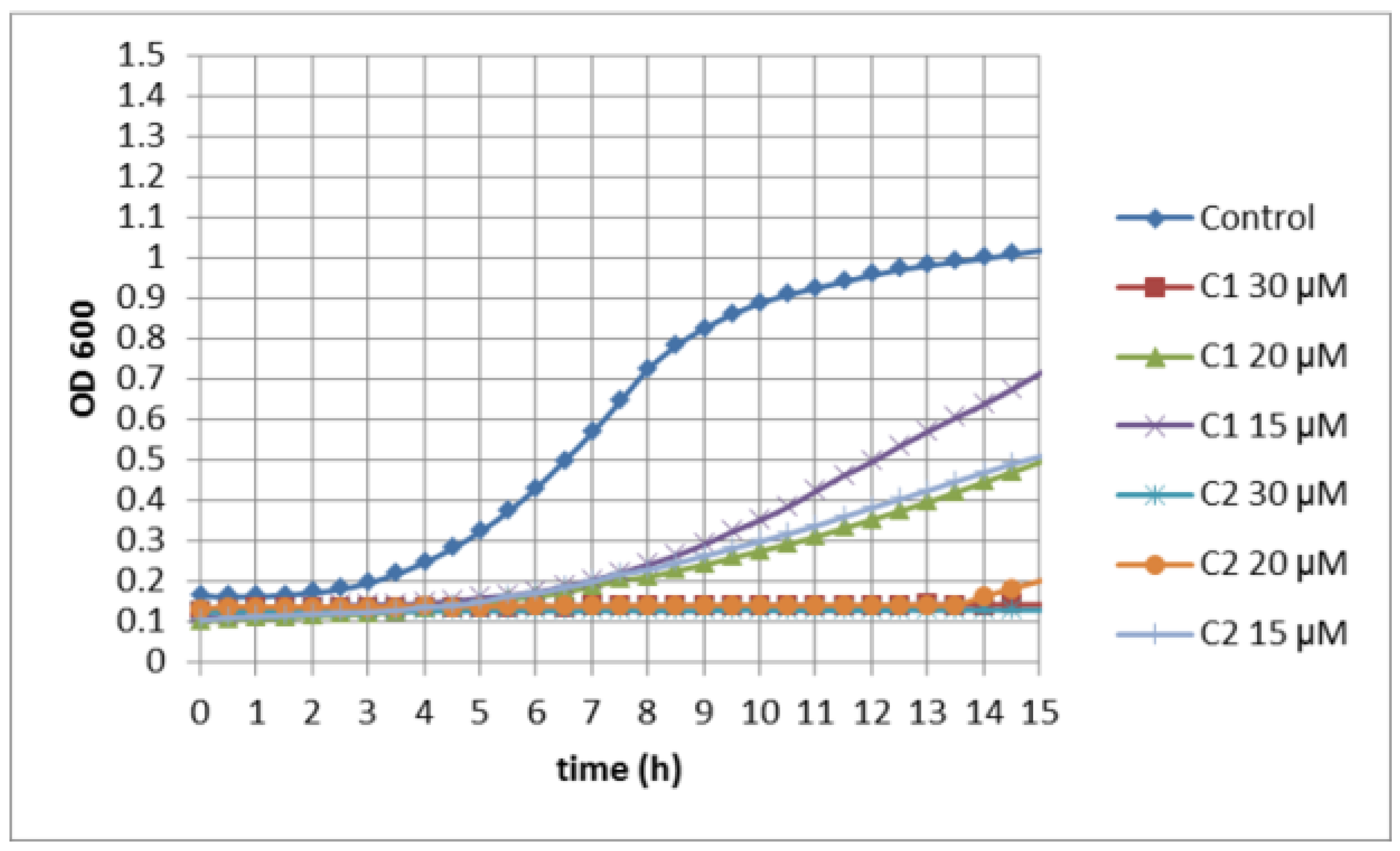
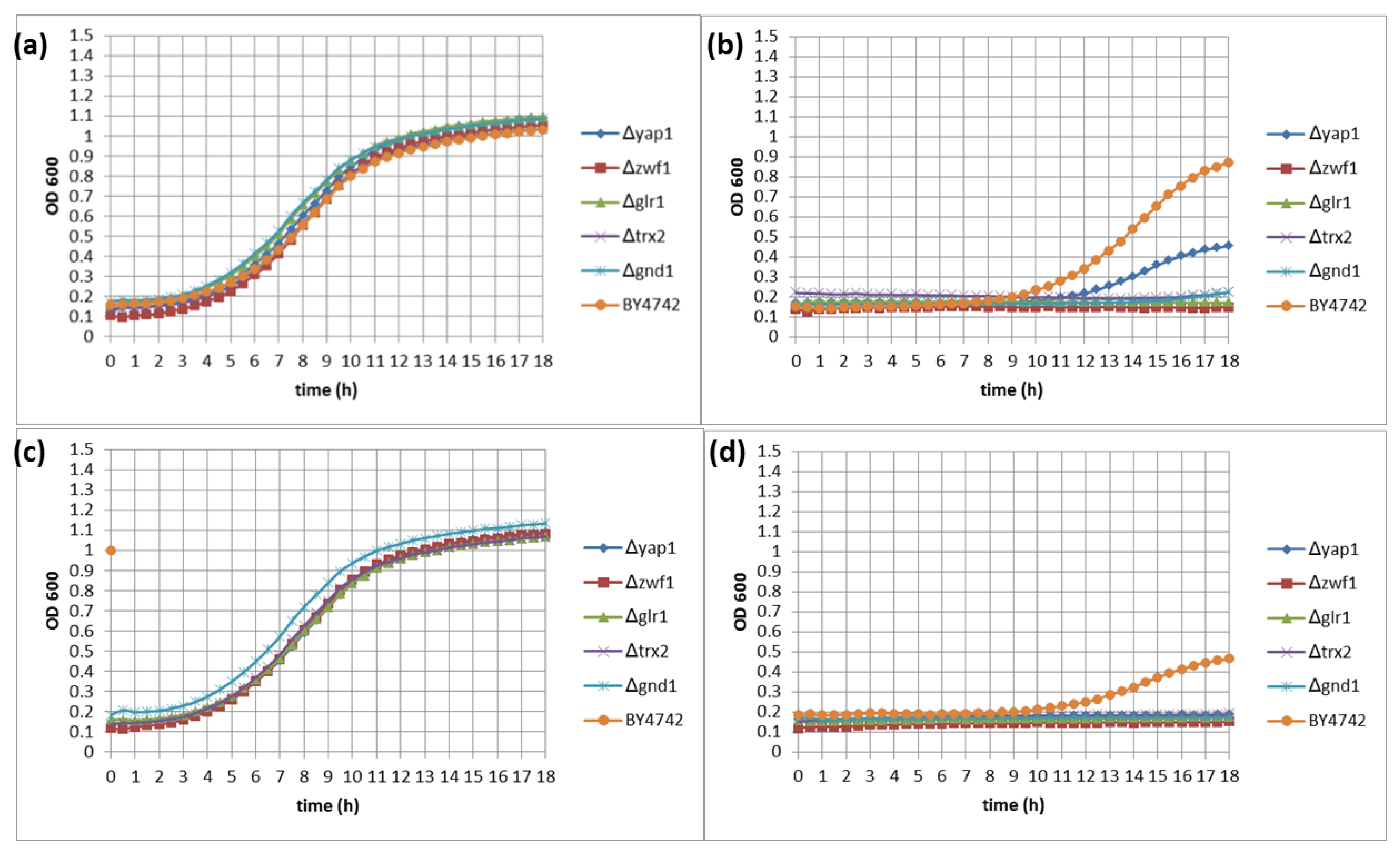
2.3. Measurements of Yeast Growth Kinetics for Quantitative Identification of Hypersensitive Strains
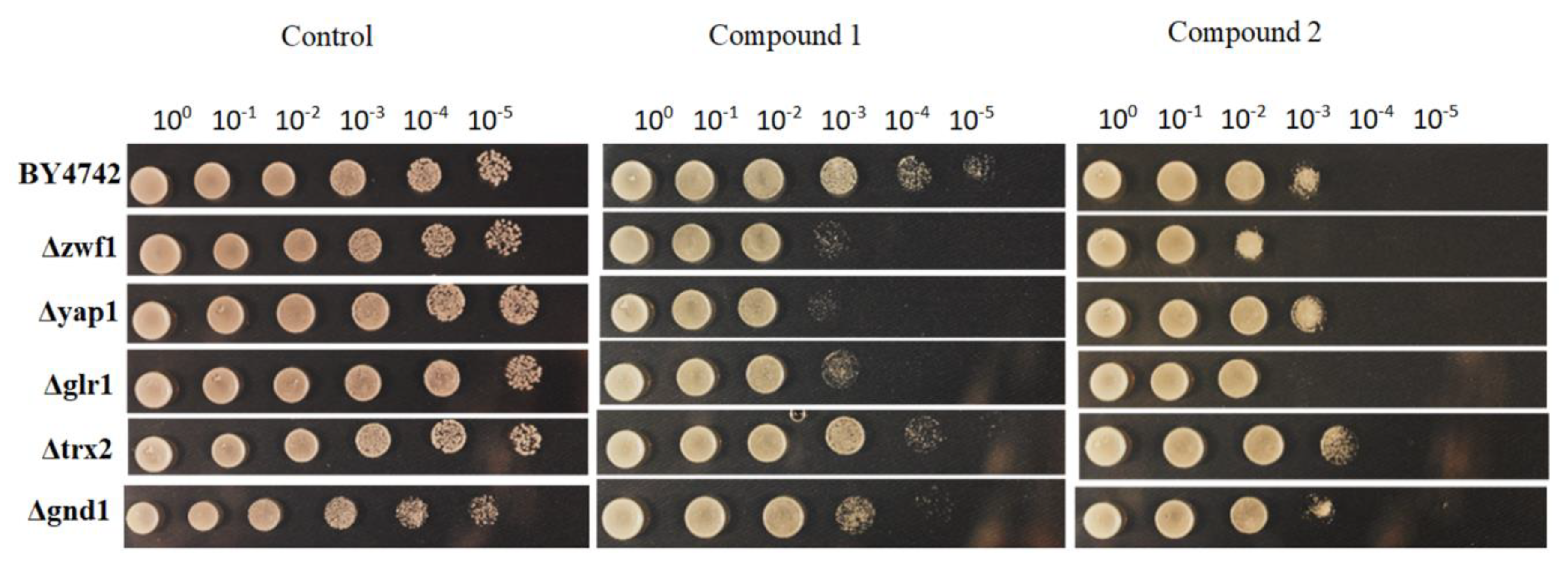
2.4. Drop Test
2.5. Plate Inhibition Zone Assay
2.6. Streak Test
2.7. Arabidopsis Seedling Root Assay
2.8. Glutathione Determination
2.9. Statistical Analysis
3. Results and Discussion
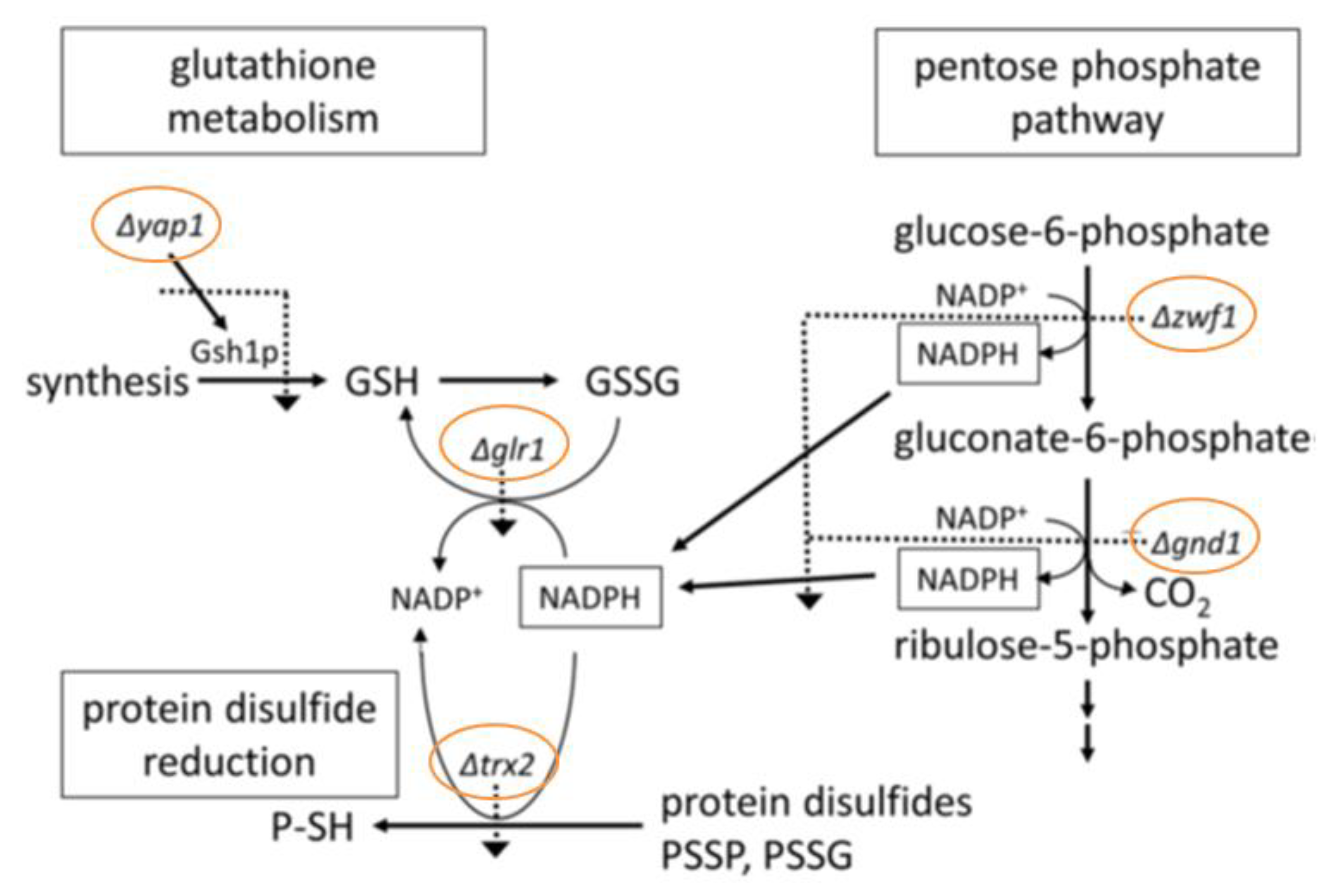
3.1. Chemogenetic Screening
3.2. Quantitative Identification of 11 Hypersensitive Strains
3.3. Activity of Selenocyanates in Drop Test
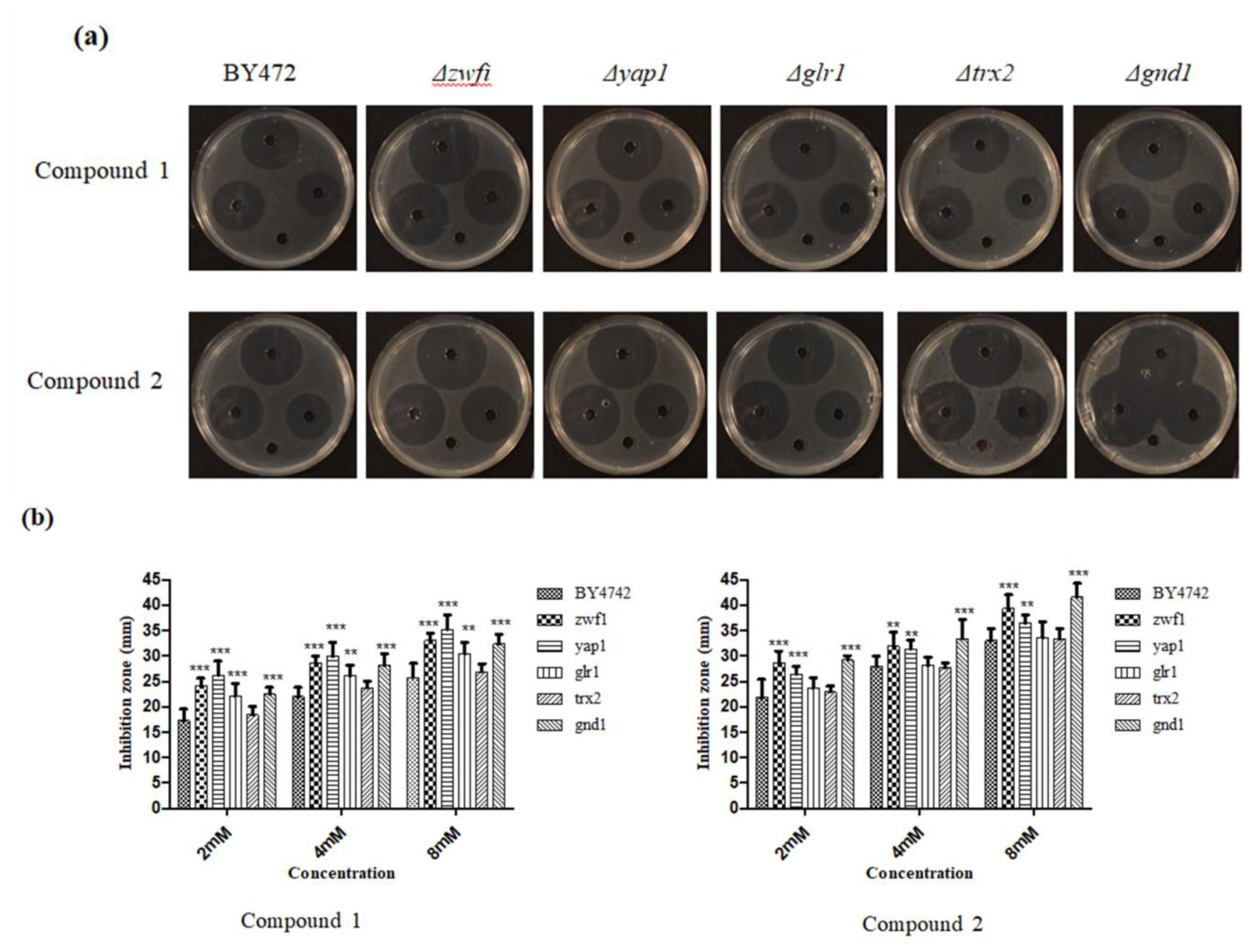
3.4. Plate Inhibition Zone Assay
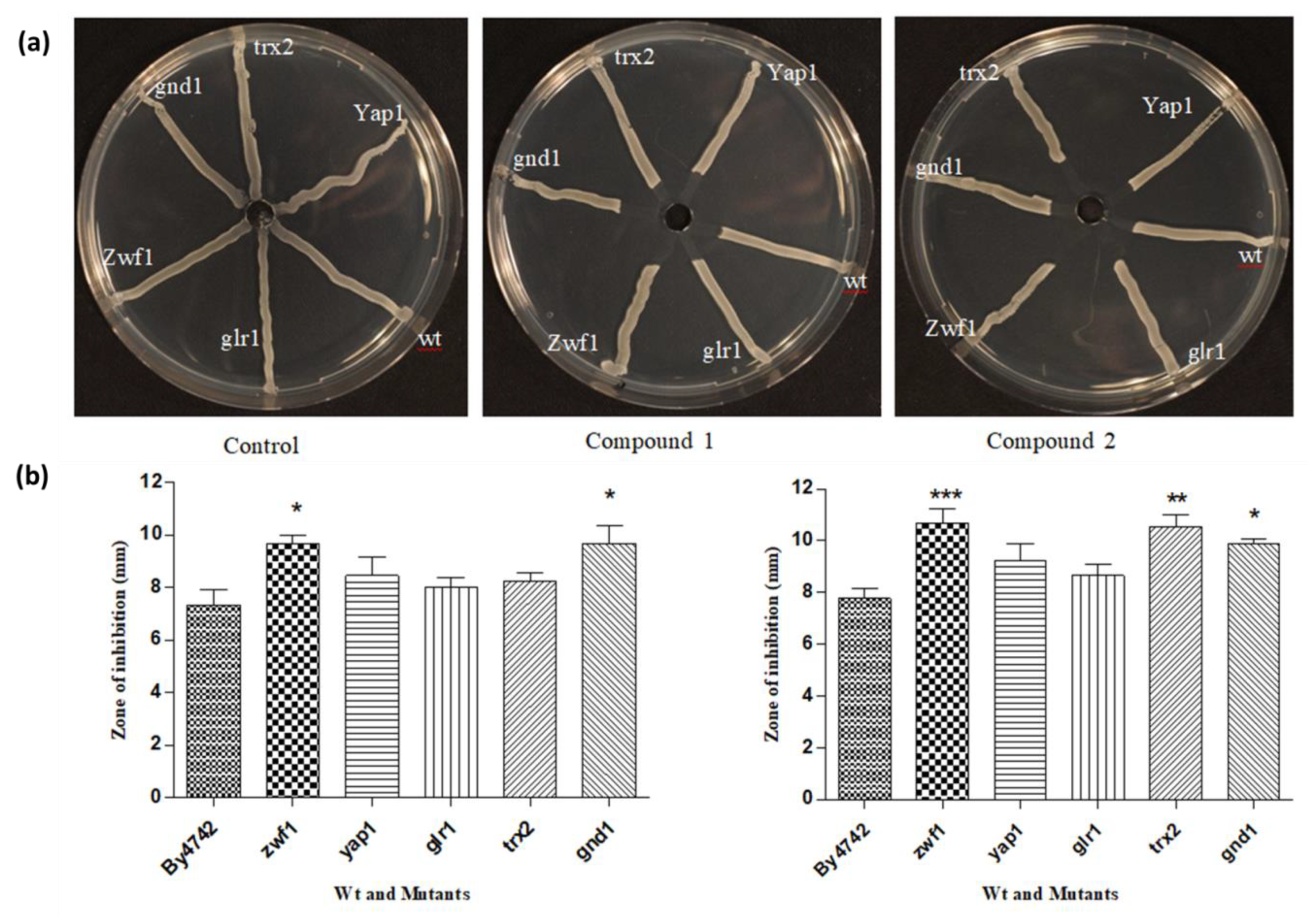
3.5. Streak Test
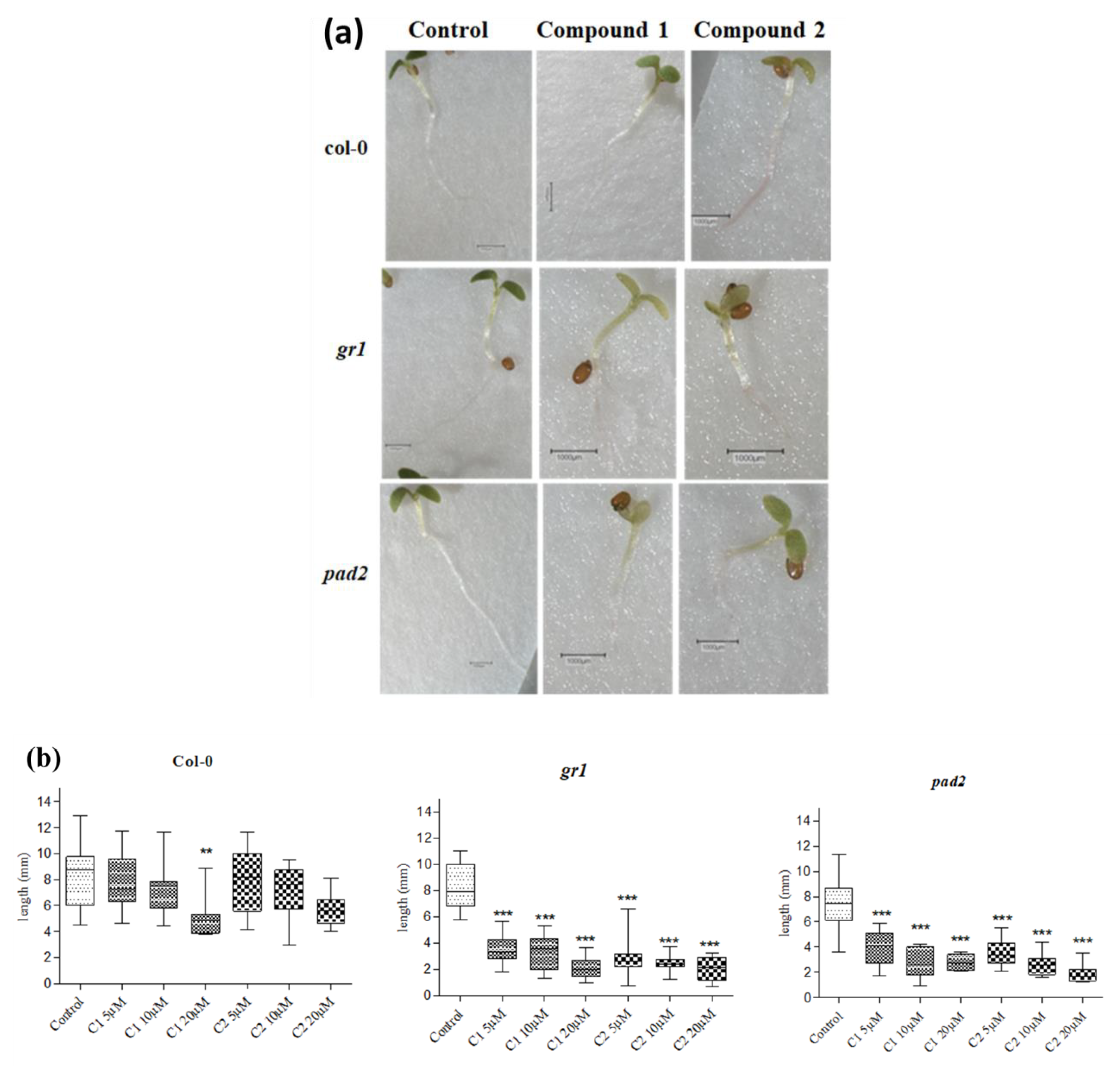
3.6. Effect of Selenocyanates on Arabidopsis Seedling Root Growth
3.7. Glutathione Determination
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jung, H.J.; Seo, Y.R. Current issues of selenium in cancer chemoprevention. Biofactors 2010, 36, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Weekley, C.M.; Harris, H.H. Which form is that? The importance of selenium speciation and metabolism in the prevention and treatment of disease. Chem. Soc. Rev. 2013, 42, 8870–8894. [Google Scholar] [CrossRef] [PubMed]
- Anwar, A.; Gould, E.; Tinson, R.; Iqbal, J.; Hamilton, C. Redox Modulation at Work: Natural Phytoprotective Polysulfanes From Alliums Based on Redox-Active Sulfur. Curr. Pharm. Rep. 2018, 4, 397. [Google Scholar] [CrossRef]
- Fry, F.H.; Holme, A.L.; Giles, N.M.; Giles, G.I.; Collins, C.; Holt, K.; Pariagh, S.; Gelbrich, T.; Hursthouse, M.B.; Gutowski, N.J.; et al. Multifunctional redox catalysts as selective enhancers of oxidative stress. Org. Biomol. Chem. 2005, 3, 2579–2587. [Google Scholar] [PubMed]
