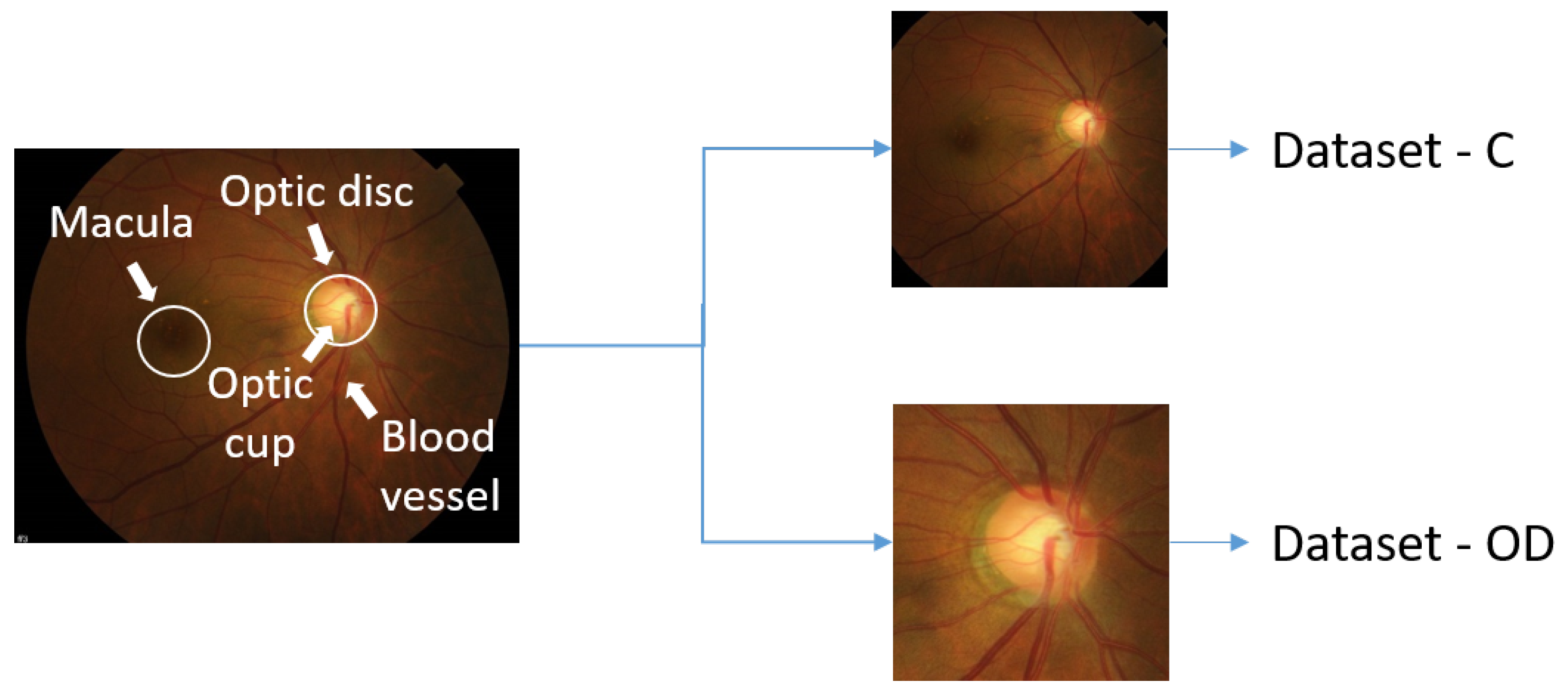
1. Introduction
Glaucoma is a progressive optic neuropathy [
1], mostly manifesting itself in the area between the optic disc and the macula. The progress of glaucoma usually remains undetected until the optic nerve gets irreversibly damaged. This damage may result in varying degrees of permanent vision loss [
2], as illustrated in
Figure 1. The earlier glaucoma is diagnosed and treated, the less patients suffer from irreversible disease progress leading to blindness.
The global prevalence of glaucoma is approximately 3–5% for people aged 40–80 years. Specifically, the number of people aged 40–80 years and affected by glaucoma worldwide was estimated to be 64 million in 2013, and this number is expected to increase to 76 million in 2020 and to 112 million in 2040 [
3].
