Multiscale Entropy Analysis with Low-Dimensional Exhaustive Search for Detecting Heart Failure
Abstract
1. Introduction
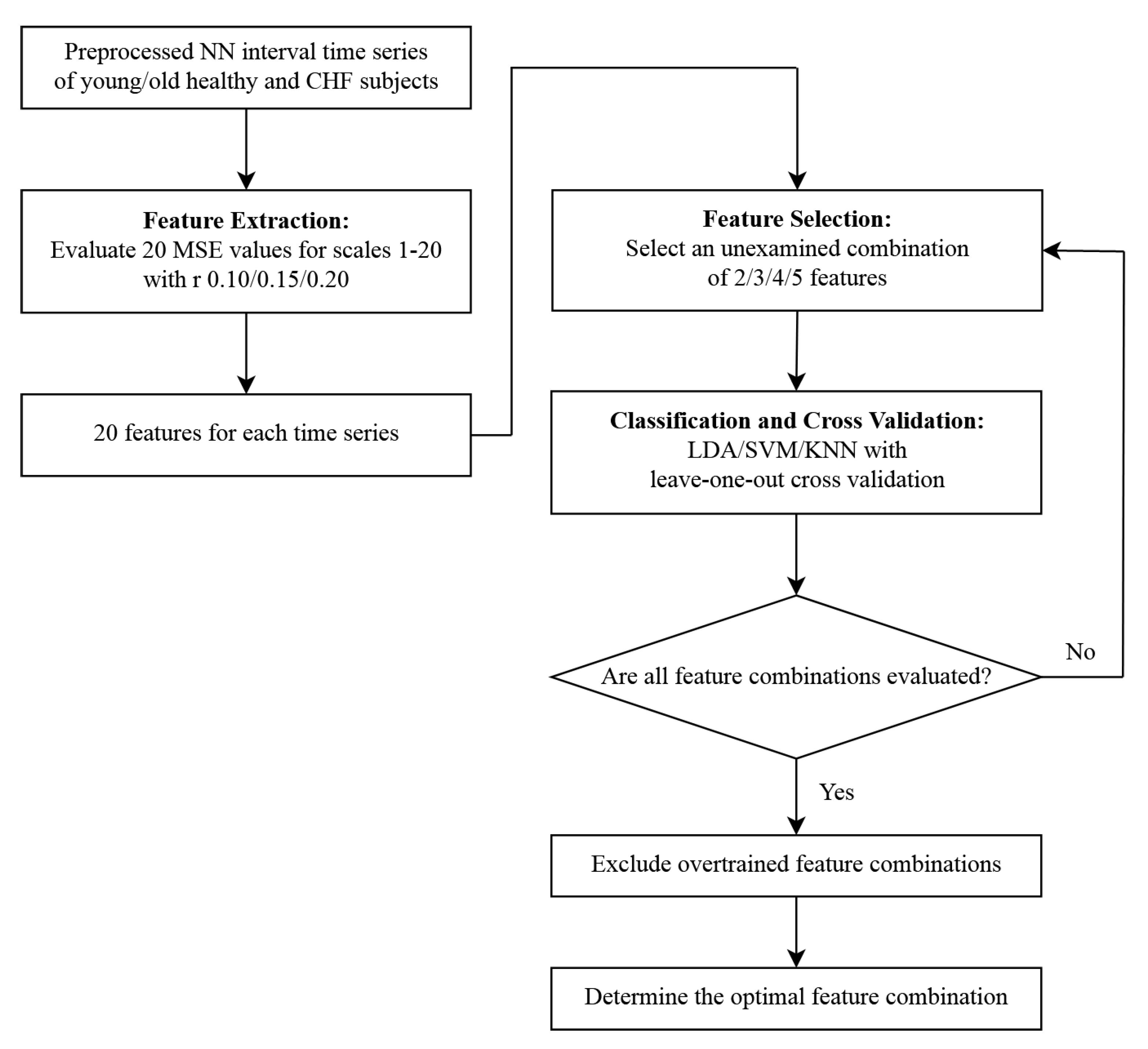
2. Materials and Methods
2.1. Multiscale Entropy
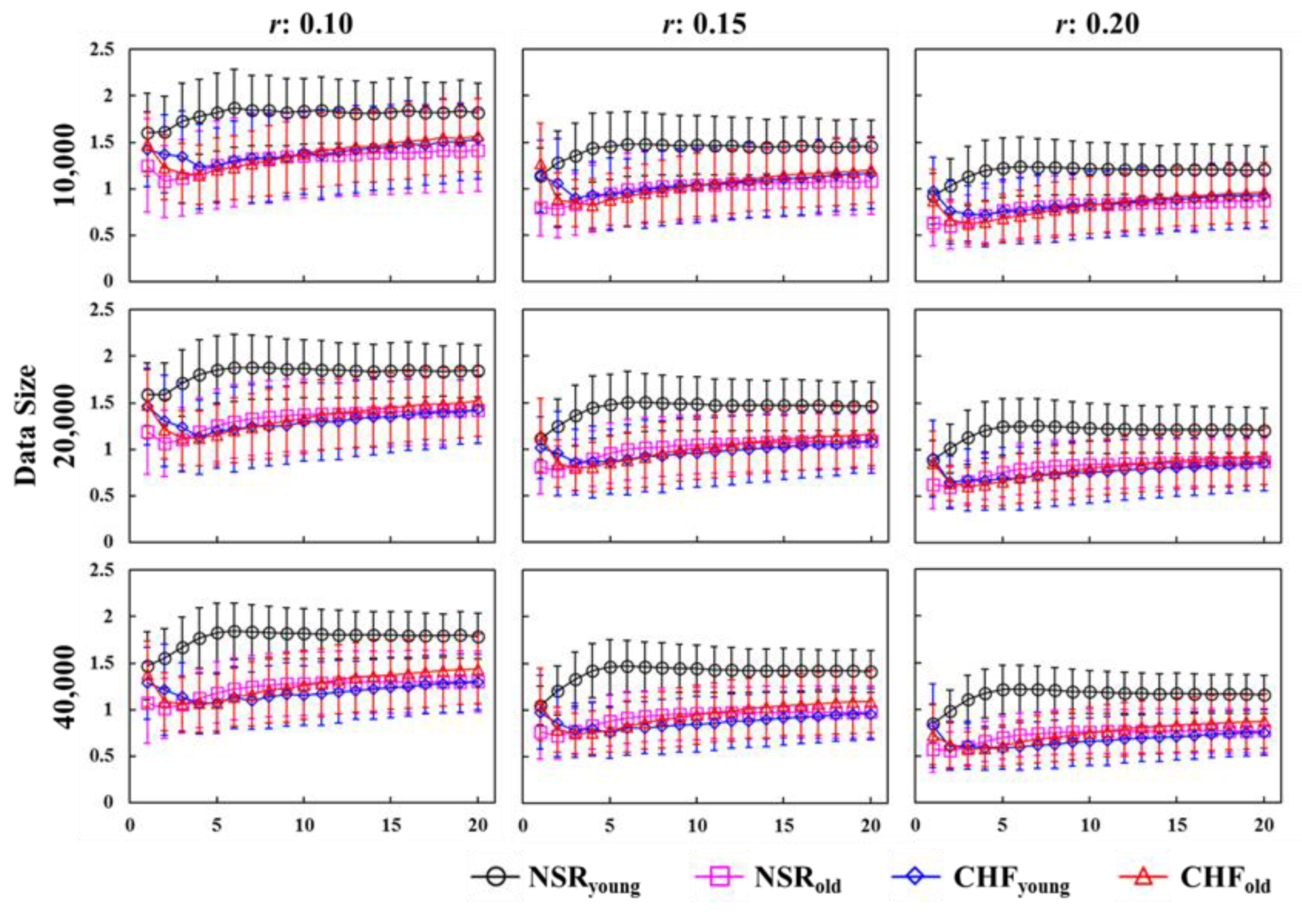
2.2. Data
2.3. Machine Learning
2.4. Performance Metrics
3. Results
3.1. Multiscale Entropy Evaluation
3.2. Discrimination between Younger NSR and CHF Participants
3.3. Discrimination between Older NSR and CHF Participants
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McMurray, J.J.; Pfeffer, M.A. Heart failure. Lancet 2005, 365, 1877–1889. [Google Scholar] [CrossRef]
- National Clinical Guideline Centre (UK). Chronic Heart Failure: National Clinical Guideline for Diagnosis and Management in Primary and Secondary Care; Royal College of Physicians: London, UK, 2010. [Google Scholar]
- Ponikowski, P.; Voors, A.A.; Anker, S.D.; Bueno, H.; Cleland, J.G.; Coats, A.J.; Falk, V.; Gonzalez-Juanatey, J.R.; Harjola, V.P.; Jankowska, E.A.; et al. 2016 esc guidelines for the diagnosis and treatment of acute and chronic heart failure: The task force for the diagnosis and treatment of acute and chronic heart failure of the european society of cardiology (esc). Developed with the special contribution of the heart failure association (hfa) of the esc. Eur. J. Heart Fail. 2016, 18, 891–975. [Google Scholar] [PubMed]
- Gottdiener, J.S.; McClelland, R.L.; Marshall, R.; Shemanski, L.; Furberg, C.D.; Kitzman, D.W.; Cushman, M.; Polak, J.; Gardin, J.M.; Gersh, B.J.; et al. Outcome of congestive heart failure in elderly persons: Influence of left ventricular systolic function—The cardiovascular health study. Ann. Int. Med. 2002, 137, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Cleland, J.G.F.; Daubert, J.; Erdmann, E.; Freemantle, N.; Gras, D.; Kappenberger, L.; Tavazzi, L.; Cleland, J.G.F.; Daubert, J.C.; Erdmann, E.; et al. The effect of cardiac resynchronization on morbidity and mortality in heart failure. N. Engl. J. Med. 2005, 352, 1539–1549. [Google Scholar] [CrossRef] [PubMed]
- Nolan, J.; Batin, P.D.; Andrews, R.; Lindsay, S.J.; Brooksby, P.; Mullen, H.; Baig, W.; Flapan, A.D.; Cowley, A.; Prescott, R.J.; et al. Prospective study of heart rate variability and mortality in chronic heart failure—Results of the United Kingdom heart failure evaluation and assessment of risk trial (UK-heart). Circulation 1998, 98, 1510–1516. [Google Scholar] [CrossRef] [PubMed]
