Computational Health Engineering Applied to Model Infectious Diseases and Antimicrobial Resistance Spread
Abstract
1. Introduction
2. Overview of Mathematical Models to Predict Infectious Diseases
3. Host–Pathogen Interactions
3.1. Modeling the Immune System
3.2. Predicting Sepsis
4. Antimicrobial-Pathogen Interactions: Overcoming Antimicrobial Resistance
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Model Type | Base on | Advantages | Limitations and Disadvantages | Examples | References |
|---|---|---|---|---|---|
| Statistical | Regression | Incidence and prevalence studies. Outbreaks studies. Simplicity. | No spatial distribution of cases. Predeterminate range of time, without time trends | Parametric and semiparametric models | [31,136,137] |
| Autoregressive | Longitudinal studies. Outbreaks studies. Repetitive studies over the time during a specific interval. | Time is stationary. Statistical testing is based on the normal distribution. | ARIMA SARIMA Eagle’s ARCH Model NMA | [32,33,136,137] | |
| Time series | Estimates the trend and seasonal effects; estimates dynamic causal effects | Prediction towards to mean. Possible prediction‘s errors. Determinist models. | [34,35,77,121,137] | ||
| Cumulative sum | Considers the deviations. Sequential. | Initially, no trends trends or seasonality are assumed. | CUSUM | [34,36,137] | |
| Moving area | Simple, cumulative, weighted or exponential. Measure the moment. | Prediction-lag. | SMA, CMA, WMA and EWMA | [137,138] | |
| Mechanistic | Deterministic | Describes the mean according to an initial defined value (condition or parameters) and allows simpler fitting | Always the same result. It may miss information. | SIR, SEIR, MSIR, MSEIR SIS, SEIS, SIR-carrier status | [39,40,41,67,103] |
| Stochastic | Contains inherent randomness being more realistic | May not be predicted precisely | Environmental and demographical | [11,42,43,44,45,75,117] | |
| Agent Based | Simulation | Focuses directly on individual variable and their interactions, reflecting relationships in a hypothetical real world considers heterogenicity | Complexity. Nonlinearity. | ABS IIR-ABM, GM-ABM | [52,83,85,86] |
| Complex Systems | The components interact among them. Study of relationships and dynamisms. | Networks, Multi-layer, System Biology | [47,48,84,111,116,125] | ||
| Intelligence | Learning and training | Learns from big-data and makes an informed decision. Accuracy | Training process | Machine Learning Deep Learning | [49,51,53,55,57,58,87,88,89,90,91,92,128,129,130,131,133,134] |
| Big data. Results reveal hidden patterns and predict future possibilities | Big data. Results rely on number of parameters. The youth of approaches. External validation studies to confirm the predictions. | ||||
| Learning and solving | AI | [50,56,98,99,100] | |||
| Successful and intelligent | Improves data management |
References
- Bruce, R.; Levin, F.M.S.; Lin, C. Resource-limited growth, competition, and predation: A model and experimental studies with bacteria and bacteriophage. Am. Nat. 1977, 111, 19. [Google Scholar] [CrossRef]
- Wolkenhauer, O.; Mesarovic, M. Feedback dynamics and cell function: Why systems biology is called systems biology. Mol. Biosyst. 2005, 1, 14–16. [Google Scholar] [CrossRef] [PubMed]
- Mesarovic, M.D.; Takahara, Y. General Systems Theory: Mathematical Foundations; Academic Press: London, UK, 1975; Volume 113, p. 267. [Google Scholar]
- Systems Biology Home Page. Available online: https://systemsbiology.org/about/what-is-systems-biology/ (accessed on 20 May 2019).