- Gandin, V.; Fernandes, A.P. Organoselenium compounds as cancer therapeutic agents. In Organoselenium Compounds in Biology and Medicine: Synthesis, Biological and Therapeutic Treatments; Royal society of Chemistry: London, UK, 2018; pp. 401–435. [Google Scholar]
- Nasim, M.J.; Ali, W.; Domínguez-Álvarez, E.; da Silva Júnior, E.N.; Saleem, R.S.Z.; Claus, J. Reactive selenium species: Redox modulation, antioxidant, antimicrobial and anticancer activities. In Organoselenium Compounds in Biology and Medicine: Synthesis, Biological and Therapeutic Treatments; The Royal Society of Chemistry: London, UK, 2018; pp. 277–302. [Google Scholar]
- Du, P.; Viswanathan, U.M.; Xu, Z.; Ebrahimnejad, H.; Hanf, B.; Burkholz, T.; Schneider, M.; Bernhardt, I.; Kirsch, G.; Jacob, C. Synthesis of amphiphilic seleninic acid derivatives with considerable activity against cellular membranes and certain pathogenic microbes. J. Hazard. Mater. 2014, 269, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Nasim, M.J.; Witek, K.; Kincses, A.; Abdin, A.Y.; Zeslawska, E.; Marc, M.A.; Gajdács, M.; Spengler, G.; Nitek, W.; Latacz, G.; et al. Pronounced activity of aromatic selenocyanates against multidrug resistant ESKAPE bacteria. N. J. Chem. 2019, 43, 6021–6031. [Google Scholar] [CrossRef]
- Sharma, A.; Sharma, A.K.; Madhunapantula, S.V.; Desai, D.; Huh, S.J.; Mosca, P.; Amin, S.; Robertson, G.P. Targeting Akt3 signaling in malignant melanoma using isoselenocyanates. Clin. Cancer Res. 2009, 15, 1674–1685. [Google Scholar] [CrossRef]
- Crampsie, M.A.; Pandey, M.K.; Desai, D.; Spallholz, J.; Amin, S.; Sharma, A.K. Phenylalkyl isoselenocyanates vs phenylalkyl isothiocyanates: Thiol reactivity and its implications. Chem. Biol. Interact. 2012, 200, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Bijlsma, M.F.; van Laarhoven, H.W. The conflicting roles of tumor stroma in pancreatic cancer and their contribution to the failure of clinical trials: A systematic review and critical appraisal. Cancer Metastasis Rev. 2015, 34, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Diaz, V.P.; Wolff, T.; Markovic, J.; Pallardo, F.V.; Foyer, C.H. A nuclear glutathione cycle within the cell cycle. Biochem. J. 2010, 431, 169–178. [Google Scholar]