- Neubauer, S. Mechanisms of disease—The failing heart—An engine out of fuel. N. Engl. J. Med. 2007, 356, 1140–1151. [Google Scholar] [CrossRef]
- Lane, R.E.; Cowie, M.R.; Chow, A.W.C. Prediction and prevention of sudden cardiac death in heart failure. Heart 2005, 91, 674–680. [Google Scholar] [CrossRef]
- Kishi, T. Heart failure as an autonomic nervous system dysfunction. J. Cardiol. 2012, 59, 117–122. [Google Scholar] [CrossRef]
- Triposkiadis, F.; Karayannis, G.; Giamouzis, G.; Skoularigis, J.; Louridas, G.; Butler, J. The sympathetic nervous system in heart failure physiology, pathophysiology, and clinical implications. J. Am. Coll. Cardiol. 2009, 54, 1747–1762. [Google Scholar] [CrossRef]
- Camm, A.J.; Malik, M.; Bigger, J.T.; Breithardt, G.; Cerutti, S.; Cohen, R.J.; Coumel, P.; Fallen, E.L.; Kennedy, H.L.; Kleiger, R.E.; et al. Heart rate variability. Standards of measurement, physiological interpretation, and clinical use. Eur. Heart J. 1996, 17, 354–381. [Google Scholar]
- Pumprla, J.; Howorka, K.; Groves, D.; Chester, M.; Nolan, J. Functional assessment of heart rate variability: Physiological basis and practical applications. Int. J. Cardiol. 2002, 84, 1–14. [Google Scholar] [CrossRef]
- Gutierrez-Tobal, G.C.; Alvarez, D.; Gomez-Pilar, J.; del Campo, F.; Hornero, R. Assessment of time and frequency domain entropies to detect sleep apnoea in heart rate variability recordings from men and women. Entropy 2015, 17, 123–141. [Google Scholar] [CrossRef]
- Costa, M.; Goldberger, A.L.; Peng, C.K. Multiscale entropy analysis of complex physiologic time series. Phys. Rev. Lett. 2002, 89, 068102. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.; Goldberger, A.L.; Peng, C.K. Multiscale entropy analysis of biological signals. Phys. Rev. E 2005, 71, 021906. [Google Scholar] [CrossRef] [PubMed]
- Pincus, S.M. Approximate entropy as a measure of system-complexity. Proc. Natl. Acad. Sci. USA 1991, 88, 2297–2301. [Google Scholar] [CrossRef] [PubMed]