- Aebersold, R.; Hood, L.E.; Watts, J.D. Equipping scientists for the new biology. Nat. Biotechnol. 2000, 18, 359. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A. New institute to study systems biology. Nat. Biotechnol. 1999, 17, 743–744. [Google Scholar] [CrossRef] [PubMed]
- Aderem, A.; Adkins, J.N.; Ansong, C.; Galagan, J.; Kaiser, S.; Korth, M.J.; Law, G.L.; McDermott, J.G.; Proll, S.C.; Rosenberger, C.; et al. A systems biology approach to infectious disease research: Innovating the pathogen-host research paradigm. MBio 2011, 2, e00325-10. [Google Scholar] [CrossRef] [PubMed]
- Kitano, H. Computational systems biology. Nature 2002, 420, 206–210. [Google Scholar] [CrossRef] [PubMed]
- Tavassoly, I.; Goldfarb, J.; Iyengar, R. Systems biology primer: The basic methods and approaches. Essays Biochem. 2018, 62, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Barrila, J.; Crabbe, A.; Yang, J.; Franco, K.; Nydam, S.D.; Forsyth, R.J.; Davis, R.R.; Gangaraju, S.; Ott, C.M.; Coyne, C.B.; et al. Modeling host-pathogen interactions in the context of the microenvironment: Three-dimensional cell culture comes of age. Infect. Immun. 2018, 86. [Google Scholar] [CrossRef] [PubMed]
- Sjoholm, K.; Kilsgard, O.; Teleman, J.; Happonen, L.; Malmstrom, L.; Malmstrom, J. Targeted proteomics and absolute protein quantification for the construction of a stoichiometric host-pathogen surface density model. Mol. Cell. Proteom. 2017, 16, S29–S41. [Google Scholar] [CrossRef] [PubMed]
- Windels, E.M.; Michiels, J.E.; Fauvart, M.; Wenseleers, T.; Van den Bergh, B.; Michiels, J. Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. ISME J. 2019. [Google Scholar] [CrossRef] [PubMed]
- Grinter, S.Z.; Zou, X. Challenges, applications, and recent advances of protein-ligand docking in structure-based drug design. Molecules 2014, 19, 10150–10176. [Google Scholar] [CrossRef] [PubMed]
- Roman, M.; Roman, D.L.; Ostafe, V.; Ciorsac, A.; Isvoran, A. Computational assessment of pharmacokinetics and biological effects of some anabolic and androgen steroids. Pharm. Res. 2018, 35, 41. [Google Scholar] [CrossRef] [PubMed]
- Kahraman, A.; Herzog, F.; Leitner, A.; Rosenberger, G.; Aebersold, R.; Malmstrom, L. Cross-link guided molecular modeling with rosetta. PLoS ONE 2013, 8, e73411. [Google Scholar] [CrossRef] [PubMed]
- Hauri, S.; Khakzad, H.; Happonen, L.; Teleman, J.; Malmstrom, J.; Malmstrom, L. Rapid determination of quaternary protein structures in complex biological samples. Nat. Commun. 2019, 10, 192. [Google Scholar] [CrossRef] [PubMed]
- Mathematical modeling. Concept paper. Ann. N. Y. Acad. Sci. 1994, 740, 271–274. [CrossRef] [PubMed]
- Domenech de Celles, M.; King, A.A.; Rohani, P. Commentary: Resolving pertussis resurgence and vaccine immunity using mathematical transmission models. Hum. Vaccin Immunother. 2018. [Google Scholar] [CrossRef] [PubMed]
- Levin, S.A.; Grenfell, B.; Hastings, A.; Perelson, A.S. Mathematical and computational challenges in population biology and ecosystems science. Science 1997, 275, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Miaou, S.P.; Song, J.J. Bayesian ranking of sites for engineering safety improvements: Decision parameter, treatability concept, statistical criterion, and spatial dependence. Accid. Anal. Prev. 2005, 37, 699–720. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.L.; Jin, Y.; Raddad, E.; McNearney, T.A.; Ni, X.; Monteith, D.; Brown, R.; Deeg, M.A.; Schnitzer, T. Applications of bayesian statistical methodology to clinical trial design: A case study of a phase 2 trial with an interim futility assessment in patients with knee osteoarthritis. Pharm. Stat. 2019, 18, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Orphanou, K.; Stassopoulou, A.; Keravnou, E. Temporal abstraction and temporal bayesian networks in clinical domains: A survey. Artif. Intell. Med. 2014, 60, 133–149. [Google Scholar] [CrossRef]
- Xing, L.; Guo, M.; Liu, X.; Wang, C.; Wang, L.; Zhang, Y. An improved bayesian network method for reconstructing gene regulatory network based on candidate auto selection. BMC Genom. 2017, 18, 844. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Stella, F.; Hommersom, A.; Lucas, P.J.F.; Boer, L.; Bischoff, E. A comparison between discrete and continuous time bayesian networks in learning from clinical time series data with irregularity. Artif. Intell. Med. 2019. [Google Scholar] [CrossRef] [PubMed]