- Duan, S.; Chen, C. S-nitrosylation/denitrosylation and apoptosis of immune cells. Cell Mol. Immunol. 2007, 4, 353–358. [Google Scholar] [PubMed]
- Selenius, M.; Hedman, M.; Brodin, D.; Gandin, V.; Rigobello, M.P.; Flygare, J.; Marzano, C.; Bindoli, A.; Brodin, O.; Bjornstedt, M.; et al. Effects of redox modulation by inhibition of thioredoxin reductase on radiosensitivity and gene expression. J. Cell Mol. Med. 2012, 16, 1593–1605. [Google Scholar] [CrossRef]
- Hu, C.; Liu, M.; Zhang, W.; Xu, Q.; Ma, K.; Chen, L.; Wang, Z.; He, S.; Zhu, H.; Xu, N. Upregulation of KLF4 by methylseleninic acid in human esophageal squamous cell carcinoma cells: Modification of histone H3 acetylation through HAT/HDAC interplay. Mol. Carcinog. 2015, 54, 1051–1059. [Google Scholar] [CrossRef]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Wang, Z.; Ganther, H.; Lu, J. Distinct effects of methylseleninic acid versus selenite on apoptosis, cell cycle, and protein kinase pathways in DU145 human prostate cancer cells. Mol. Cancer 2002, 1, 1059–1066. [Google Scholar]
- Thirunavukkarasu, C.; Premkumar, K.; Sheriff, A.K.; Sakthisekaran, D. Sodium selenite enhances glutathione peroxidase activity and DNA strand breaks in hepatoma induced by N-nitrosodiethylamine and promoted by phenobarbital. Mol. Cell Biochem. 2008, 310, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Seitomer, E.; Balar, B.; He, D.; Copeland, P.R.; Kinzy, T.G. Analysis of Saccharomyces cerevisiae null allele strains identifies a larger role for DNA damage versus oxidative stress pathways in growth inhibition by selenium. Mol. Nutr. Food Res. 2008, 52, 1305–1315. [Google Scholar] [CrossRef]