- Richman, J.S.; Moorman, J.R. Physiological time-series analysis using approximate entropy and sample entropy. Am. J. Physiol.-Heart Circ. 2000, 278, H2039–H2049. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.T.; Zhuang, J.; Yu, W.X.; Wang, Z.Z. Measuring complexity using fuzzyen, apen, and sampen. Med. Eng. Phys. 2009, 31, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.D.; Cheng, J.S.; Yang, Y.; Luo, S.R. A rolling bearing fault diagnosis method based on multi-scale fuzzy entropy and variable predictive model-based class discrimination. Mech. Mach. Theory 2014, 78, 187–200. [Google Scholar] [CrossRef]
- Li, M.A.; Liu, H.N.; Zhu, W.; Yang, J.F. Applying improved multiscale fuzzy entropy for feature extraction of mi-eeg. Appl. Sci. 2017, 7, 92. [Google Scholar] [CrossRef]
- Rostaghi, M.; Azami, H. Dispersion entropy: A measure for time-series analysis. IEEE Signal Proc. Lett. 2016, 23, 610–614. [Google Scholar] [CrossRef]
- Azami, H.; Rostaghi, M.; Abasolo, D.; Escudero, J. Refined composite multiscale dispersion entropy and its application to biomedical signals. IEEE Trans. Biomed. Eng. 2017, 64, 2872–2879. [Google Scholar]
- Zhang, Y.D.; Tong, S.G.; Cong, F.Y.; Xu, J. Research of feature extraction method based on sparse reconstruction and multiscale dispersion entropy. Appl. Sci. 2018, 8, 888. [Google Scholar] [CrossRef]
- Fazan, F.S.; Brognara, F.; Fazan, R.; Murta, L.O.; Silva, L.E.V. Changes in the complexity of heart rate variability with exercise training measured by multiscale entropy-based measurements. Entropy 2018, 20, 47. [Google Scholar] [CrossRef]
- Li, P.; Liu, C.Y.; Li, K.; Zheng, D.C.; Liu, C.C.; Hou, Y.L. Assessing the complexity of short-term heartbeat interval series by distribution entropy. Med. Biol. Eng. Comput. 2015, 53, 77–87. [Google Scholar] [CrossRef]
- Lee, D.Y.; Choi, Y.S. Multiscale distribution entropy analysis of short-term heart rate variability. Entropy 2018, 20, 952. [Google Scholar] [CrossRef]
- Bandt, C.; Pompe, B. Permutation entropy: A natural complexity measure for time series. Phys. Rev. Lett. 2002, 88, 174102. [Google Scholar] [CrossRef]
- Ouyang, G.X.; Li, J.; Liu, X.Z.; Li, X.L. Dynamic characteristics of absence eeg recordings with multiscale permutation entropy analysis. Epilepsy Res. 2013, 104, 246–252. [Google Scholar] [CrossRef]