- Adabor, E.S.; Acquaah-Mensah, G.K.; Oduro, F.T. Saga: A hybrid search algorithm for bayesian network structure learning of transcriptional regulatory networks. J. Biomed. Inform. 2015, 53, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Shi, X. A mixture copula bayesian network model for multimodal genomic data. Cancer Inform. 2017, 16, 1176935117702389. [Google Scholar] [CrossRef]
- MacLehose, R.F.; Hamra, G.B. Applications of bayesian methods to epidemiologic research. Curr. Epidemiol. Rep. 2014, 1, 103–109. [Google Scholar] [CrossRef]
- Haddawy, P.; Hasan, A.; Kasantikul, R.; Lawpoolsri, S.; Sa-Angchai, P.; Kaewkungwal, J.; Singhasivanon, P. Spatiotemporal bayesian networks for malaria prediction. Artif. Intell. Med. 2018, 84, 127–138. [Google Scholar] [CrossRef]
- Lang, J.; Jin, L.; Yao, Y. Comparative efficacy of interventions for reducing injection and sexual risk behaviours to prevent hiv in injection drug users: Protocol for bayesian network meta-analysis. BMJ Open 2019, 9, e022811. [Google Scholar] [CrossRef]
- Mir, D.; Graf, T.; Esteves de Matos Almeida, S.; Pinto, A.R.; Delatorre, E.; Bello, G. Inferring population dynamics of hiv-1 subtype c epidemics in eastern africa and southern brazil applying different bayesian phylodynamics approaches. Sci. Rep. 2018, 8, 8778. [Google Scholar] [CrossRef]
- Medu, O.; Anderson, M.; Enns, A.; Wright, J.; Dunlop, T.; Kapaj, S.; Opondo, J. Predictors of pertussis outbreak in urban and rural municipalities of saskatchewan, canada. Can. J. Public Health 2018, 109, 362–368. [Google Scholar] [CrossRef]
- Zahirul Islam, M.; Rutherford, S.; Phung, D.; Uzzaman, M.N.; Baum, S.; Huda, M.M.; Asaduzzaman, M.; Talukder, M.R.R.; Chu, C. Correlates of climate variability and dengue fever in two metropolitan cities in bangladesh. Cureus 2018, 10, e3398. [Google Scholar] [CrossRef]
- Aryee, G.; Kwarteng, E.; Essuman, R.; Nkansa Agyei, A.; Kudzawu, S.; Djagbletey, R.; Owusu Darkwa, E.; Forson, A. Estimating the incidence of tuberculosis cases reported at a tertiary hospital in ghana: A time series model approach. BMC Public Health 2018, 18, 1292. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.; Park, H.W.; Choi, Y.H.; Kim, J.; Munkhdalai, L.; Musa, I.; Ryu, K.H. A simulation-based study on the comparison of statistical and time series forecasting methods for early detection of infectious disease outbreaks. Int. J. Environ. Res. Public Health 2018, 15. [Google Scholar] [CrossRef] [PubMed]
- La Martire, G.; Robin, C.; Oubaya, N.; Lepeule, R.; Beckerich, F.; Leclerc, M.; Barhoumi, W.; Toma, A.; Pautas, C.; Maury, S.; et al. De-escalation and discontinuation strategies in high-risk neutropenic patients: An interrupted time series analyses of antimicrobial consumption and impact on outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1931–1940. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.L.; Anneducharme, C.; Bucher, B.J. Development and implementation of a surveillance network system for emerging infectious diseases in the caribbean (aricaba). Online J. Public Health Inform. 2011, 3. [Google Scholar] [CrossRef] [PubMed]
- Mayfield, H.J.; Lowry, J.H.; Watson, C.H.; Kama, M.; Nilles, E.J.; Lau, C.L. Use of geographically weighted logistic regression to quantify spatial variation in the environmental and sociodemographic drivers of leptospirosis in fiji: A modelling study. Lancet Planet. Health 2018, 2, e223–e232. [Google Scholar] [CrossRef]
- Siettos, C.I.; Russo, L. Mathematical modeling of infectious disease dynamics. Virulence 2013, 4, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Almocera, A.E.S.; Hernandez-Vargas, E.A. Coupling multiscale within-host dynamics and between-host transmission with recovery (sir) dynamics. Math. Biosci. 2019, 309, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, Y.; Xiao, Y.; Li, M.Y. Global dynamics of a discrete age-structured sir epidemic model with applications to measles vaccination strategies. Math. Biosci. 2018, 308, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Chanprasopchai, P.; Tang, I.M.; Pongsumpun, P. Sir model for dengue disease with effect of dengue vaccination. Comput. Math. Methods Med. 2018, 2018, 9861572. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, C.; Carlson, J.M. Optimizing real-time vaccine allocation in a stochastic sir model. PLoS ONE 2016, 11, e0152950. [Google Scholar] [CrossRef] [PubMed]