As glaucoma progresses, glaucomatous optic disc changes (e.g., Optic Nerve Head (ONH) rim thinning or notching) and parapapillary retinal nerve fiber defects become representative morphological patterns [
4,
5,
6]. As those patterns can be captured by fundus images, an ophthalmologist is able to diagnose glaucoma by manual screening of these images. However, due to individual diversity in optic disc and retina morphology [
7,
8,
9], a human diagnosis usually does not only require fundus images, but also other types of medical images (Retinal Nerve Fiber Layer (RNFL), Optical Coherence Tomography (OCT) disc/macula, perimetry) in order to achieve high accuracy, sensitivity, and specificity values. Furthermore, the whole process of image capturing and manual screening is typically labor intensive and time consuming. Therefore, various Computer-Aided Diagnosis (CAD) techniques have been developed for identifying glaucoma, with the aim of improving the visual interpretation of fundus images by ophthalmologists. The latter type of images is commonly available in hospitals world-wide, given that their acquisition is relatively inexpensive compared to the acquisition of other types of medical images.
In the area of computer-aided glaucoma diagnosis, we can identify two major research directions: (1) glaucoma detection and (2) Optic Disc (OD) segmentation. When scanning the literature on glaucoma detection, we can observe that the accuracy of classification may vary significantly. In particular, as shown in
Table 1, the classification accuracy may range from 80%–96%, depending on the feature extraction method and the type of classifier used.
Features are often generated by higher order spectral transforms [
10,
11,
12], wavelet transforms [
13,
14], and/or thresholding [
15,
16]. It is also common to apply one or more feature extraction methods to OD images in order to compute the Cup-to-Disc Ratio (CDR) and/or the ISNT (Inferior, Superior, Nasal, Temporal) rule. The extracted features can then be fed into classifiers like
k-Nearest Neighbors (
k-NN), naive Bayes, Artificial Neural Networks (ANNs), Support Vector Machines (SVMs), Sequential Minimal Optimizations (SMOs), and Random Forests (RFs). The use of Convolutional Neural Networks (CNNs) has recently also become popular [
17].
Based on
Table 1, we can point out two additional concerns. First, many studies only make use of a limited number of images, with the number of images often varying between 50 and 200. This makes the classifiers used prone to fitting only to the given image distributions. As a result, it is highly likely that the proposed approaches do not generalize well to other sets of fundus images. Second, many studies do not provide a comprehensive quantitative analysis in terms of accuracy, sensitivity, and specificity.
When scanning the literature on OD segmentation, we can observe efforts on ONH localization using local contrast enhancement, brightest image pixel selection, and the circular Hough transform [
18]. However, the use of brightness does not always allow finding the ONH, thus sometimes leading to the wrong diagnosis result. Another popular method makes use of Principal Component Analysis (PCA) [
19] as a feature extractor. Moreover, algorithms such as the normalized correlation coefficient [
20] and pyramidal decomposition [
21] have been introduced for ONH localization. These approaches are relatively more accurate than their predecessors, while gradually reducing the time complexity. Nevertheless, when the ONH is not clearly visible in an input image, then the accuracy drops significantly [
22].
A recent effort on ONH localization leveraged CNNs [
27], using U-Net and polar transformations. However, this approach only segments the optic disc and the optic cup for measuring the CDR, given that the latter is considered to be an important factor for glaucoma diagnosis. We can therefore identify two drawbacks. First, from a clinical point of view, there is the possibility of having the wrong diagnosis outcome, given that a deviating CDR does not necessarily point to the presence of glaucoma. Second, the aforementioned approach only works in a supervised learning context. In other words, if segmentation annotations are not available for a particular dataset, then the approach at hand cannot be applied.
In this paper, we propose a novel approach towards computer-aided identification of glaucoma, making it possible to both diagnose and localize this eye disease in fundus images with a high effectiveness. More precisely, we can summarize our contributions as follows:
High diagnosis effectiveness: Our predictive model comes with a high diagnosis accuracy of 96%, as well as a high sensitivity of 96% and a high specificity of 100% for Dataset-OD, a set of center-cropped fundus images highlighting the optic disc. Current state-of-the-art techniques for computer-aided diagnosis using deep learning [
17] have an accuracy of 94%, a sensitivity of 92%, and a specificity of 96%. Furthermore, our predictive model is able to achieve an AUC score of 99%.