- Gao, Y.D.; Villecco, F.; Li, M.; Song, W.Q. Multi-scale permutation entropy based on improved lmd and hmm for rolling bearing diagnosis. Entropy 2017, 19, 176. [Google Scholar] [CrossRef]
- Shannon, C.E. The mathematical theory of communication (reprinted). MD Comput. 1997, 14, 306–317. [Google Scholar]
- Hsu, C.F.; Wei, S.Y.; Huang, H.P.; Hsu, L.; Chi, S.; Peng, C.K. Entropy of entropy: Measurement of dynamical complexity for biological systems. Entropy 2017, 19, 550. [Google Scholar] [CrossRef]
- Hsu, C.F.; Lin, P.Y.; Chao, H.H.; Hsu, L.; Chi, S. Average entropy: Measurement of disorder for cardiac rr interval signals. Physica A 2019, 529, 1215333. [Google Scholar] [CrossRef]
- Sena, P.; Attianese, P.; Pappalardo, M.; Villecco, F. FIDELITY: Fuzzy Inferential Diagnostic Engine for on-Line Support to Physicians. In Proceedings of the 4th International Conference on Biomedical Engineering in Vietnam, Ho Chi Minh City, Vietnam, 8–10 January 2012; Toi, V., Toan, N., Dang Khoa, T., Lien Phuong, T., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 396–400. [Google Scholar]
- Acharya, U.R.; Fujita, H.; Sudarshan, V.K.; Oh, S.L.; Muhammad, A.; Koh, J.E.W.; Tan, J.H.; Chua, C.K.; Chua, K.P.; Tan, R.S. Application of empirical mode decomposition (emd) for automated identification of congestive heart failure using heart rate signals. Neural Comput. Appl. 2017, 28, 3073–3094. [Google Scholar] [CrossRef]
- Melillo, P.; Fusco, R.; Sansone, M.; Bracale, M.; Pecchia, L. Discrimination power of long-term heart rate variability measures for chronic heart failure detection. Med. Biol. Eng. Comput. 2011, 49, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Pecchia, L.; Melillo, P.; Sansone, M.; Bracale, M. Discrimination power of short-term heart rate variability measures for chf assessment. IEEE Trans. Inf. Technol. Biomed. 2011, 15, 40–46. [Google Scholar] [CrossRef]
- Pecchia, L.; Melillo, P.; Bracale, M. Remote health monitoring of heart failure with data mining via cart method on hrv features. IEEE Trans. Biomed. Eng. 2011, 58, 800–804. [Google Scholar] [CrossRef]