- Ball, F.; Neal, P. A general model for stochastic sir epidemics with two levels of mixing. Math. Biosci. 2002, 180, 73–102. [Google Scholar] [CrossRef]
- Ouboter, T.; Meester, R.; Trapman, P. Stochastic sir epidemics in a population with households and schools. J. Math. Biol. 2016, 72, 1177–1193. [Google Scholar] [CrossRef] [PubMed]
- Miao, A.; Zhang, J.; Zhang, T.; Pradeep, B. Threshold dynamics of a stochastic sir model with vertical transmission and vaccination. Comput. Math. Methods Med. 2017, 2017, 4820183. [Google Scholar] [CrossRef] [PubMed]
- Price, D.J.; Breuze, A.; Dybowski, R.; Mastroeni, P.; Restif, O. An efficient moments-based inference method for within-host bacterial infection dynamics. PLoS Comput. Biol. 2017, 13, e1005841. [Google Scholar] [CrossRef] [PubMed]
- Barnard, R.C.; Kiss, I.Z.; Berthouze, L.; Miller, J.C. Edge-based compartmental modelling of an sir epidemic on a dual-layer static-dynamic multiplex network with tunable clustering. Bull. Math. Biol. 2018, 80, 2698–2733. [Google Scholar] [CrossRef]
- Savini, L.; Candeloro, L.; Conte, A.; De Massis, F.; Giovannini, A. Development of a forecasting model for brucellosis spreading in the italian cattle trade network aimed to prioritise the field interventions. PLoS ONE 2017, 12, e0177313. [Google Scholar] [CrossRef]
- Chae, S.; Kwon, S.; Lee, D. Predicting infectious disease using deep learning and big data. Int. J. Environ. Res. Public Health 2018, 15. [Google Scholar] [CrossRef]
- Wong, Z.S.Y.; Zhou, J.; Zhang, Q. Artificial intelligence for infectious disease big data analytics. Infect. Dis. Health 2019, 24, 44–48. [Google Scholar] [CrossRef]
- Leite, D.M.C.; Brochet, X.; Resch, G.; Que, Y.A.; Neves, A.; Pena-Reyes, C. Computational prediction of inter-species relationships through omics data analysis and machine learning. BMC Bioinform. 2018, 19, 420. [Google Scholar] [CrossRef]
- Cockrell, R.C.; An, G. Examining the controllability of sepsis using genetic algorithms on an agent-based model of systemic inflammation. PLoS Comput. Biol. 2018, 14, e1005876. [Google Scholar] [CrossRef]
- Forbes, J.D.; Chen, C.Y.; Knox, N.C.; Marrie, R.A.; El-Gabalawy, H.; de Kievit, T.; Alfa, M.; Bernstein, C.N.; Van Domselaar, G. A comparative study of the gut microbiota in immune-mediated inflammatory diseases-does a common dysbiosis exist? Microbiome 2018, 6, 221. [Google Scholar] [CrossRef] [PubMed]
- Kaul, R.B.; Evans, M.V.; Murdock, C.C.; Drake, J.M. Spatio-temporal spillover risk of yellow fever in Brazil. Parasit Vectors 2018, 11, 488. [Google Scholar] [CrossRef] [PubMed]
- Sharaha, U.; Rodriguez-Diaz, E.; Sagi, O.; Riesenberg, K.; Lapidot, I.; Segal, Y.; Bigio, I.J.; Huleihel, M.; Salman, A. Detection of extended-spectrum beta-lactamase-producing escherichia coli using infrared microscopy and machine-learning algorithms. Anal. Chem. 2019. [Google Scholar] [CrossRef] [PubMed]
- Bailey, A.; Ledeboer, N.; Burnham, C.D. Clinical microbiology is growing up: The total laboratory automation revolution. Clin. Chem. 2018. [Google Scholar] [CrossRef] [PubMed]
- Lupolova, N.; Dallman, T.J.; Holden, N.J.; Gally, D.L. Patchy promiscuity: Machine learning applied to predict the host specificity of salmonella enterica and escherichia coli. Microb. Genom. 2017, 3, e000135. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, N.E.; Gardner, P.P.; Barquist, L. Machine learning identifies signatures of host adaptation in the bacterial pathogen salmonella enterica. PLoS Genet. 2018, 14, e1007333. [Google Scholar] [CrossRef] [PubMed]
- Gestal, M.C.; Whitesides, L.T.; Harvill, E.T. Integrated signaling pathways mediate bordetella immunomodulation, persistence, and transmission. Trends Microbiol. 2019, 27, 118–130. [Google Scholar] [CrossRef]
- Tomar, N.; De, R.K. Immunoinformatics: A brief review. In Immunoinformatics; De, R.K., Tomar, N., Eds.; Springer New York: New York, NY, USA, 2014; pp. 23–55. [Google Scholar]
- Thakar, J.; Pilione, M.; Kirimanjeswara, G.; Harvill, E.T.; Albert, R. Modeling systems-level regulation of host immune responses. PLoS Comput. Biol. 2007, 3, e109. [Google Scholar] [CrossRef]
