Abstract
L-phenylseptin (L-Phes) and D-phenylseptin (D-Phes) are amphibian antimicrobial peptides isolated from the skin secretion of Hypsiboas punctatus. In the N-termini, L-Phes and D-Phes contain three consecutive phenylalanine residues, l-Phe-l-Phe-l-Phe and l-Phe-d-Phe-l-Phe, respectively. They are known to exhibit antimicrobial activity against Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa, and Xanthomonas axonopodis pv. Glycines. However, their mechanism of action and the role of the D-amino acid residue have not been elucidated yet. In this study, the interactions of both peptides with 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) were investigated by means of quartz crystal microbalance, circular dichroism, vibrational circular dichroism, 31P solid-state NMR, and molecular dynamics simulation. Both peptides have similar binding constants to the DMPC lipid bilayers, in the order of 106 M−1, and form an α-helix structure in the DMPC lipid bilayers. Both the peptides induce similar changes in the dynamics of DMPC lipids. Thus, in spite of the difference in the conformations caused by the chirality at the N-terminus, the peptides showed similar behavior in the membrane-bound state, experimentally and computationally.
1. Introduction
Antimicrobial peptides are promising candidates for new antibiotics and have broad-spectrum activity against gram-negative and gram-positive bacteria, fungi, and parasites [1,2]. Generally, these peptides are amphipathic in nature and tend to adopt α-helices in the membrane. They act on and destroy the microbial cell wall through several mechanisms, such as forming pores and channels and exerting detergent-like activity [3,4]. The membrane interactions play a major role in the peptide functions. However, even before their antimicrobial activity, the peptides may get degraded by the target organisms [5]. Therefore, incorporation of one or more d-amino acids into these peptides has received significant attention in recent decades [6].
Since the first d-amino acid was found in a protein about 100 years ago, many peptides have been discovered to have d-amino acids, which fall beyond the realm of the central dogma [7,8]. Frog skin is a precious source of d-amino acid containing antimicrobial peptides. This includes dermorphin [9], several deltorphin analogous [10,11,12,13,14], bombinins [15,16,17], and phenylseptins [18]. Interestingly, they coexist with their all l-forms and possess a d-amino acid at the second position in their sequences. The presence of d-amino acids in these peptides dramatically increases their activity [17,19].
Dermorphin contains d-alanine at position 2 and is the first example of d-amino acid containing peptides isolated from vertebrates [9,19]. Dermorphin not only exhibits higher efficiency and potency, but also elicits lesser side effects than morphine does [19]. Another example is bombinin H4, which exhibits greater binding affinity and hydrophobicity compared to its all l-isomer, bombinin H2 [15,17,20]. Bombinin H4 binds to the Leishmania-mimetic membrane 5-fold stronger than bombinin H2 does [17]. Recently, MD simulations revealed the side chain cooperation between the residues, l- Ile and d-allo-Ile in bombinin H4, which helps to insert into the model membrane. N-terminal of bombinin H4 displayed higher stability in the membrane than bombinin H2 did [21]. Electrophysiological experiments confirmed that bombinin H4 quickly forms small channels in the membrane, which enabled it to act on the model membrane rapidly [22].
Like the above-mentioned instances, l- and d-phenylseptin (l-Phes and d-Phes) peptides, isolated from the skin secretion of Hypsiboas punctatus, displayed antimicrobial activity against Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa, and Xanthomonas axonopodis pv. Glycines [18]. D-Phes exhibited 2- and 8-fold potency than L-Phes did against Staphylococcus aureus and Xanthomonas axonopodis pv. glycines, respectively [18]. Both peptides consist of 18 amino acids with the following sequences. In contrast to L-Phes, which has three L-Phe residues at positions 1-3, D-Phes contains a d-phenylalanine at the second position.
- L-Phenylseptin (L-Phes):
Phe-l-Phe-Phe-Asp-Thr-Leu-Lys-Asn-Leu-Ala-Gly-Lys-Val-Ile-Gly-Ala-Leu-Thr-NH2
- D-Phenylseptin (D-Phes):
Phe-d-Phe-Phe-Asp-Thr-Leu-Lys-Asn-Leu-Ala-Gly-Lys-Val-Ile-Gly-Ala-Leu-Thr-NH2
These peptides contain positively charged N-termini and two lysine residues, and the hydrophobic residues occur near the C-termini. Three consecutive Phe residues in D-Phes were conformationally more stable than that of L-Phes [18]. Ion mobility mass spectrometry revealed that L-Phes displayed two major conformations while D-Phes showed a major and a minor one [18]. The membrane interactions and mechanism of action of the peptides have not been well documented. Moreover, the role of d-amino acid in the function is unclear.
In this work, we performed a comparative analysis of the membrane interactions of the L-Phes and D-Phes peptides with the 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) membrane using quartz crystal microbalance (QCM), circular dichroism (CD), vibrational circular dichroism (VCD), solid-state nuclear magnetic resonance (NMR), and molecular dynamics (MD) simulations. The combination of these techniques are widely applied to monitor and study the membrane interactions, secondary structures, and the structural dynamics of the antimicrobial peptides [21,23,24].
2. Materials and Methods
2.1. Synthesis of Peptides
L-Phes and D-Phes peptides were chemically synthesized by microwave-assisted solid-phase peptide chemistry, using an automatic Initiator+ Alstra peptide synthesizer (Biotage, Japan), respectively. These peptides were purified by reverse phase HPLC (Shimazu Prominence) equipped with an ODS C18 column (Kinetex Axia) (Figure A1). The purity of each peptide was >98% without production of any diastereomers. The peptide was confirmed by MALDI-TOF-MS (Bruker Daltonics) (found [M+H]+ = 1954.2 and [M+Na]+ = 1976.2 in Figure A2). L-Phes and D-Phes peptides were dissolved in 20 mM Tris 100 mM NaCl buffer at pH 7.4.