- Kamath, C. A new approach to detect congestive heart failure using teager energy nonlinear scatter plot of r-r interval series. Med. Eng. Phys. 2012, 34, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.N.; Lee, M.Y. Bispectral analysis and genetic algorithm for congestive heart failure recognition based on heart rate variability. Comput. Biol. Med. 2012, 42, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Melillo, P.; De Luca, N.; Bracale, M.; Pecchia, L. Classification tree for risk assessment in patients suffering from congestive heart failure via long-term heart rate variability. IEEE J. Biomed. Health 2013, 17, 727–733. [Google Scholar] [CrossRef]
- Liu, G.Z.; Wang, L.; Wang, Q.; Zhou, G.M.; Wang, Y.; Jiang, Q. A new approach to detect congestive heart failure using short-term heart rate variability measures. PLoS ONE 2014, 9, e93399. [Google Scholar] [CrossRef] [PubMed]
- Narin, A.; Isler, Y.; Ozer, M. Investigating the performance improvement of HRV indices in CHF using feature selection methods based on backward elimination and statistical significance. Comput. Biol. Med. 2014, 45, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Kuntamalla, S.; Lekkala, R.G.R. Reduced data dualscale entropy analysis of HRV signals for improved congestive heart failure detection. Meas. Sci. Rev. 2014, 14, 294–301. [Google Scholar] [CrossRef]
- Huang, X.L.; Ning, X.B.; Wang, X.L. Multiscale analysis of heart beat interval increment series and its clinical significance. Chin. Sci. Bull. 2009, 54, 3784–3789. [Google Scholar] [CrossRef]
- Wessel, N.; Schirdewan, A.; Kurths, J. Intermittently decreased beat-to-beat variability in congestive heart failure. Phys. Rev. Lett. 2003, 91, 119801. [Google Scholar] [CrossRef] [PubMed]
- Thuraisingham, R.A.; Gottwald, G.A. On multiscale entropy analysis for physiological data. Physica A 2006, 366, 323–332. [Google Scholar] [CrossRef]
- Costa, M.; Healey, J.A. Multiscale entropy analysis of complex heart rate dynamics: Discrimination of age and heart failure effects. Comput. Cardiol. 2003, 30, 705–708. [Google Scholar]
- Chen, Y.; Yang, H. A comparative analysis of alternative approaches for quantifying nonlinear dynamics in cardiovascular system. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 2599–2602. [Google Scholar]
- Liu, C.Y.; Gao, R. Multiscale entropy analysis of the differential rr interval time series signal and its application in detecting congestive heart failure. Entropy 2017, 19, 251. [Google Scholar]