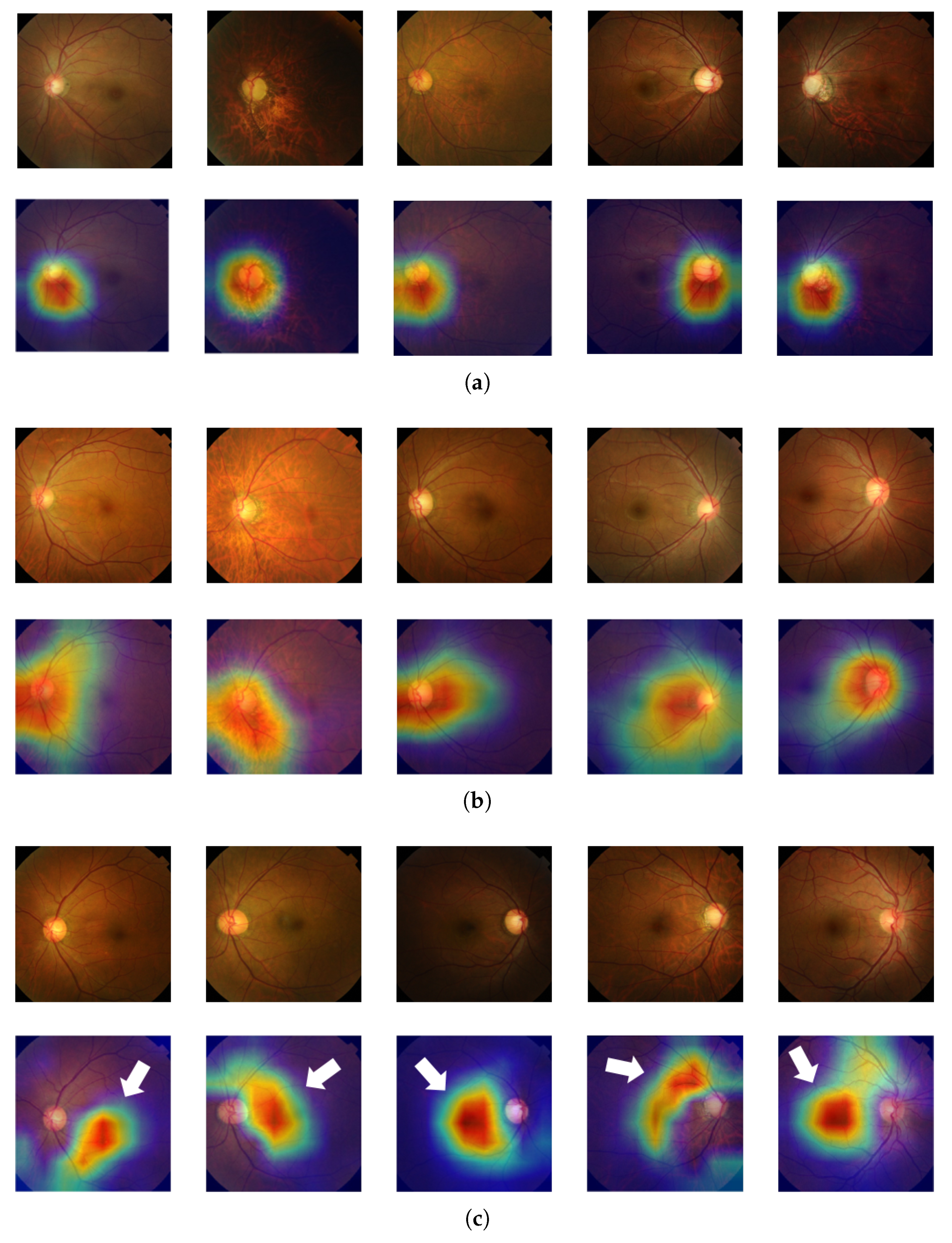
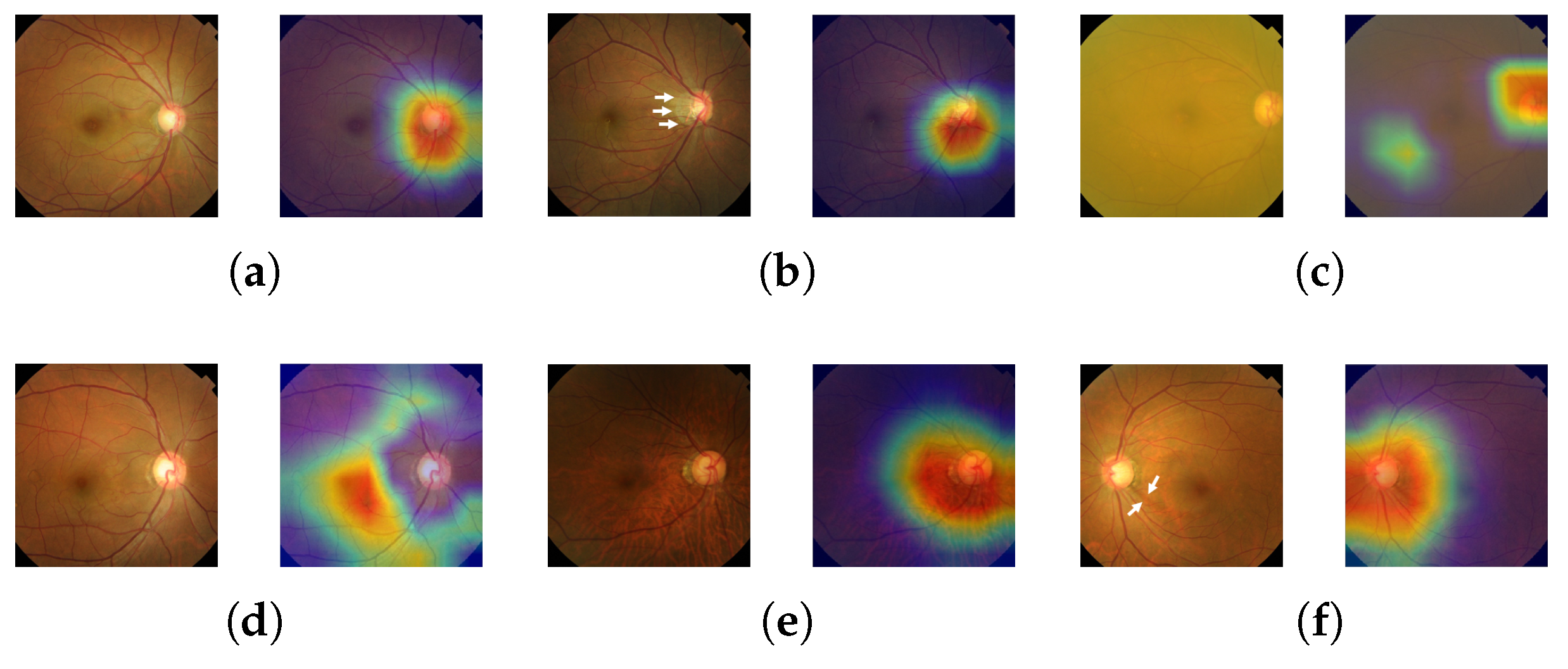
Glaucomatous area localization: Our predictive model does not only come with a high diagnosis accuracy; it is also able to localize the glaucomatous area, thus helping ophthalmologists obtain a diagnosis that is more trustworthy.
Probability of glaucoma diagnosis: Upon diagnosis of glaucoma, we provide a probability that represents the diagnostic confidence.
Use of 1903 and 220 fundus images for training and testing, respectively: The dataset used was curated by ophthalmologists working at Samsung Medical Center in Seoul, Korea. More details about this dataset, which can be made available upon request, can be found in
Section 3. Furthermore, we also validated the effectiveness of our predictive model using an external dataset [
28].
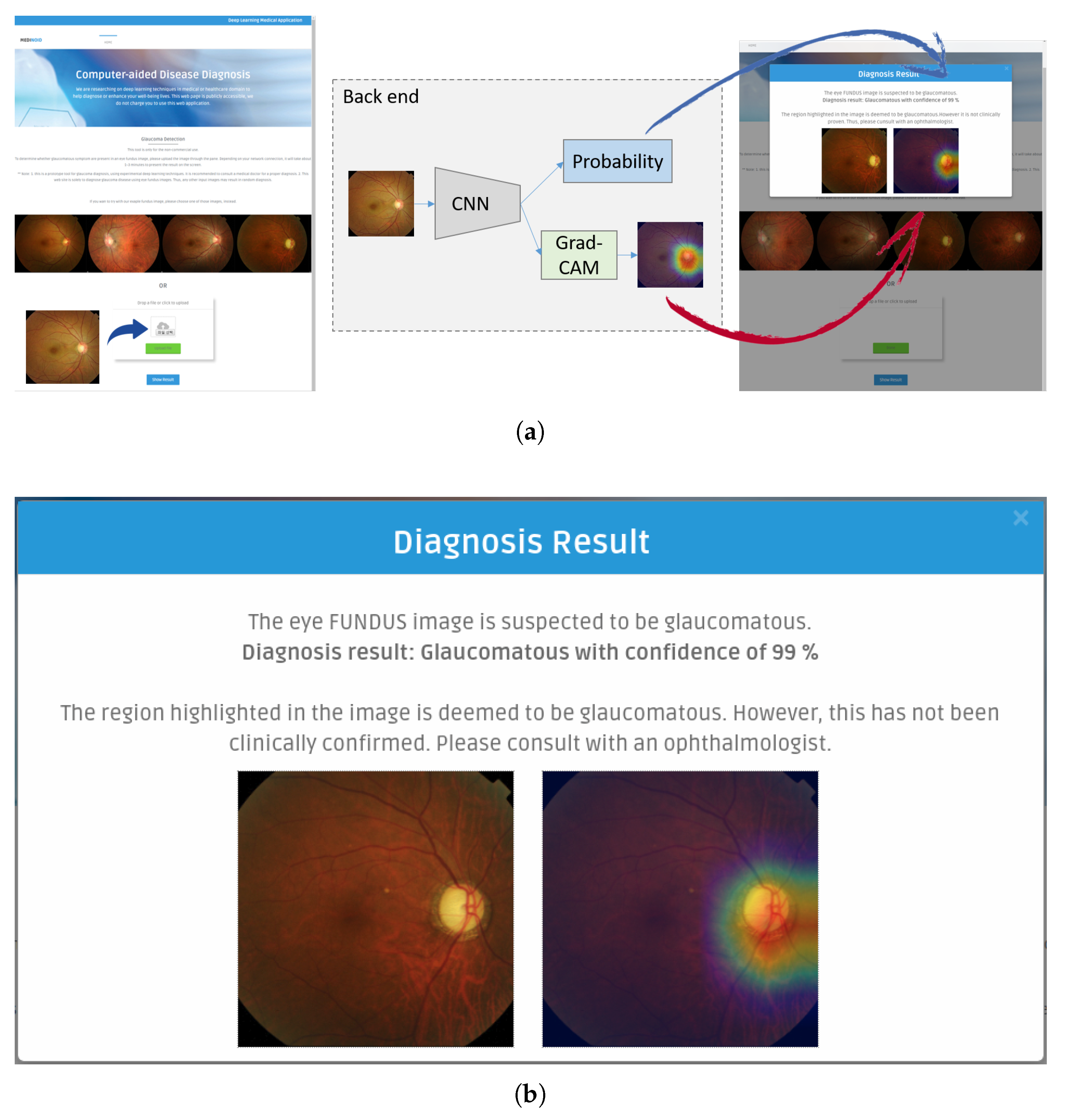
Medinoid: We have developed Medinoid (
http://www.medinoid.org), a publicly-available prototype web application for computer-aided diagnosis and localization of glaucoma in fundus images, integrating our predictive model. This web application is intuitive to use for medical doctors, as well as for people who have difficulties in accessing human experts and/or specialized medical imaging equipment.
Note that this paper is an extension of work previously presented at a conference [