2.2. Phospholipids
DMPC (1,2-dimyristoyl-sn-glycero-3-phosphocholine) and DHPC (1,2-dihexanoic-sn-glycero-3-phosphocholine) were purchased form Avanti Polar Lipids.
2.3. Quartz Crystal Microbalance
The binding constants (Ka) of phenylseptin peptides to DMPC membrane were estimated by changes in the frequency (F, Hz) of QCM using an Affinix QNμ (INITIUM, Japan) with a 27-MHz resonator. The relationship between the changes in mass (m, g) and F is based on Sauerbrey’s Equation (1), [25] as follows:
where F0 is the reference frequency of the QCM (27 MHz, A is the area of the gold electrode (4.9 × 10−2 cm2), ρq is the density of the quartz (2.65 g cm−3), and the shear modulus of quartz μq (2.95 x 106 N cm−2). DMPC lipid bilayers were immobilized on the Au electrode of the QCM sensor. QCM measurements were conducted at 30 °C in 500 μL of Tris buffer pH 7.4. One microliter of peptide solution at 100 μM was injected into the QCM electrode.
2.4. Far-Ultraviolet Circular Dichroism (Far-UV CD)
Bicelle solution (20 mM Tris buffer at pH 7.4) was prepared by hydration of a mixture of DMPC and 1,2-dihexanoic-sn-glycero-3-phosphocholine (DHPC) (Avanti polar lipids) at the molar ratio of 1:1. The peptide was dissolved in the suspension, the peptide-lipid (P/L) molar ratio was adjusted to 1:20. Far-ultraviolet circular dichroism (Far-UV CD) spectra of the peptides in the bicelle dispersion were acquired on a J-725 spectrometer (JASCO, Japan) at 30 °C.
2.5. Vibrational Circular Dichroism (VCD)
Peptide samples for vibrational circular dichroism (VCD) experiments were prepared by mixing a peptide and KBr at a ratio of 1:100 and turning the mixture into a transparent 10mm diameter pellet. VCD and infrared (IR) spectroscopic properties were analyzed using a JASCO PRESTO-S-2016 VCD/LD spectrometer. The IR intensity was adjusted to approximately 1.0. The sample cell was rotated along the direction of monitoring at 0° and 45° to confirm the reliability. The signals were accumulated for 10,000 scans for each sample.
2.6. P Solid-State NMR
The mixture of peptide and DMPC was dissolved in chloroform and subsequently evaporated to make a thin film. The peptide/lipid molar ratio was adjusted to 1:20. This film was carefully hydrated with Tris-NaCl buffer (pH 7.4), and then the lipids were incubated overnight at 40 °C for 1 day. Solid-state NMR experiments were performed on Bruker AVANCE III 600MHz spectrometer equipped with a double resonance standard-bore probe at 31P resonance frequencies of 242.95MHz. High-power proton dipolar decoupling (DD) under static conditions (31P DD-static MAS) were applied to measure 31P NMR signals at 40 °C for fully hydrated samples. 31P chemical shift values were determined in reference to 85% H3PO4 at 0.00ppm. The NMR data were processed and analyzed using the Bruker TOPSPIN program.
2.7. Molecular Dynamics Simulations
We performed two different types of simulations of the peptides with the membrane using GROMACS 5.1.4 [26] with CHARMM36 force field [27]. We initially modelled the α-helical conformations of the peptides using Swiss PDB Viewer program [28]. The structures with protonated N-termini (NH3+) and amidated C-termini (NH2) were considered in this work at the neutral pH condition. The initial structures of the simulations and the system contents were included in Figure A3 and Table A1.
First, we performed 50 ns simulations of L-Phes and D-Phes, which were started from the fully inserted conformations in DMPC membrane. We chose the most populated clusters of the peptides in the last 25 ns of the simulations as the main conformations of the peptides in the membrane. The conformations were applied as the initial structures of the peptides for the next two types of simulations. Second, we placed the peptides in the water region around 3 nm away from the membrane surface to observe the initial interaction difference between L-Phes and D-Phes with the model membrane. Third, C-terminal regions of the peptides were inserted into the membrane to elucidate the interface actions of the peptides.
Initial structures were constructed with CHARMM-GUI membrane builder [29]. We used the standard minimization, equilibrations, and production settings of the membrane builder [30]. Periodic boundary condition was applied in all directions. The time step was 2 fs and the data were saved in every 2 ps. Verlet cut-off scheme was employed for neighbor searching using grid cells and updated every 20 steps [31]. The linear constraint solver (LINCS) algorithm constrained all bonds [32]. Lennard–Jones interactions were switched off at 10–12Å with a force-based switching function [33]. The particle mesh Ewald method controlled the long-range electrostatics [34,35]. Nose–Hoover thermostat maintained the temperature using a time constant of 0.1 ps at 313 K [36,37]. Semi-isotropic Parrinello–Rahman barostat maintained the pressure at 1 bar with a time constant of 5.0 ps and compressibility of 4.5 × 10−5 bar−1 [38]. The analyses were performed using standard tools of the GROMACS package. The secondary structures of the peptides assigned by DSSP program [39]. The structures were displayed using visual molecular dynamics (VMD) [40]. Plots were displayed using Grace software [41].