- Awan, I.; Aziz, W.; Shah, I.H.; Habib, N.; Alowibdi, J.S.; Saeed, S.; Nadeem, M.S.A.; Shah, S.A.A. Studying the dynamics of interbeat interval time series of healthy and congestive heart failure subjects using scale based symbolic entropy analysis. PLoS ONE 2018, 13, e0196823. [Google Scholar] [CrossRef]
- Pincus, S.M.; Goldberger, A.L. Physiological time-series analysis—What does regularity quantify. Am. J. Physiol. 1994, 266, H1643–H1656. [Google Scholar] [CrossRef]
- Zhao, L.N.; Wei, S.S.; Zhang, C.Q.; Zhang, Y.T.; Jiang, X.G.; Liu, F.; Liu, C.Y. Determination of sample entropy and fuzzy measure entropy parameters for distinguishing congestive heart failure from normal sinus rhythm subjects. Entropy 2015, 17, 6270–6288. [Google Scholar] [CrossRef]
- Lake, D.E.; Richman, J.S.; Griffin, M.P.; Moorman, J.R. Sample entropy analysis of neonatal heart rate variability. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002, 283, R789–R797. [Google Scholar] [CrossRef]
- Goldberger, A.L.; Amaral, L.A.N.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. Physiobank, physiotoolkit, and physionet—Components of a new research resource for complex physiologic signals. Circulation 2000, 101, E215–E220. [Google Scholar] [CrossRef]
- Cascio, D.; Taormina, V.; Cipolla, M.; Bruno, S.; Fauci, F.; Raso, G. A multi-process system for HEp-2 cells classification based on SVM. Pattern Recognit. Lett. 2016, 82, 56–63. [Google Scholar] [CrossRef]
- Benammar Elgaaied, A.; Cascio, D.; Bruno, S.; Ciaccio, M.C.; Cipolla, M.; Fauci, A.; Morgante, R.; Taormina, V.; Gorgi, Y.; Marrakchi Triki, R.; et al. Computer-assisted classification patterns in autoimmune diagnostics: The AIDA Project. BioMed Res. Int. 2016, 2016, 2073076. [Google Scholar] [CrossRef]
- Hua, Z.; Chen, C.; Zhang, R.; Liu, G.; Wen, W. Diagnosing various severity levels of congestive heart failure based on long-term HRV signal. Appl. Sci. 2019, 9, 2544. [Google Scholar]
| LDA | ||||||||||||
| 2D | 3D | 4D | 5D | |||||||||
| r | 0.10 | 0.15 | 0.20 | 0.10 | 0.15 | 0.20 | 0.10 | 0.15 | 0.20 | 0.10 | 0.15 | 0.20 |
| 40,000 | 95.5 | 90.9 | 93.2 | 95.5 | 95.5 | 95.5 | 97.7 | 95.5 | 97.7 1 | 97.7 | 97.7 | 97.7 |