29], leveraging deeper models so as to be able to increase the effectiveness of diagnosis and additional datasets so as to be able to better evaluate generalization power. Furthermore, we present more in-depth experimental results and introduce Medinoid, a web application for both glaucoma diagnosis and localization.
The remainder of this paper is organized as follows. In
Section 2, we provide an in-depth overview of our approach towards glaucoma diagnosis and localization. In
Section 3, we outline our experimental setup, and in
Section 4, we subsequently discuss the results obtained. Finally, we provide conclusions and directions for future research in
Section 5.
2. Methodology
Our proposed approach encompasses two major computer vision tasks, namely image classification and localization. The first task outputs the diagnosis result (normal or glaucomatous) and the diagnostic confidence. Upon diagnosis of glaucoma, the second task outputs the most suspicious area in the given fundus image.
At the core of the classification task is a CNN architecture that acts as a feature extractor, returning a predicted output for the class i. The softmax function is then applied to calculate the probability of each class . We used the resulting probability of the predicted output as a measure of the diagnosis confidence, given that the softmax function normalizes the output into a distribution of K probabilities.
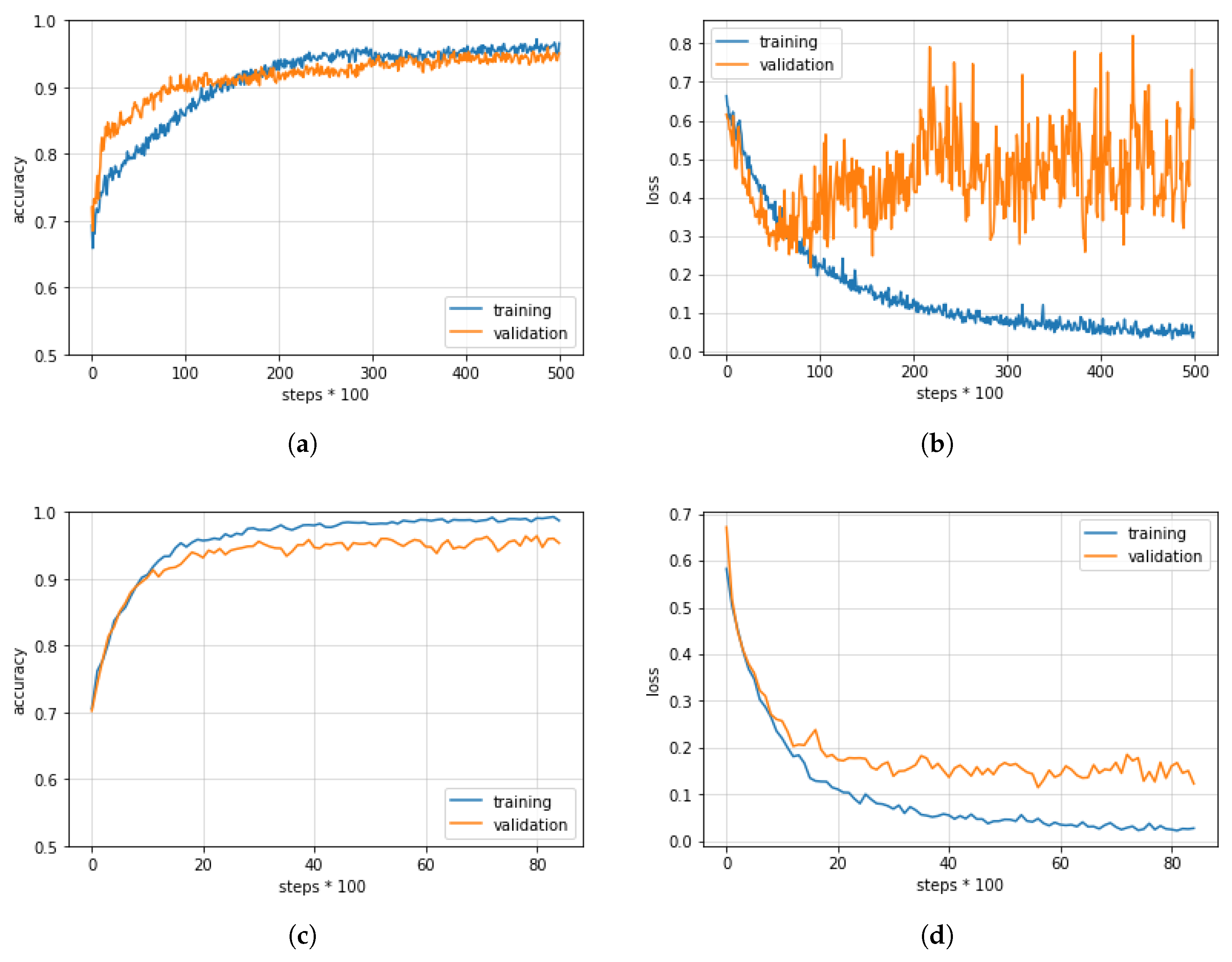