3. Results and Discussions
3.1. QCM
QCM can estimate the affinity of membrane-bound peptide with immobilized membrane [41,42]. To investigate the affinity of phenylseptin peptides with DMPC bilayers, we performed QCM measurements. Table 1 summarizes the values of binding constants Ka of L-Phes and D-Phes. The order (106 M−1) suggested that the peptides have a certain affinity for the DMPC membrane. The binding constant values also showed that D-Phes has a slightly stronger affinity to DMPC membrane. These results indicated that the differences in the N-terminal structure might be manifest as differences in the affinity and antimicrobial activity. Additionally, L-Phes and D-Phes differed in their hydrophobicity, as evident from the different HPLC retention times of L-Phes and D-Phes (Figure A1). This is consistent with the result of de Magalhães and colleagues [18]. The higher hydrophobicity of D-Phes could be also related to the conformations adopted by the N-terminal region, which in turn, could affect the interactions of the phenylalanine residues with the model membrane.

Table 1.
The binding constants Ka of peptides to 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) using quartz crystal microbalance (QCM).
3.2. Far-UV CD Spectroscopy
Far-UV CD spectra are strongly dependent on the secondary structure of peptides [43,44]. CD spectra of L-Phes and D-Phes in DMPC/DHPC bicelle (DMPC:DHPC=1:1) are shown in Figure 1. Bicelle is used as a model systems of lipid bilayers to investigate the interaction peptide and the membranes [42,45]. The DMPC/DHPC=1:1 forms a small discoidal membrane that is able to avoid a light scattering of lipid systems. The spectral patterns show that both peptides mainly formed α-helices in DMPC/DHPC bicelles [43]. However, the magnitude of molar ellipticity of the positive band at 195 nm and the negative bands at 210 nm and 225 nm of D-Phes was lesser than those of L-Phes. This indicated that the helical content of D-Phes is slightly lesser than that of L-Phes, which could be due to the presence of D-Phe2 residue in D-Phes.
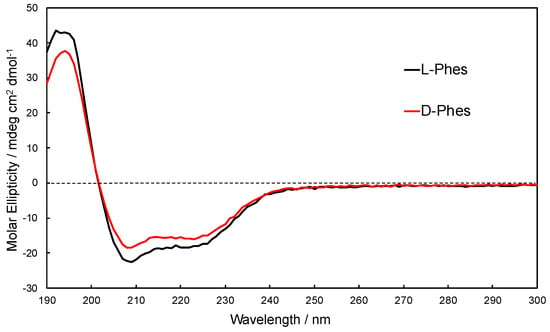
Figure 1.
Far-UV CD spectra of L-Phes (black line) and D-Phes (red line) into DMPC/1,2-dihexanoic-sn-glycero-3-phosphocholine (DHPC) bicelles.
3.3. VCD and IR Spectroscopy
VCD is sensitive a technique used to detect and characterize chiral molecules [46,47,48]. To investigate chiral effects of the Phe2 residue on the peptide structure, a comparison of VCD/IR spectra between L-Phes and D-Phes in the KBr pellets was performed as shown in Figure 2a,b. The bands of amide I and amide II vibrations appeared mainly at around 1650 and 1525 cm−1, respectively. It has been shown that typical VCD signal of α-helix exhibits a couplet at 1660 (negative)/1640 (positive) cm−1 [46]. In the VCD spectra of L-Phes and D-Phes (Figure 2a), the couplet of amide I bands was observed at 1667 (negative sign) and 1648 (positive sign) cm−1. Amide II band was observed at 1514 cm−1 (negative sign). This showed that both Phes peptides take helical structure. However, the intensity of the amide I couplet and amide II of D-Phes was significantly decreased, indicating that the conformation of N-terminal part is different between the two peptides; while the N-terminus of L-Phes adopts a helical conformation, the same region in D-Phes is disordered. Previously, we have reported the differences in the VCD spectra of bombinin H4, containing D-allo-isoleucine at the second residue, and its diastereomer H2, and showed differences in the structure of the N-terminal region [21]. Thus, our CD and VCD experiments of the phenylseptin showed that both peptides form similar α-helical structures, however in the N-terminal region, the conformation is quite different for the two peptides.
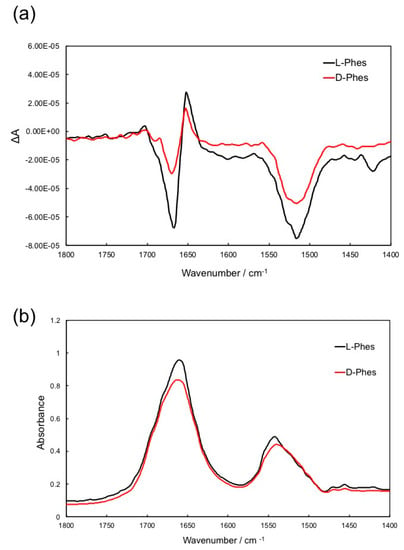
Figure 2.
(a) Vibrational circular dichroism (VCD) and (b) IR spectra of L-Phes (black) and D-Phes (red) in KBr pellets.
3.4. 31P Solid-State NMR
31P solid-state NMR spectroscopy is a useful technique for detection of dynamics of lipid molecules and phase behaviors [41,42,49]. Figure 3 shows 31P static NMR spectra of the DMPC lipid bilayers at 40 °C. First, an axial symmetric powder pattern of liquid-crystalline phase of the membrane was obtained because gel-to-liquid crystalline phase transition temperature of DMPC was around 23 °C (Figure 3, grey line). The values of 31P chemical shift anisotropy ∆δ (δ∕∕ - δ⊥) of the DMPC with L-Phes and D-Phes were lesser as compared with that obtained with only DMPC membrane. The spectral patterns of both peptides are very similar. This indicated that both peptides interact with DMPC bilayers similarly, corresponding with the interaction affinity as estimated by QCM.