- De Miranda, J.X.; Andrade, F.O.; Conti, A.; Dagli, M.L.; Moreno, F.S.; Ong, T.P. Effects of selenium compounds on proliferation and epigenetic marks of breast cancer cells. J. Trace Elem. Med. Biol. 2014, 28, 486–491. [Google Scholar] [CrossRef]
- Hughes, T.; Andrews, B.; Boone, C. Old drugs, new tricks: Using genetically sensitized yeast to reveal drug targets. Cell 2004, 116, 5–7. [Google Scholar] [CrossRef]
- Giaever, G.; Shoemaker, D.D.; Jones, T.W.; Liang, H.; Winzeler, E.A.; Astromoff, A.; Davis, R.W. Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat. Genet. 1999, 21, 278–283. [Google Scholar] [CrossRef]
- Smith, A.M.; Ammar, R.; Nislow, C.; Giaever, G. A survey of yeast genomic assays for drug and target discovery. Pharmacol. Ther. 2010, 127, 156–164. [Google Scholar] [CrossRef]
- Giaever, G.; Chu, A.M.; Ni, L.; Connelly, C.; Riles, L.; Veronneau, S.; Dow, S.; Lucau-Danila, A.; Anderson, K.; Andre, B.; et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 2002, 418, 387–391. [Google Scholar] [CrossRef]
- Baldauf, S.L.; Palmer, J.D. Animals and fungi are each other’s closest relatives: Congruent evidence from multiple proteins. Proc. Natl. Acad. Sci. USA 1993, 90, 11558–11562. [Google Scholar] [CrossRef]
- Cross, F.R.; Buchler, N.E.; Skotheim, J.M. Evolution of networks and sequences in eukaryotic cell cycle control. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 366, 3532–3544. [Google Scholar] [CrossRef]
- Botstein, D.; Chervitz, S.A.; Cherry, J.M. Yeast as a model organism. Science 1997, 277, 1259–1260. [Google Scholar] [CrossRef]
- Botstein, D.; Fink, G.R. Yeast: An experimental organism for 21st Century biology. Genetics 2011, 189, 695–704. [Google Scholar] [CrossRef]
- Leontiev, R.; Slusarenko, A.J. Finding the starting point for mode-of-action studies of novel selenium compounds: Yeast as a genetic toolkit. Curr. Org. Synth. 2017, 14, 1102–1108. [Google Scholar] [CrossRef]