In our research, through a thorough comparison of various CNN architectures and datasets, we focused on obtaining a high diagnosis accuracy and a localization technique that is clinically meaningful. More details about the different CNN architectures used and the localization technique employed can be found below.
2.1. Convolutional Neural Networks
Given their high effectiveness, CNNs are currently the most widely-used technique in image classification [
30,
31,
32]. Their strength stems from the use of convolutional filters that are composed of neurons, also known as receptive fields. Inspired by biological processes, the neurons convolve over local regions in the input layer and respond to certain patterns, as the visual cortex does to stimuli in a local space. This is an important characteristic because, unlike the full connectivity of ordinary neural networks, this local connectivity enables CNNs to handle high-dimensional inputs such as natural images by substantially reducing the number of parameters to compute. In addition, CNNs are able to learn and classify hierarchical features directly from raw input images (end-to-end learning), thus not needing extraction of hand-crafted features. As CNNs have been demonstrated to be highly effective across various general image classification tasks, including the ImageNet Large-Scale Visual Recognition Challenge (ILSVRC), which comes with more than 1.2 millions images distributed over 1000 classes [
33], medical image analysis using CNNs followed quickly [
34], targeting use cases such as diabetic retinopathy diagnosis [
35] and chest pathology analysis [
36,
37].
To achieve our goal of effective glaucoma diagnosis, we explored three representative CNNs: (1) VGG Networks (VGGNets) [
38], (2) Inception Networks (InceptionNets) [
31], and (3) Residual Networks (ResNets) [
39]. Compared with their predecessors such as LeNetand AlexNet, these networks are all based on deeper architectures, all achieving higher accuracies while maintaining lower error rates. A comparison of the accuracies achieved by the different types of networks can be found in
Table 2.
The aforementioned CNNs were able to obtain high levels of accuracy thanks to the availability of ImageNet. This implies that, to achieve similar levels of effectiveness for our use case of glaucoma diagnosis, we need a dataset of similar size to ImageNet or we need to modify the CNNs in question so that they are able to mitigate overfitting issues caused by the usage of a limited amount of data [
34]. As such, starting from the CNNs in question, we incorporated several regularization techniques, including early stopping, data augmentation, dropout layers, and transfer learning [
41], with the aim of achieving better model generalization.