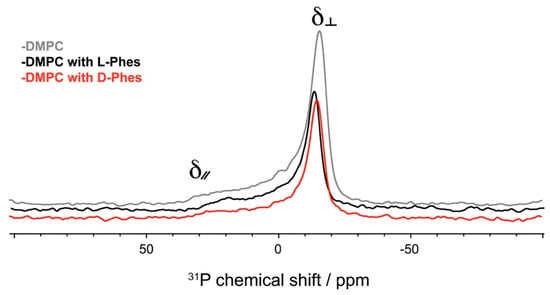
Figure 3.
31P solid-state NMR spectra of DMPC (grey), DMPC with L-Phes (black), and DMPC with D-Phes (red) under static condition.
3.5. Molecular Dynamics Simulations
Previously, it has been reported that the peptides L-Phes and D-Phes adopt α-helical conformations in a mixture of acetonitrile and water, using solution NMR technique [18]. Here, we evaluated the structural stability and interactions of the peptides with DMPC membrane with three different types of simulations. The initial structures were shown in Figure A3, and Table A1 displays the contents of systems. First, we simulated the helical structures inside the model membrane. Both peptides exhibited the stable helical conformation during these simulations, and the first and last snapshots of the simulations are shown in Figure A4a. We identified the most populated structures during the last 25 ns of the simulations, which are shown in Figure A4b. We applied the structures as the initial conformations for the next simulations.
Second, we placed the peptides in the water region of the membrane system to examine differences between L-Phes and D-Phes in their initial interactions with DMPC membrane. During these simulations, we did not observe stable interactions between the membrane and the peptides. This indicated that the peptides need longer simulation time to show initial insertions into DMPC membrane. Perhaps, the peptides need to establish an initial electrostatic interaction with negatively charged phospholipids, or alternatively, the concentrations of the peptides may need to be increased in the membrane system to stimulate the process. The last snapshots and secondary structure profiles are shown in the Figure 4. The relative positions of the peptides were almost similar with their initial position along the z-axis. Although both peptides partially lost their helical conformations during the last-half of the simulations, L-Phes exhibited the higher helicity than D-Phes. Especially, the second position of L-Phes mostly formed α-helical secondary structures compared to the random coil structure of D-Phes. These observations were consistent with our CD experiments, which found the slightly higher helicity for L-Phes.
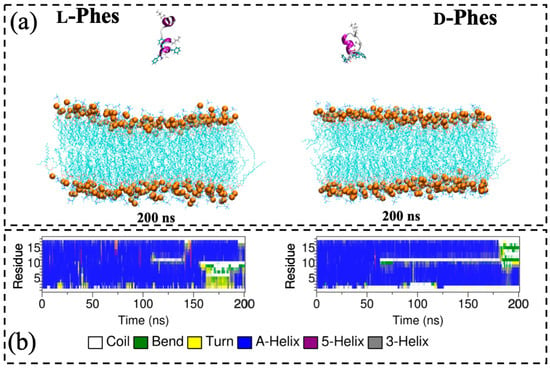
Figure 4.
The last snapshots of the second type of simulations of L- (left) and D-Phes (right) (a) and corresponding secondary structure profiles during the simulations (b). Water molecules and ions were omitted for the clarity. Charged and three phenylalanine residues are shown by licorice model. Orange balls show phosphorus atoms of DMPC membrane.
Third, we performed the pseudo-transmembrane simulations with three different replicas. We inserted C-terminus of the peptides into DMPC membrane, because the charged residues of the peptides are near to N-terminus. We extensively analyzed these simulations to understand the mechanism of action and membrane interactions of the peptides at the membrane interface. The series of snapshots are displayed in Figure 5. Both peptides took their positions within 50 ns of simulations, which were the same as their last snapshots.
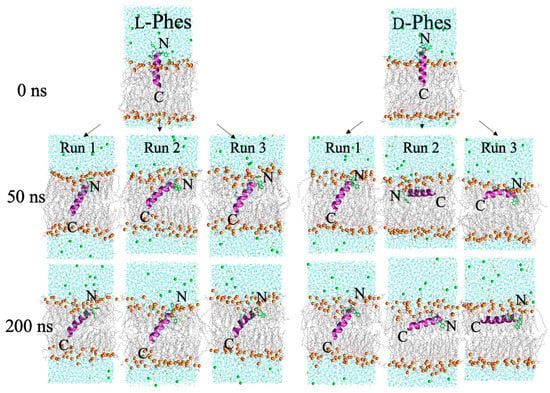
Figure 5.
The last snapshots of the third type of simulations of L- (left) and D-Phes (right). Water molecules and ions were omitted for the clarity. Charged and three-phenylalanine residues are shown by licorice model. Orange balls show phosphorus atoms of DMPC membrane. Green and yellow balls represent the sodium and chloride ions, respectively.
Figure 6 displays the secondary structure profiles during the simulations of L-Phes and D-Phes. The second residue of L-Phes mostly formed α-helical secondary structure during all three runs. In contrast, the second residue of D-Phes presented the random coil conformations in all three runs. These results agreed with the secondary structure results of CD and VCD experiments. This suggested that the membrane interactions of N-terminal side could be different between L-Phes and D-Phes. The pseudo-transmembrane position of L-Phes transferred into transmembrane state in the three different runs. In contrast, only one simulation showed the fully inserted D-Phes. During the simulations, L-Phes peptide tilted around 39° in all three simulations. When D-Phes transferred to transmembrane state (Run 1), the average tilt angle was equal to around 39°. This indicated that when the peptides get inserted, they would tilt similarly in the model membrane. Second and third runs of D-Phes displayed around 82° of the tilt angles. The average values of the tilt angles in the last 100 ns simulations are summarized in Table A2. These data possibly indicate the heterogeneity of membrane-bound states of D-Phes at the interface of DMPC membrane. Otherwise, the difference could stem from the higher hydrophobicity of L-D-L phenylalanine residues of D-Phes.