- Schweppe, D.K.; Harding, C.; Chavez, J.D.; Wu, X.; Ramage, E.; Singh, P.K.; Manoil, C.; Bruce, J.E. Host-microbe protein interactions during bacterial infection. Chem. Biol. 2015, 22, 1521–1530. [Google Scholar] [CrossRef]
- Rohrs, J.A.; Wang, P.; Finley, S.D. Understanding the dynamics of t-cell activation in health and disease through the lens of computational modeling. JCO Clin. Cancer Inform. 2019, 3, 1–8. [Google Scholar] [CrossRef]
- Hawse, W.F.; Morel, P.A. An immunology primer for computational modelers. J. Pharmacokinet. Pharmacodyn. 2014, 41, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, E.K.; Kwiatkowski, D.P.; Sabeti, P.C. Natural selection and infectious disease in human populations. Nat. Rev. Genet. 2014, 15, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.M.; Tato, C.M.; Furman, D. Systems immunology: Just getting started. Nat. Immunol. 2017, 18, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, E.; Toledano, V.; Morilla, F.; Hernandez-Jimenez, E.; Cubillos-Zapata, C.; Varela-Serrano, A.; Casas-Martin, J.; Avendano-Ortiz, J.; Aguirre, L.A.; Arnalich, F.; et al. A system dynamics model to predict the human monocyte response to endotoxins. Front. Immunol. 2017, 8, 915. [Google Scholar] [CrossRef] [PubMed]
- Musser, J.M.; DeLeo, F.R. Toward a genome-wide systems biology analysis of host-pathogen interactions in group a streptococcus. Am. J. Pathol. 2005, 167, 1461–1472. [Google Scholar] [CrossRef]
- Aziz, R.K.; Kansal, R.; Aronow, B.J.; Taylor, W.L.; Rowe, S.L.; Kubal, M.; Chhatwal, G.S.; Walker, M.J.; Kotb, M. Microevolution of group a streptococci in vivo: Capturing regulatory networks engaged in sociomicrobiology, niche adaptation, and hypervirulence. PLoS ONE 2010, 5, e9798. [Google Scholar] [CrossRef]
- Karlsson, C.A.Q.; Jarnum, S.; Winstedt, L.; Kjellman, C.; Bjorck, L.; Linder, A.; Malmstrom, J.A. Streptococcus pyogenes infection and the human proteome with a special focus on the immunoglobulin g-cleaving enzyme ides. Mol. Cell. Proteom. 2018, 17, 1097–1111. [Google Scholar] [CrossRef]
- Sjoholm, K.; Karlsson, C.; Linder, A.; Malmstrom, J. A comprehensive analysis of the streptococcus pyogenes and human plasma protein interaction network. Mol. Biosyst. 2014, 10, 1698–1708. [Google Scholar] [CrossRef]
- Malmstrom, L.; Bakochi, A.; Svensson, G.; Kilsgard, O.; Lantz, H.; Petersson, A.C.; Hauri, S.; Karlsson, C.; Malmstrom, J. Quantitative proteogenomics of human pathogens using dia-ms. J. Proteom. 2015, 129, 98–107. [Google Scholar] [CrossRef]
- Malmstrom, E.; Kilsgard, O.; Hauri, S.; Smeds, E.; Herwald, H.; Malmstrom, L.; Malmstrom, J. Large-scale inference of protein tissue origin in gram-positive sepsis plasma using quantitative targeted proteomics. Nat. Commun. 2016, 7, 10261. [Google Scholar] [CrossRef]
- Gog, J.R.; Murcia, A.; Osterman, N.; Restif, O.; McKinley, T.J.; Sheppard, M.; Achouri, S.; Wei, B.; Mastroeni, P.; Wood, J.L.; et al. Dynamics of salmonella infection of macrophages at the single cell level. J. R. Soc. Interface 2012, 9, 2696–2707. [Google Scholar] [CrossRef] [PubMed]
- Grant, A.J.; Restif, O.; McKinley, T.J.; Sheppard, M.; Maskell, D.J.; Mastroeni, P. Modelling within-host spatiotemporal dynamics of invasive bacterial disease. PLoS Biol. 2008, 6, e74. [Google Scholar] [CrossRef] [PubMed]
- Restif, O.; Goh, Y.S.; Palayret, M.; Grant, A.J.; McKinley, T.J.; Clark, M.R.; Mastroeni, P. Quantification of the effects of antibodies on the extra- and intracellular dynamics of salmonella enterica. J. R. Soc. Interface 2013, 10, 20120866. [Google Scholar] [CrossRef] [PubMed]
- Golumbeanu, M.; Desfarges, S.; Hernandez, C.; Quadroni, M.; Rato, S.; Mohammadi, P.; Telenti, A.; Beerenwinkel, N.; Ciuffi, A. Proteo-transcriptomic dynamics of cellular response to hiv-1 infection. Sci. Rep. 2019, 9, 213. [Google Scholar] [CrossRef] [PubMed]
- Ford, S.A.; Kao, D.; Williams, D.; King, K.C. Microbe-mediated host defence drives the evolution of reduced pathogen virulence. Nat. Commun. 2016, 7, 13430. [Google Scholar] [CrossRef] [PubMed]
- Barreiro, L.B.; Quintana-Murci, L. From evolutionary genetics to human immunology: How selection shapes host defence genes. Nat. Rev. Genet. 2010, 11, 17–30. [Google Scholar] [CrossRef]