- Leontiev, R.; Hohaus, N.; Jacob, C.; Gruhlke, M.C.H.; Slusarenko, A.J. A Comparison of the Antibacterial and Antifungal Activities of Thiosulfinate Analogues of Allicin. Sci. Rep. 2018, 8, 6763. [Google Scholar] [CrossRef]
- Griffith, O.W. Determination of glutathione and glutathione disulfide using glutathione reductase and 2-vinylpyridine. Anal. Biochem. 1980, 106, 207–212. [Google Scholar] [CrossRef]
- Anderson, M.E. Determination of glutathione and glutathione disulfide in biological samples. Methods Enzym. 1985, 113, 548–555. [Google Scholar]
- Walker, G.M. Yeast Physiology and Biotechnology; John Wiley & Sons: Chichester, UK, 1998. [Google Scholar]
- Baetz, K.; McHardy, L.; Gable, K.; Tarling, T.; Reberioux, D.; Bryan, J.; Andersen, R.J.; Dunn, T.; Hieter, P.; Roberge, M. Yeast genome-wide drug-induced haploinsufficiency screen to determine drug mode of action. Proc. Natl. Acad. Sci. USA 2004, 101, 4525–4530. [Google Scholar] [CrossRef]
- Botet, J.; Rodriguez-Mateos, M.; Ballesta, J.P.; Revuelta, J.L.; Remacha, M. A chemical genomic screen in Saccharomyces cerevisiae reveals a role for diphthamidation of translation elongation factor 2 in inhibition of protein synthesis by sordarin. Antimicrob. Agents Chemother 2008, 52, 1623–1629. [Google Scholar] [CrossRef]
- Deutschbauer, A.M.; Jaramillo, D.F.; Proctor, M.; Kumm, J.; Hillenmeyer, M.E.; Davis, R.W.; Nislow, C.; Giaever, G. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics 2005, 169, 1915–1925. [Google Scholar] [CrossRef]
- Morano, K.A.; Grant, C.M.; Moye-Rowley, W.S. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics 2012, 190, 1157–1195. [Google Scholar] [CrossRef]