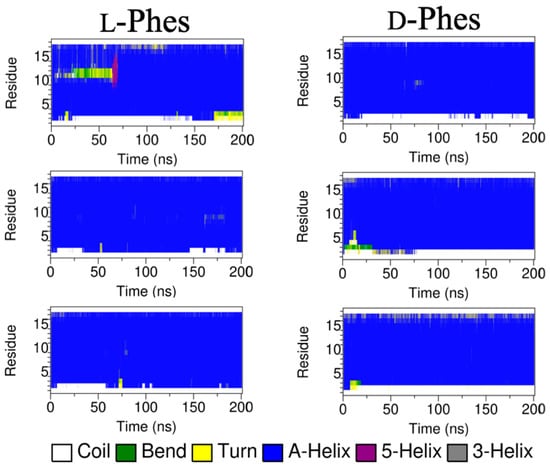
Figure 6.
Secondary structure profiles during the third type of simulations (MD3).
We surveyed the Lennard–Jones interaction term of the three phenylalanine residues with the hydrophobic tail of DMPC membrane to estimate hydrophobicity of N-terminal parts of the peptides. Figure 7 shows the average interaction energies between the three phenylalanine residues and the hydrophobic tails of DMPC membrane. The three Phe residues of L-Phes exhibited slightly more rapid and favorable interaction with the membrane than those of D-Phes did in the first 30 ns. Then, the residues of D-Phes showed slightly more interaction compared to the residues of L-Phes during the period from 30 ns to 200 ns. These data suggest that the higher hydrophobicity of D-Phes, which was also observed in our HPLC results, facilitated interactions with the membrane.
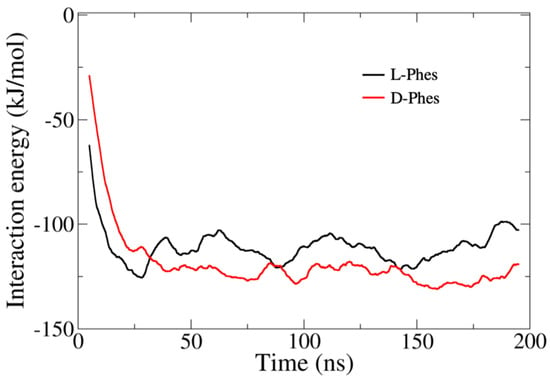
Figure 7.
Lennard–Jones interaction components between the three phenylalanine residues and the hydrophobic tails of DMPC membrane. Three different runs of the peptides averaged, and the running average values are shown in this figure for every 1 ns.
Furthermore, we analyzed the center of mass distances between the phenyl rings during the last 100 ns simulations. There was a lack of difference between the distributions of the first two rings. Figure 8 shows the distribution of the center of mass distances between the second and third phenyl rings. The distribution of D-Phes was smaller than that of L-Phes. The distances in L-Phes showed broader distribution than 6 Å. When the distance between two phenyl rings was less than 6 Å, π-π stacking could occur between them [50]. In Figure 8, we observed a population of D-Phes smaller than 6 Å. When the peptides form oligomeric state in solution [3], the second and the third rings of d-Phes could with stack each other. Stacking of the phenyl rings are important for the self-assembly of short peptides, and phenylalanine tripeptides consisting of l- and d- amino acids have been shown to self-assemble into nanostructures [51]. Although we did not consider peptide–peptide interactions in this work, the higher probability of the packing in D-Phes could affect the association of the peptides and thus affect its interactions with membrane.
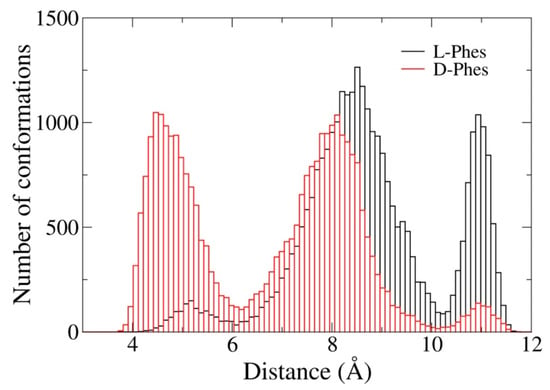
Figure 8.
Distribution of the center of mass distance between second and third phenyl rings during the last 100 ns of all three runs.
4. Conclusions
In this work, we comparatively analyzed the membrane interactions of L-Phes and D-Phes antimicrobial peptides with DMPC model membrane, experimentally and computationally. From the different HPLC retention times, we found D-Phes to have slightly higher hydrophobicity than L-Phes. Our experiments and calculations agreed that the two peptide isomers adopted α-helical secondary structures. MD simulations showed that the second position of L-Phes and D-Phes mostly formed α-helical and random coil secondary structures, respectively. The MD simulations found a mutually stable contact between d-Phe2 and l-Phe3 in D-Phes peptide in the DMPC bilayer. The contacts could influence the membrane interactions and explain its stronger antimicrobial activity compared to L-Phes.
Author Contributions
I.K., B.M., and Y.M. conceived and designed the experiments; Y.M. prepared peptide and performed Far-UV CD and QCM experiments. I.K. and Y.M. performed 31P solid-state NMR experiment. H.S. performed VCD experiments. B.M. and K.U. performed MD simulations. B.M. and I.K wrote the paper.