In what follows, we explain the main characteristics of each model, with
Table 3 summarizing the modifications we made.
VGGNets: Developed at and named after Oxford’s Visual Geometry Group (VGG), VGGNets [
38] were the first models to combine small receptive fields with a size of
with multiple CNN layers. VGGNets are constructed using several convolutional blocks that contain two to three convolutional layers, followed by a max pooling layer. VGG-16 is currently the most representative network, having a relatively low number of parameters, but a higher top 1 classification accuracy compared to VGG-19. Like ResNets, VGGNets require input images to be normalized, resulting in zero mean and unit variance. Furthermore, VGGNets and ResNets make use of the same input size.
VGG-16 in its original form uses Stochastic Gradient Descent (SGD) as an optimizer, also employing learning rate decay. In our research, however, we made use of the Adaptive Moment estimation (ADAM) optimizer [
42], so as to be able to overcome a number of drawbacks of SGD, including a slow convergence and a high error fluctuation.
ResNets: Microsoft Research introduced ResNets in 2016 [
39]. These models obtained the first place in the ImageNet and the COCO2015 competitions, covering the tasks of image classification, semantic segmentation, and object detection. The core idea behind ResNets is the use of a residual network, which leverages skip connections to jump over some layers, reducing the impact of vanishing gradients, as there are fewer layers through which to propagate. As such, the use of residual networks makes it possible to train networks that are deeper, obtaining lower training errors and higher levels of accuracy. Depending on the number of residual networks, ResNets come with certain variations. In our research, we used a ResNet with 152 layers (i.e., ResNet-152), given that this network, among the different ResNets available, is able to obtain some of the highest accuracy levels.
InceptionNets: Unlike VGGNets and ResNets, InceptionNets [
31] make it possible to better focus on the location of features that are important for the purpose of classification. Since salient parts can have different sizes per image, choosing the right receptive field size is difficult. In addition, in deep networks, there is the issue of overfitting and the issue of vanishing gradients. Therefore, Google Research introduced a new module, called “inception”, having several receptive fields with a different size [
43]. Specifically, filter sizes of
,
, and
were used, making the network wider. After max pooling, each output is concatenated and then sent to the next inception module. Over the course of time, inception modules have been modified and improved, leading to more powerful InceptionNets. Inception-ResNet-v2 and Inception-v4 are currently the most effective networks. By default, InceptionNets take as input images with a size of
, with pixel values belonging to [−1, 1]. The optimizer used is also different from the previous networks: InceptionNets typically make use of the Root Mean Squared Propagation (RMSProp) optimizer [
44].
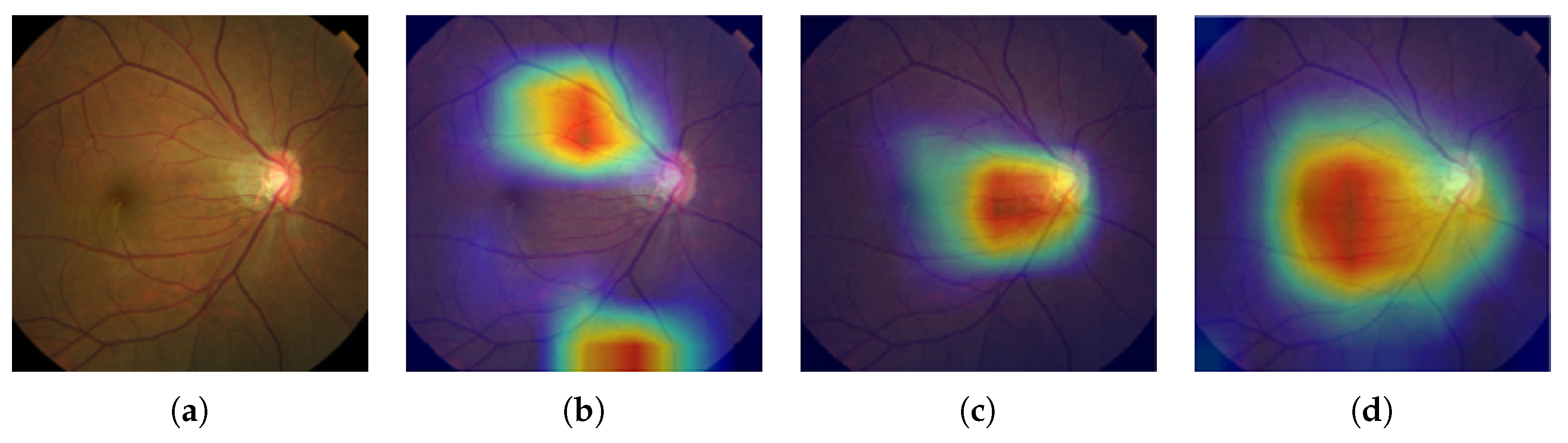
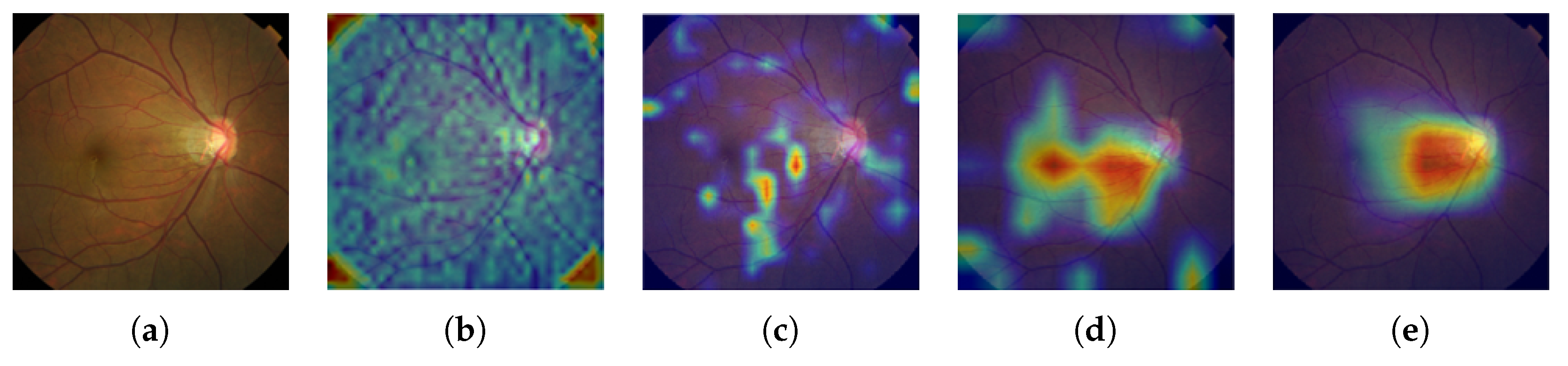
2.2. Grad-CAM