- Gruhlke, M.C.H.; Schlembach, I.; Leontiev, R.; Uebachs, A.; Gollwitzer, P.U.G.; Weiss, A.; Delaunay, A.; Toledano, M.; Slusarenko, A.J. Yap1p, the central regulator of the S. cerevisiae oxidative stress response, is activated by allicin, a natural oxidant and defence substance of garlic. Free Radic. Biol. Med. 2017, 108, 793–802. [Google Scholar] [CrossRef]
- Lee, J.; Godon, C.; Lagniel, G.; Spector, D.; Garin, J.; Labarre, J.; Toledano, M.B. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 1999, 274, 16040–16046. [Google Scholar] [CrossRef]
- Gruhlke, M.C.; Slusarenko, A.J. The biology of reactive sulfur species (RSS). Plant Physiol. Biochem. 2012, 59, 98–107. [Google Scholar] [CrossRef]
- Ankri, S.; Mirelman, D. Antimicrobial properties of allicin from garlic. Microbes Infect. 1999, 1, 125–129. [Google Scholar] [CrossRef]
- Müller, A.; Eller, J.; Albrecht, F.; Prochnow, P.; Kuhlmann, K.; Bandow, J.E.; Slusarenko, A.J.; Leichert, L.I. Allicin Induces Thiol Stress in Bacteria through S-Allylmercapto Modification of Protein Cysteines. J. Biol. Chem. 2016, 291, 11477–11490. [Google Scholar] [CrossRef]
- Outten, C.E.; Culotta, V.C. A novel NADH kinase is the mitochondrial source of NADPH in Saccharomyces cerevisiae. EMBO J. 2003, 22, 2015–2024. [Google Scholar] [CrossRef]
- Lopez-Mirabal, H.R.; Thorsen, M.; Kielland-Brandt, M.C.; Toledano, M.B.; Winther, J.R. Cytoplasmic glutathione redox status determines survival upon exposure to the thiol-oxidant 4,4’-dipyridyl disulfide. Fems Yeast Res. 2007, 7, 391–403. [Google Scholar] [CrossRef]
- Temple, M.D.; Perrone, G.G.; Dawes, I.W. Complex cellular responses to reactive oxygen species. Trends Cell Biol. 2005, 15, 319–326. [Google Scholar] [CrossRef]
- Gulshan, K.; Rovinsky, S.A.; Coleman, S.T.; Moye-Rowley, W.S. Oxidant-specific folding of Yap1p regulates both transcriptional activation and nuclear localization. J. Biol. Chem. 2005, 280, 40524–40533. [Google Scholar] [CrossRef]