- Didelot, X.; Walker, A.S.; Peto, T.E.; Crook, D.W.; Wilson, D.J. Within-host evolution of bacterial pathogens. Nat. Rev. Microbiol. 2016, 14, 150–162. [Google Scholar] [CrossRef]
- Chao, D.L.; Davenport, M.P.; Forrest, S.; Perelson, A.S. Stochastic stage-structured modeling of the adaptive immune system. In Proceedings of the IEEE Computational Systems Bioinformatics Conference, Stanford, CA, USA, 11–14 August 2003; Volume 2, pp. 124–131. [Google Scholar]
- Kumar, R.; Clermont, G.; Vodovotz, Y.; Chow, C.C. The dynamics of acute inflammation. J. Theor. Biol. 2004, 230, 145–155. [Google Scholar] [CrossRef]
- Seal, J.B.; Alverdy, J.C.; Zaborina, O.; An, G. Agent-based dynamic knowledge representation of pseudomonas aeruginosa virulence activation in the stressed gut: Towards characterizing host-pathogen interactions in gut-derived sepsis. Theor. Biol. Med. Model. 2011, 8, 33. [Google Scholar] [CrossRef]
- Shi, Z.; Wu, C.H.; Ben-Arieh, D.; Simpson, S.Q. Mathematical model of innate and adaptive immunity of sepsis: A modeling and simulation study of infectious disease. Biomed. Res. Int. 2015, 2015, 504259. [Google Scholar] [CrossRef]
- Shi, Z.; Chapes, S.K.; Ben-Arieh, D.; Wu, C.H. An agent-based model of a hepatic inflammatory response to salmonella: A computational study under a large set of experimental data. PLoS ONE 2016, 11, e0161131. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.E.; Bell, C.S.; McClain, M.S.; Cover, T.L.; Giorgio, T.D. Dynamic computational model of symptomatic bacteremia to inform bacterial separation treatment requirements. PLoS ONE 2016, 11, e0163167. [Google Scholar] [CrossRef] [PubMed]
- Lamping, F.; Jack, T.; Rubsamen, N.; Sasse, M.; Beerbaum, P.; Mikolajczyk, R.T.; Boehne, M.; Karch, A. Development and validation of a diagnostic model for early differentiation of sepsis and non-infectious sirs in critically ill children—A data-driven approach using machine-learning algorithms. BMC Pediatr. 2018, 18, 112. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Liu, T.; Shepherd, S.; Paiva, W. Using statistical and machine learning methods to evaluate the prognostic accuracy of sirs and qsofa. Healthc. Inform. Res. 2018, 24, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gallo, J.E.; Fonseca-Ruiz, N.J.; Celi, L.A.; Duitama-Munoz, J.F. A machine learning-based model for 1-year mortality prediction in patients admitted to an intensive care unit with a diagnosis of sepsis. Med. Intensiv. 2018. [Google Scholar] [CrossRef] [PubMed]
- Meiring, C.; Dixit, A.; Harris, S.; MacCallum, N.S.; Brealey, D.A.; Watkinson, P.J.; Jones, A.; Ashworth, S.; Beale, R.; Brett, S.J.; et al. Optimal intensive care outcome prediction over time using machine learning. PLoS ONE 2018, 13, e0206862. [Google Scholar] [CrossRef]
- Masino, A.J.; Harris, M.C.; Forsyth, D.; Ostapenko, S.; Srinivasan, L.; Bonafide, C.P.; Balamuth, F.; Schmatz, M.; Grundmeier, R.W. Machine learning models for early sepsis recognition in the neonatal intensive care unit using readily available electronic health record data. PLoS ONE 2019, 14, e0212665. [Google Scholar] [CrossRef]
- Taylor, R.A.; Moore, C.L.; Cheung, K.H.; Brandt, C. Predicting urinary tract infections in the emergency department with machine learning. PLoS ONE 2018, 13, e0194085. [Google Scholar] [CrossRef]
- Vellido, A.; Ribas, V.; Morales, C.; Ruiz Sanmartin, A.; Ruiz Rodriguez, J.C. Machine learning in critical care: State-of-the-art and a sepsis case study. Biomed. Eng. Online 2018, 17, 135. [Google Scholar] [CrossRef]
- Delahanty, R.J.; Alvarez, J.; Flynn, L.M.; Sherwin, R.L.; Jones, S.S. Development and evaluation of a machine learning model for the early identification of patients at risk for sepsis. Ann. Emerg. Med. 2019. [Google Scholar] [CrossRef]
- Van Wyk, F.; Khojandi, A.; Mohammed, A.; Begoli, E.; Davis, R.L.; Kamaleswaran, R. A minimal set of physiomarkers in continuous high frequency data streams predict adult sepsis onset earlier. Int. J. Med. Inform. 2019, 122, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Saqib, M.; Sha, Y.; Wang, M.D. Early prediction of sepsis in emr records using traditional ml techniques and deep learning lstm networks. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 4038–4041. [Google Scholar] [CrossRef]