In practice, most medical images do not have any localization information, making it impossible to apply a state-of-the-art image segmentation approach like U-Net [
30]. Therefore, we adopted Gradient-weighted Class Activation Mapping (Grad-CAM) [
45] to localize glaucoma, a weakly-supervised learning approach, forgoing the need to give explicit segmentation information to a network. Grad-CAM generalizes the Class Activation Mapping (CAM) method originally proposed in [
46], integrating gradient weights. CAM visualizes what CNNs look at when they classify the input, highlighting activations in the feature maps of the last convolutional layer, using the predicted output and backpropagation.
To produce the final localization map
of the glaucomatous area, we first calculate the gradients of the predicted output
for the class glaucoma with respect to the feature maps
of the last convolutional layer, having width
u and height
v, where
, and with
K denoting the total number of feature maps. The result
then goes through a global-average pooling process, calculating the weights
of the glaucoma class as follows:
Equation (
1) can be interpreted as the importance of each feature map for the glaucomatous class. The weights and the corresponding feature maps are linearly combined and subsequently given as an input to a Rectified Linear Unit (ReLU) [
47], with the latter only selecting the positive activations. The overall process can be formally summarized as follows:
5. Conclusions and Future Research
In this paper, we introduced a new computer-aided approach towards glaucoma diagnosis and localization. The predictive CNN-based model we developed, namely ResNet-152-M, obtained the best diagnosis scores among all metrics used. Moreover, using Grad-CAM, a weakly-supervised localization method, our predictive model was able to highlight where a glaucomatous area can be found in a given input image. This demonstrates that the application of deep learning tools to medical images, even though the number of images available for training is typically small, can help doctors in diagnosing glaucoma in a more effective and efficient way. Lastly, we presented Medinoid, a web application that integrates our predictive model into its backend, making it for instance possible to diagnose glaucoma in a constrained medical environment.
If a set of training images is too small or if a set of training images is not representing well the general nature of the classes, then it is easy for a deep learning model to be biased towards the training set. In this context, we were for instance able to make the following observation: the experiments that made use of an external dataset produced lower accuracy scores than the experiments that made use of our own dataset. As a result, to mitigate model bias, future research will focus on incorporating various types of fundus images, for instance taken by other medical equipment using different angles, so as to be able to include more diversity. Furthermore, in future research, we plan to leverage our experience with deep learning-based diagnosis and localization of glaucoma in the context of other diseases, particularly paying attention to disease diagnosis and localization using 3D medical imagery.