- Kuge, S.; Jones, N.; Nomoto, A. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 1997, 16, 1710–1720. [Google Scholar] [CrossRef]
- Yan, C.; Lee, L.H.; Davis, L.I. Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 1998, 17, 7416–7429. [Google Scholar] [CrossRef]
- Delaunay, A.; Isnard, A.D.; Toledano, M.B. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 2000, 19, 5157–5166. [Google Scholar] [CrossRef]
- Ahmad, KA.; Clement, M.V.; Pervaiz, S. Pro-oxidant activity of low doses of resveratrol inhibits hydrogen peroxide-induced apoptosis. Ann. N. Y. Acad. Sci. 2003, 1010, 365–373. [Google Scholar] [CrossRef]
- Escote, X.; Miranda, M.; Menoyo, S.; Rodriguez-Porrata, B.; Carmona-Gutierrez, D.; Jungwirth, H.; Madeo, F.; Cordero, R.R.; Mas, A.; Tinahones, F.; et al. Resveratrol induces antioxidant defence via transcription factor Yap1p. Yeast 2012, 29, 251–263. [Google Scholar] [CrossRef]
- Vernoux, T.; Wilson, R.C.; Seeley, K.A.; Reichheld, J.P.; Muroy, S.; Brown, S.; Maughan, S.C.; Cobbett, C.S.; Van, M.M.; Inze, D.; et al. The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 2000, 12, 97–110. [Google Scholar] [CrossRef]
- Parisy, V.; Poinssot, B.; Owsianowski, L.; Buchala, A.; Glazebrook, J.; Mauch, F. Identification of PAD2 as a gamma-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant. J. 2007, 49, 159–172. [Google Scholar] [CrossRef]
- Mhamdi, A.; Hager, J.; Chaouch, S.; Queval, G.; Han, Y.; Taconnat, L.; Saindrenan, P.; Gouia, H.; Issakidis-Bourguet, E.; Renou, J.P.; et al. Arabidopsis GLUTATHIONE REDUCTASE1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol. 2010, 153, 1144–1160. [Google Scholar] [CrossRef] [PubMed]
| ORF | Gene | Biological/Molecular Function |
|---|---|---|
| YNL241C | ZWF1 | glucose-6-phosphate dehydrogenase activity |
| YML007W | YAP1 | DNA-binding transcription factor activity |
| YPL091W | GLR1 | glutathione-disulfide reductase activity |