- Donovan, M.J.; Fernandez, G.; Scott, R.; Khan, F.M.; Zeineh, J.; Koll, G.; Gladoun, N.; Charytonowicz, E.; Tewari, A.; Cordon-Cardo, C. Development and validation of a novel automated gleason grade and molecular profile that define a highly predictive prostate cancer progression algorithm-based test. Prostate Cancer Prostatic Dis. 2018, 21, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Kamaleswaran, R.; Akbilgic, O.; Hallman, M.A.; West, A.N.; Davis, R.L.; Shah, S.H. Applying artificial intelligence to identify physiomarkers predicting severe sepsis in the picu. Pediatr. Crit. Care Med. 2018, 19, e495–e503. [Google Scholar] [CrossRef] [PubMed]
- Nemati, S.; Holder, A.; Razmi, F.; Stanley, M.D.; Clifford, G.D.; Buchman, T.G. An interpretable machine learning model for accurate prediction of sepsis in the icu. Crit. Care Med. 2018, 46, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Komorowski, M.; Celi, L.A.; Badawi, O.; Gordon, A.C.; Faisal, A.A. The artificial intelligence clinician learns optimal treatment strategies for sepsis in intensive care. Nat. Med. 2018, 24, 1716–1720. [Google Scholar] [CrossRef]
- Torres-Sangiao, E.; Holban, A.M.; Gestal, M.C. Advanced nanobiomaterials: Vaccines, diagnosis and treatment of infectious diseases. Molecules 2016, 21. [Google Scholar] [CrossRef]
- Wang, L.; Ruan, S. Modeling nosocomial infections of methicillin-resistant staphylococcus aureus with environment contamination. Sci. Rep. 2017, 7, 580. [Google Scholar] [CrossRef]
- Sharma, A.; Hill, A.; Kurbatova, E.; van der Walt, M.; Kvasnovsky, C.; Tupasi, T.E.; Caoili, J.C.; Gler, M.T.; Volchenkov, G.V.; Kazennyy, B.Y.; et al. Estimating the future burden of multidrug-resistant and extensively drug-resistant tuberculosis in india, the philippines, russia, and south africa: A mathematical modelling study. Lancet Infect. Dis. 2017, 17, 707–715. [Google Scholar] [CrossRef]
- Pinterest Home Page. Available online: https://www.pinterest.es (accessed on 25 February 2019).
- Shutterstock Home Page. Available online: https://www.shutterstock.com (accessed on 25 February 2019).
- Estrela, S.; Brown, S.P. Community interactions and spatial structure shape selection on antibiotic resistant lineages. PLoS Comput. Biol. 2018, 14, e1006179. [Google Scholar] [CrossRef]
- Volz, E.M.; Didelot, X. Modeling the growth and decline of pathogen effective population size provides insight into epidemic dynamics and drivers of antimicrobial resistance. Syst. Biol. 2018, 67, 719–728. [Google Scholar] [CrossRef]
- Yu, G.; Baeder, D.Y.; Regoes, R.R.; Rolff, J. Predicting drug resistance evolution: Insights from antimicrobial peptides and antibiotics. Proc. Biol. Sci. 2018, 285. [Google Scholar] [CrossRef]
- Baquero, F. From pieces to patterns: Evolutionary engineering in bacterial pathogens. Nat. Rev. Microbiol. 2004, 2, 510–518. [Google Scholar] [CrossRef] [PubMed]
- Canton, R.; Ruiz-Garbajosa, P. Co-resistance: An opportunity for the bacteria and resistance genes. Curr. Opin. Pharmacol. 2011, 11, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.; Capilla, R.; Naya, F.; Futami, R.; Coque, T.; Moya, A.; Fernandez-Lanza, V.; Canton, R.; Sempere, J.M.; Llorens, C.; et al. Simulating multilevel dynamics of antimicrobial resistance in a membrane computing model. MBio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Lim-Wilby, M. Molecular docking. Methods Mol. Biol. 2008, 443, 365–382. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Zhou, W.; Fan, Y.; Zhao, J.; Zhu, L.; Lu, S.; Lu, T.; Chen, Y.; Liu, H. A combined drug discovery strategy based on machine learning and molecular docking. Chem. Biol. Drug. Des. 2019. [Google Scholar] [CrossRef] [PubMed]
- Bonten, M.J.; Austin, D.J.; Lipsitch, M. Understanding the spread of antibiotic resistant pathogens in hospitals: Mathematical models as tools for control. Clin. Infect. Dis. 2001, 33, 1739–1746. [Google Scholar] [CrossRef]
- Spicknall, I.H.; Foxman, B.; Marrs, C.F.; Eisenberg, J.N. A modeling framework for the evolution and spread of antibiotic resistance: Literature review and model categorization. Am. J. Epidemiol. 2013, 178, 508–520. [Google Scholar] [CrossRef]
- Cen, X.; Feng, Z.; Zheng, Y.; Zhao, Y. Bifurcation analysis and global dynamics of a mathematical model of antibiotic resistance in hospitals. J. Math. Biol. 2017, 75, 1463–1485. [Google Scholar] [CrossRef]