Funding
This work was partly supported by JSPS KAKENHI, Grant Number to I. K. (JP16H00828 and JP18H02387), to H. S. (JP16H00840 and JP17H03044), and JST MIRAI grants to H. S. and I. K. (JPMJMI18GC).
Acknowledgments
MD simulations were performed on computer clusters or supercomputers of the Research Center for Computational Science, Okazaki, Japan. The authors thank Akira Naito at Yokohama National University for the valuable discussions. The authors thank Shirakata at Graduate School of Engineering Science, Yokohama National University for rechecking the sample. B.M acknowledges the scholarship of Mongolian-Japan Engineering Education Development program (M-JEED project, J11B16) and the financial support of “The Global Doctoral Program for Academic Career Support” program of Yokohama National University.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
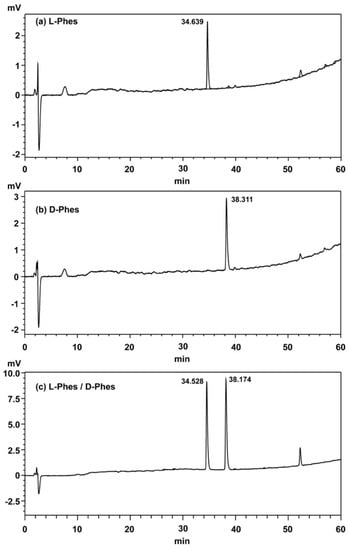
Figure A1.
HPLC charts with C18 ODS column of (a) L-Phes, (b) D-Phes, (c) L-Phes, and D-Phes.
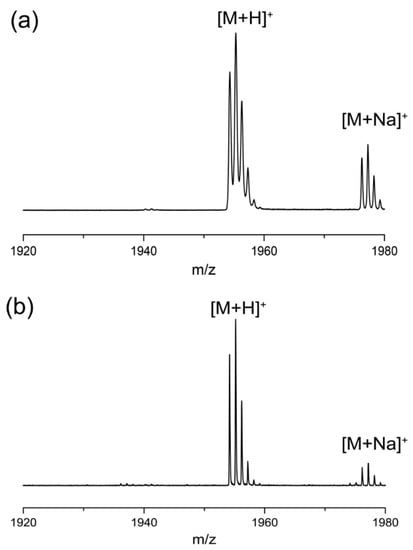
Figure A2.
MALDI-TOF MS spectra of the isotopic patterns of (a) L-Phes and (b) D-Phes ([M+H]+ = 1953.2, [M+Na]+ = 1976.2).
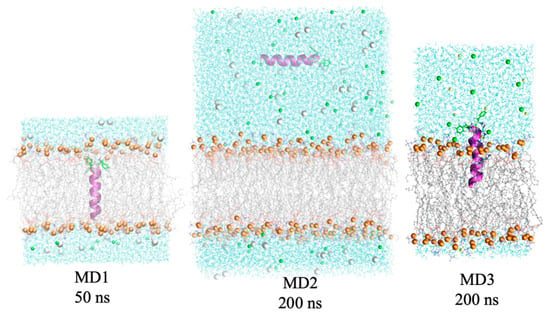
Figure A3.
Examples of the initial structures and representations of the three different types of simulations.
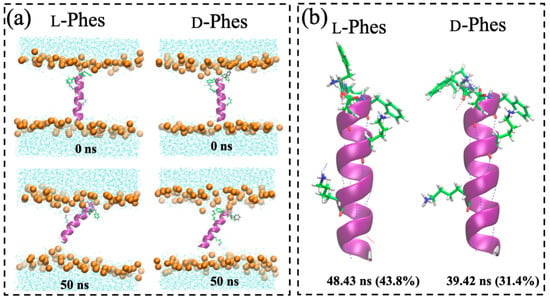
Figure A4.
The first and last snapshots in the first simulations systems (a); the most populated structures of L- Phes and D-Phes in the first simulations (b). Tail groups of DMPC membrane were omitted for the clarity.

Table A1.
Information of simulation systems.
Table A1.
Information of simulation systems.
| Simulation ID | Peptide | Time, ns | Number of contents | |||
|---|---|---|---|---|---|---|
| DMPC | Water | Na+ | Cl− | |||
| MD1 | L-Phes | 50 | 128 | 3691 | 9 | 11 |
| D-Phes | 50 | 128 | 3697 | 9 | 11 | |
| MD2 | L-Phes | 200 | 240 | 17865 | 48 | 50 |
| D-Phes | 200 | 240 | 17698 | 47 | 49 | |
| MD3 | L-Phes | 200 (x3) 1 | 66 | 2959 | 7 | 9 |
| D-Phes | 200 (x3) 1 | 66 | 3119 | 8 | 10 | |
1 The number of replicas were written in parenthesis.

Table A2.
Average tilt angle in the last 100 ns simulations (MD3).
Table A2.
Average tilt angle in the last 100 ns simulations (MD3).