| YGR209C | TRX2 | disulfide oxidoreductase activity |
| YHR183W | GND1 | phosphogluconate dehydrogenase (decarboxylating) activity |
| YOR375C | GDH1 | glutamate dehydrogenase (NADP+) activity |
| YDL085W | NDE2 | NADH dehydrogenase activity |
| YDR197W | CBS2 | translation regulator activity |
| YJR009C | TDH2 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) activity |
| YGR234W | YHB1 | nitric oxide reductase activity |
| YML056C | IMD4 | IMP dehydrogenase activity |
| Experiment 1 | Experiment 2 | |||||
|---|---|---|---|---|---|---|
| Untreated Cells | Treated Cells (Compound 1) | Treated Cells (Compound 2) | Untreated Cells | Treated Cells (Compound 1) | Treated Cells (Compound 2) | |
| Total cellular conc. of glutathione [mM] | 1.730 ± 0.115 | 1.170 ± 0.020 | 1.120 ± 0.025 | 2.050 ± 0.130 | 1.176 ± 0.0530 | 1.150 ± 0.000 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarfraz, M.; Nasim, M.J.; Jacob, C.; Gruhlke, M.C.H. Yeast Chemogenetic Screening as a Tool to Unravel the Antifungal Mode of Action of Two Selected Selenocyanates. Appl. Sci. 2019, 9, 3728. https://doi.org/10.3390/app9183728
Sarfraz M, Nasim MJ, Jacob C, Gruhlke MCH. Yeast Chemogenetic Screening as a Tool to Unravel the Antifungal Mode of Action of Two Selected Selenocyanates. Applied Sciences. 2019; 9(18):3728. https://doi.org/10.3390/app9183728
Chicago/Turabian StyleSarfraz, Muhammad, Muhammad Jawad Nasim, Claus Jacob, and Martin C.H. Gruhlke. 2019. "Yeast Chemogenetic Screening as a Tool to Unravel the Antifungal Mode of Action of Two Selected Selenocyanates" Applied Sciences 9, no. 18: 3728. https://doi.org/10.3390/app9183728
APA StyleSarfraz, M., Nasim, M. J., Jacob, C., & Gruhlke, M. C. H. (2019). Yeast Chemogenetic Screening as a Tool to Unravel the Antifungal Mode of Action of Two Selected Selenocyanates. Applied Sciences, 9(18), 3728. https://doi.org/10.3390/app9183728