| 20,000 | 86.4 | 86.4 | 86.4 | 86.4 | 86.4 | 88.6 | 88.6 | 88.6 | 90.9 1 | 88.6 | 88.6 | 90.9 |
| 10,000 | 86.4 | 86.4 | 89.6 | 88.6 | 90.9 | 90.9 | 88.6 | 93.2 | 93.2 1 | 90.9 | 93.2 | 93.2 |
| Mean ± SD | 83.6 ± 4.7 | 90.9 ± 3.6 | 92.7 ± 3.5 | 93.2 ± 3.6 | ||||||||
| SVM | ||||||||||||
| 2D | 3D | 4D | 5D | |||||||||
| r | 0.10 | 0.15 | 0.20 | 0.10 | 0.15 | 0.20 | 0.10 | 0.15 | 0.20 | 0.10 | 0.15 | 0.20 |
| 40,000 | 93.2 | 90.9 | 90.9 | 93.2 | 90.9 | 90.9 | 90.9 | 90.9 | 90.9 | 90.9 | 90.9 | 90.9 |
| 20,000 | 88.6 | 88.6 | 88.6 | 88.6 | 88.6 | 88.6 | 88.6 | 88.6 | 88.6 | 88.6 | 88.6 | 88.6 |
| 10,000 | 81.8 | 79.5 | 84.1 | 86.4 | 88.6 | 88.6 | 88.6 | 88.6 | 88.6 | 88.6 | 88.6 | 88.6 |
| Mean ± SD | 87.4 ± 4.3 | 89.1 ± 2.1 | 88.6 ± 1.9 | 88.6 ± 1.8 | ||||||||
| KNN | ||||||||||||
| 2D | 3D | 4D | 5D | |||||||||
| r | 0.10 | 0.15 | 0.20 | 0.10 | 0.15 | 0.20 | 0.10 | 0.15 | 0.20 | 0.10 | 0.15 | 0.20 |
| 40,000 | 88.6 | 88.6 | 90.9 | 90.9 | 88.6 | 90.9 | 90.9 | 88.6 | 90.9 | 88.6 | 88.6 | 90.9 |
| 20,000 | 86.4 | 86.4 | 86.4 | 86.4 | 84.1 | 88.6 | 88.6 | 86.4 | 86.4 | 88.6 | 86.4 | 86.4 |
| 10,000 | 81.8 | 84.1 | 84.1 | 81.8 | 84.1 | 86.4 | 84.1 | 86.4 | 86.4 | 88.6 | 86.4 | 86.4 |
| Mean ± SD | 86.4 ± 2.6 | 86.9 ± 3.0 | 87.6 ± 2.2 | 87.9 ± 1.5 | ||||||||
| Data Sample Sizes | Scales | Acc | Sen | Spe | PPV | NPV |
|---|---|---|---|---|---|---|
| 40,000 | {4, 5, 6, 9} | 97.7 | 94.4 | 100.0 | 100.0 | 96.3 |
| 20,000 | {1, 6, 9, 13} | 90.9 | 83.3 | 96.2 | 93.8 | 89.3 |
| {1, 6, 9, 18} | 90.9 | 83.3 | 96.2 | 93.8 | 89.3 | |
| {1, 6, 9, 19} | 90.9 | 83.3 | 96.2 | 93.8 | 89.3 | |
| 10,000 | {3, 4, 11, 16} | 93.2 | 88.9 | 96.2 | 94.1 | 92.6 |
| LDA | ||||||||||||
| 2D | 3D | 4D | 5D | |||||||||
| r | 0.10 | 0.15 | 0.20 | 0.10 | 0.15 | 0.20 | 0.10 | 0.15 | 0.20 | 0.10 | 0.15 | 0.20 |
| 40,000 | 88.7 | 88.7 | 90.1 | 90.1 | 94.4 2 | 91.5 | 91.5 | 93.0 | 93.0 | 91.5 | 93.0 | 93.0 |
| 20,000 | 83.1 | 84.5 | 84.5 | 87.3 | 90.1 2 | 88.7 | 88.7 | 90.1 | 87.3 | 88.7 | 90.1 | 88.7 |
| 10,000 | 83.1 | 84.5 | 84.5 | 90.1 | 93.0 2 | 90.1 | 88.7 | 93.0 | 93.0 | 90.1 | 93.0 | 93.0 |
| Mean ± SD | 85.8 ± 2.5 | 90.6 ± 2.0 | 90.9 ± 2.1 | 91.2 ± 1.7 | ||||||||
| SVM | ||||||||||||
| 2D | 3D | 4D | 5D | |||||||||
| r | 0.10 | 0.15 | 0.20 | 0.10 | 0.15 | 0.20 | 0.10 | 0.15 | 0.20 | 0.10 | 0.15 | 0.20 |