| MDs | L-Phes | D-Phes |
|---|---|---|
| Run 1 | 39.1 ± 6.0 | 38.8 ± 7.0 |
| Run 2 | 38.8 ± 7.0 | 83.1 ± 4.4 |
| Run 3 | 38.5 ± 6.2 | 83.3 ± 4.5 |
| Average | 38.8 ± 6.4 | 68.4 ± 21.6 |
References
- Rozek, T.; Wegener, K.L.; Bowie, J.H.; Olver, I.N.; Carver, J.A.; Wallace, J.C.; Tyler, M.J. The antibiotic and anticancer active aurein peptides from the Australian bell frogs Litoria aurea and Litoria raniformis. Eur. J. Biochem. 2000, 267, 5330–5341. [Google Scholar] [CrossRef] [PubMed]
- Hicks, R.P.; Russell, A.L. Application of unnatural amino acids to the de novo design of selective antibiotic peptides. In Unnatural Amino Acids: Methods and Protocols; Pollegioni, L., Servi, S., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 135–167. [Google Scholar]
- Shai, Y. Mode of action of membrane active antimicrobial peptides. Biopolymers 2002, 66, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Marr, A.K.; McGwire, B.S.; McMaster, W.R. Modes of action of leishmanicidal antimicrobial peptides. Future Microbiol. 2012, 7, 1047–1059. [Google Scholar] [CrossRef] [PubMed]
- Bowdish, D.M.; Davidson, D.J.; Hancock, R. A re-evaluation of the role of host defence peptides in mammalian immunity. Curr. Prot. Pept. Sci. 2005, 6, 35–51. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, M.; Qiu, S.; Wang, J.; Peng, J.; Zhao, P.; Zhu, R.; Wang, H.; Li, Y.; Wang, K.; et al. Antimicrobial activity and stability of the D-amino acid substituted derivatives of antimicrobial peptide polybia-MPI. AMB Express 2016, 6, 122. [Google Scholar] [CrossRef] [PubMed]
- Ollivaux, C.; Soyez, D.; Toullec, J.-Y. Biogenesis of D-amino acid containing peptides/proteins: Where, when and how? J. Pept. Sci. 2014, 20, 595–612. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Sheeley, S.; Sweedler, J.V. Analysis of endogenous D-amino acid-containing peptides in Metazoa. Bioanal. Rev. 2009, 1, 7–24. [Google Scholar] [CrossRef]
- Glaser, T.; Hübner, K.; Castiglione, R.; de Hamprecht, B. Dermorphins, opioid peptides from amphibian skin, act on opioid receptors of mouse neuroblastoma x rat glioma hybrid cells. J. Neurochem. 1981, 37, 1613–1617. [Google Scholar] [CrossRef]
- Kreil, G. d-Amino acids in animal peptides. Annu. Rev. Biochem. 1997, 66, 337–345. [Google Scholar] [CrossRef]
- Tancredi, T.; Temussi, P.A.; Picone, D.; Amodeo, P.; Tomatis, R.; Salvadori, S.; Marastoni, M.; Santagada, V.; Balboni, G. New insights on μ/δ selectivity of opioid peptides: Conformational analysis of deltorphin analogues. Biopolymers 1991, 31, 751–760. [Google Scholar] [CrossRef]
- Barra, D.; Mignogna, G.; Simmaco, M.; Pucci, P.; Severini, C.; Falconieri-Erspamer, G.; Negri, L.; Erspamer, V. [d-Leu2] Deltorphin, A 17 amino acid opioid peptide from the skin of the Brazilian hylid frog, Phyllomedusa burmeisteri. Peptides 1994, 15, 199–202. [Google Scholar] [CrossRef]
- Mor, A.; Delfour, A.; Sagan, S.; Amiche, M.; Pradelles, P.; Rossier, J.; Nicolas, P. Isolation of dermenkephalin from amphibian skin, a high-affinity (δ-selective opioid heptapeptide containing a D-amino acid residue. FEBS Lett. 1989, 255, 269–274. [Google Scholar] [CrossRef]
- Erspamer, V.; Melchiorri, P.; Falconieri-Erspamer, G.; Negri, L.; Corsi, R.; Severini, C.; Barra, D.; Simmaco, M.; Kreil, G. Deltorphins: A family of naturally occurring peptides with high affinity and selectivity for delta opioid binding sites. Proc. Natl. Acad. Sci. USA 1989, 86, 5188–5192. [Google Scholar] [CrossRef] [PubMed]
- Mignogna, G.; Simmaco, M.; Kreil, G.; Barra, D. Antibacterial and haemolytic peptides containing D-alloisoleucine from the skin of Bombina variegata. EMBO J. 1993, 12, 4829–4832. [Google Scholar] [CrossRef] [PubMed]
- Mangoni, M.L.; Grovale, N.; Giorgi, A.; Mignogna, G.; Simmaco, M.; Barra, D. Structure-function relationships in bombinins H, antimicrobial peptides from Bombina skin secretions. Peptides 2000, 21, 1673–1679. [Google Scholar] [CrossRef]
- Mangoni, M.L.; Papo, N.; Saugar, J.M.; Barra, D.; Shai, Y.; Simmaco, M.; Rivas, L. Effect of natural L- to D-amino acid conversion on the organization, membrane binding, and biological function of the antimicrobial peptides bombinins H. Biochemistry 2006, 45, 4266–4276. [Google Scholar] [CrossRef] [PubMed]
- de Magalhães, M.T.Q.; Barbosa, E.A.; Prates, M.V.; Verly, R.M.; Munhoz, V.H.O.; de Araújo, I.E.; Bloch, C., Jr. conformational and functional effects induced by D- and L-amino acid epimerization on a single gene encoded peptide from the skin secretion of Hypsiboas punctatus. PLoS ONE 2013, 8, e59255. [Google Scholar] [CrossRef] [PubMed]
- Melchiorri, P.; Negri, L. The dermorphin peptide family. Gen. Pharm. 1996, 27, 1099–1107. [Google Scholar] [CrossRef]
- Simmaco, M.; Barra, D.; Chiarini, F.; Noviello, L.; Melchiorri, P.; Kreil, G.; Richter, K. A family of bombinin-related peptides from the skin of Bombina variegata. Eur. J. Biochem. 1991, 199, 217–222. [Google Scholar] [CrossRef]
- Mijiddorj, B.; Kaneda, S.; Sato, H.; Kitahashi, Y.; Javkhlantugs, N.; Naito, A.; Ueda, K.; Kawamura, I. The role of D-allo-isoleucine in the deposition of the anti-leishmania peptide bombinin H4 as revealed by 31P solid-state NMR, VCD spectroscopy, and MD simulation. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 789–798. [Google Scholar] [CrossRef]
- Sekiya, Y.; Shimizu, K.; Kitahashi, Y.; Ohyama, A.; Kawamura, I.; Kawano, R. Electrophysiological analysis of membrane disruption by bombinin and its isomer using the lipid bilayer system. ACS Appl. Bio Mater. 2019, 2, 1542–1548. [Google Scholar] [CrossRef]
- Nagao, T.; Mishima, D.; Javkhlantugs, N.; Wang, J.; Ishioka, D.; Yokota, K.; Norisada, K.; Kawamura, I.; Ueda, K.; Naito, A. Structure and Orientation of antibiotic peptide alamethicin in phospholipid bilayers as revealed by chemical shift oscillation analysis of solid state nuclear magnetic resonance and molecular dynamics simulation. Biochim. Biophys. Acta Biomembr. 2015, 1848, 2789–2798. [Google Scholar] [CrossRef] [PubMed]
- Zerweck, J.; Strandberg, E.; Bürck, J.; Reichert, J.; Wadhwani, P.; Kukharenko, O.; Ulrich, A.S. Homo- and heteromeric interaction strengths of the synergistic antimicrobial peptides PGLa and Magainin 2 in membranes. Eur. Biophys. J. 2016, 45, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Sauerbrey, G. The use of quartz oscillators for weighing thin layers and for microweighing. Z. Phys. 1959, 155, 206–222. [Google Scholar] [CrossRef]
- Abraham, M.J.; van der Spoel, D.; Lindahl, E.; Hess, B. The GROMACS Development Team; GROMACS User Manual Version 5.1.2. Available online: www.gromacs.org (accessed on 27 June 2019).
- Pastor, R.W.; MacKerell, A.D., Jr. Development of the CHARMM force field for lipids. J. Phys. Chem. Lett. 2011, 2, 1526–1532. [Google Scholar] [CrossRef] [PubMed]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Jo, S.; Lim, J.B.; Klauda, J.B.; Im, W. CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophys. J. 2009, 97, 50–58. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Steinbach, P.J.; Brooks, B.R. New spherical-cutoff methods for long-range forces in macromolecular simulation. J. Comput. Chem. 1994, 15, 667–683. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Nosé, S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 1984, 52, 255–268. [Google Scholar] [CrossRef]
- Hoover, W.G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A 1985, 31, 1695–1697. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Tsutsumi, A.; Javkhlantugs, N.; Kira, A.; Umeyama, M.; Kawamura, I.; Nishimura, K.; Ueda, K.; Naito, A. structure and orientation of bovine lactoferrampin in the mimetic bacterial membrane as revealed by solid-state NMR and molecular dynamics simulation. Biophys. J. 2012, 103, 1735–1743. [Google Scholar] [CrossRef]
- Kira, A.; Javkhlantugs, N.; Miyamori, T.; Sasaki, Y.; Eguchi, M.; Kawamura, I.; Ueda, K.; Naito, A. Interaction of extracellular loop II of kappa-opioid receptor (196-228) with opioid peptide dynorphin in membrane environments as revealed by solid state nuclear magnetic resonance, quartz crystal microbalance and molecular dynamics simulation. J. Phys. Chem. B. 2014, 118, 9604–9612. [Google Scholar] [CrossRef]
- Hiraoki, T.; Brown, S.B.; Stevenson, K.J.; Vogel, H.J. Structural comparison between oxidized and reduced Escherichia Coli thioredoxin. Proton NMR and CD studies. Biochemistry 1988, 27, 5000–5008. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J.; Fasman, G.D. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry 1969, 8, 4108–4116. [Google Scholar] [CrossRef] [PubMed]
- Durr, H.N.U.; Gildenberg, M.; Ramamoorthy, A. The magic of bicelles lights up membrane protein structure. Chem. Rev. 2012, 112, 6054–6074. [Google Scholar] [CrossRef]
- Kurouski, D. Advances of vibrational circular dichroism (VCD) in bioanalytical chemistry. A review. Anal. Chim. Acta 2017, 990, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Nafie, L.A.; Keiderling, T.A.; Stephens, P.J. Vibrational circular dichroism. J. Am. Chem. Soc. 1976, 98, 2715–2723. [Google Scholar] [CrossRef]
- Sato, H.; Kawamura, I.; Yamagishi, A.; Sato, F. Solid-state vibrational circular dichroism spectra of isoleusine and its related compounds: Effects of interplay between two chiral centers. Chem. Lett. 2017, 46, 449–452. [Google Scholar] [CrossRef]
- Naito, A.; Nagao, T.; Norisada, K.; Mizuno, T.; Tuzi, S.; Saitô, H. Conformation and dynamics of melittin bound to magnetically oriented lipid bilayers by solid-state 31P and 13C NMR spectroscopy. Biophys. J. 2000, 78, 2405–2417. [Google Scholar] [CrossRef]
- Sinnokrot, M.O.; Valeev, E.F.; Sherrill, C.D. Estimates of the ab initio limit for π−π interactions: The benzene dimer. J. Am. Chem. Soc. 2002, 124, 10887–10893. [Google Scholar] [CrossRef]
- Ozawa, Y.; Sato, H.; Kayano, Y.; Yamaki, N.; Izato, Y.; Miyake, A.; Naito, A.; Kawamura, I. Self-assembly of tripeptides into γ-turn nanostructures. Phys. Chem. Chem. Phys. 2019, 21, 10879–10883. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).