| 40,000 | 85.9 | 87.3 | 85.9 | 90.1 | 88.7 | 87.3 | 90.1 | 90.1 | 87.3 | 91.5 | 90.1 | 87.3 |
| 20,000 | 80.3 | 81.7 | 81.7 | 83.1 | 88.7 | 83.1 | 85.9 | 88.7 | 83.1 | 87.3 | 88.7 | 85.9 |
| 10,000 | 80.3 | 81.7 | 78.9 | 87.3 | 90.1 | 88.7 | 87.3 | 91.5 | 91.5 | 88.7 | 91.5 | 90.1 |
| Mean ± SD | 82.6 ± 2.8 | 87.5 ± 2.5 | 88.4 ± 2.6 | 89.0 ± 1.8 | ||||||||
| KNN | ||||||||||||
| 2D | 3D | 4D | 5D | |||||||||
| r | 0.10 | 0.15 | 0.20 | 0.10 | 0.15 | 0.20 | 0.10 | 0.15 | 0.20 | 0.10 | 0.15 | 0.20 |
| 40,000 | NA 3 | NA | NA | 84.5 | 84.5 | 85.9 | 85.9 | 88.7 | 85.9 | 85.9 | 85.9 | 85.9 |
| 20,000 | 77.5 | NA | NA | 81.7 | NA | NA | 83.1 | 76.1 | 81.7 | 84.5 | NA | 83.1 |
| 10,000 | 77.5 | 78.9 | 85.9 | 84.5 | 84.5 | 84.5 | 83.1 | 87.3 | 88.7 | 84.5 | 88.7 | 90.1 |
| Mean ± SD | 79.9 ± 3.5 | 84.3 ± 1.2 | 84.5 ± 3.8 | 86.1 ± 2.1 | ||||||||
| Data Sample Sizes | Scales | Acc | Sen | Spe | PPV | NPV |
|---|---|---|---|---|---|---|
| 40,000 | {3, 6, 12} | 94.4 | 84.0 | 100.0 | 100.0 | 92.0 |
| 20,000 | {1, 6, 12} | 90.1 | 76.0 | 97.8 | 95.0 | 88.2 |
| {1, 6, 13} | 90.1 | 76.0 | 97.8 | 95.0 | 88.2 | |
| {1, 6, 18} | 90.1 | 76.0 | 97.8 | 95.0 | 88.2 | |
| {2, 6, 9} | 90.1 | 72.0 | 100.0 | 100.0 | 86.8 | |
| 10,000 | {1, 9, 18} | 93.0 | 80.0 | 100.0 | 100.0 | 90.2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chao, H.-H.; Yeh, C.-W.; Hsu, C.F.; Hsu, L.; Chi, S. Multiscale Entropy Analysis with Low-Dimensional Exhaustive Search for Detecting Heart Failure. Appl. Sci. 2019, 9, 3496. https://doi.org/10.3390/app9173496
Chao H-H, Yeh C-W, Hsu CF, Hsu L, Chi S. Multiscale Entropy Analysis with Low-Dimensional Exhaustive Search for Detecting Heart Failure. Applied Sciences. 2019; 9(17):3496. https://doi.org/10.3390/app9173496
Chicago/Turabian StyleChao, Hsuan-Hao, Chih-Wei Yeh, Chang Francis Hsu, Long Hsu, and Sien Chi. 2019. "Multiscale Entropy Analysis with Low-Dimensional Exhaustive Search for Detecting Heart Failure" Applied Sciences 9, no. 17: 3496. https://doi.org/10.3390/app9173496
APA StyleChao, H.-H., Yeh, C.-W., Hsu, C. F., Hsu, L., & Chi, S. (2019). Multiscale Entropy Analysis with Low-Dimensional Exhaustive Search for Detecting Heart Failure. Applied Sciences, 9(17), 3496. https://doi.org/10.3390/app9173496