- Boelle, P.Y.; Thomas, G. Resistance to antibiotics: Limit theorems for a stochastic sis model structured by level of resistance. J. Math. Biol. 2016, 73, 1353–1378. [Google Scholar] [CrossRef][Green Version]
- Yoshida, M.; Hinkley, T.; Tsuda, S.; Abul-Haija, Y.M.; McBurney, R.T.; Kulikov, V.; Jennifer, S.; Mathieson, J.S.; Reyes, S.G.; Castro, M.D.; et al. Using evolutionary algorithms and machine learning to explore sequence space for the discovery of antimicrobial peptides. Chem 2018, 10. [Google Scholar] [CrossRef]
- Nocedo-Mena, D.; Cornelio, C.; Camacho-Corona, M.D.R.; Garza-Gonzalez, E.; Waksman de Torres, N.; Arrasate, S.; Sotomayor, N.; Lete, E.; Gonzalez-Diaz, H. Modeling antibacterial activity with machine learning and fusion of chemical structure information with microorganism metabolic networks. J. Chem. Inf. Model. 2019, 59, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Sharaha, U.; Rodriguez-Diaz, E.; Sagi, O.; Riesenberg, K.; Salman, A.; Bigio, I.J.; Huleihel, M. Fast and reliable determination of escherichia coli susceptibility to antibiotics: Infrared microscopy in tandem with machine learning algorithms. J. Biophotonics 2019, e201800478. [Google Scholar] [CrossRef] [PubMed]
- Arepyeva, M.A.; Kolbin, A.S.; Sidorenko, S.V.; Lawson, R.; Kurylev, A.A.; Balykina, Y.E.; Mukhina, N.V.; Spiridonova, A.A. A mathematical model for predicting the development of bacterial resistance based on the relationship between the level of antimicrobial resistance and the volume of antibiotic consumption. J. Glob. Antimicrob. Resist. 2017, 8, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Ternent, L.; Dyson, R.J.; Krachler, A.M.; Jabbari, S. Bacterial fitness shapes the population dynamics of antibiotic-resistant and -susceptible bacteria in a model of combined antibiotic and anti-virulence treatment. J. Theor. Biol. 2015, 372, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Dasbasi, B.; Ozturk, I. Mathematical modelling of bacterial resistance to multiple antibiotics and immune system response. Springerplus 2016, 5, 408. [Google Scholar] [CrossRef] [PubMed]
- Levin, B.R.; Baquero, F.; Ankomah, P.P.; McCall, I.C. Phagocytes, antibiotics, and self-limiting bacterial infections. Trends Microbiol. 2017, 25, 878–892. [Google Scholar] [CrossRef] [PubMed]
- Caudill, L.; Lawson, B. A unified inter-host and in-host model of antibiotic resistance and infection spread in a hospital ward. J. Theor. Biol. 2017, 421, 112–126. [Google Scholar] [CrossRef]
- Gillespie, J.J.; Wattam, A.R.; Cammer, S.A.; Gabbard, J.L.; Shukla, M.P.; Dalay, O.; Driscoll, T.; Hix, D.; Mane, S.P.; Mao, C.; et al. Patric: The comprehensive bacterial bioinformatics resource with a focus on human pathogenic species. Infect. Immun. 2011, 79, 4286–4298. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef]
- Nguyen, M.; Brettin, T.; Long, S.W.; Musser, J.M.; Olsen, R.J.; Olson, R.; Shukla, M.; Stevens, R.L.; Xia, F.; Yoo, H.; et al. Developing an in silico minimum inhibitory concentration panel test for klebsiella pneumoniae. Sci. Rep. 2018, 8, 421. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.; Long, S.W.; McDermott, P.F.; Olsen, R.J.; Olson, R.; Stevens, R.L.; Tyson, G.H.; Zhao, S.; Davis, J.J. Using machine learning to predict antimicrobial mics and associated genomic features for nontyphoidal salmonella. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef] [PubMed]
- Babajide Mustapha, I.; Saeed, F. Bioactive molecule prediction using extreme gradient boosting. Molecules 2016, 21. [Google Scholar] [CrossRef] [PubMed]
- Arango-Argoty, G.; Garner, E.; Pruden, A.; Heath, L.S.; Vikesland, P.; Zhang, L. Deeparg: A deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome 2018, 6, 23. [Google Scholar] [CrossRef] [PubMed]
- Nicoloff, H.; Hjort, K.; Levin, B.R.; Andersson, D.I. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat. Microbiol. 2019, 4, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Kavvas, E.S.; Catoiu, E.; Mih, N.; Yurkovich, J.T.; Seif, Y.; Dillon, N.; Heckmann, D.; Anand, A.; Yang, L.; Nizet, V.; et al. Machine learning and structural analysis of mycobacterium tuberculosis pan-genome identifies genetic signatures of antibiotic resistance. Nat. Commun. 2018, 9, 4306. [Google Scholar] [CrossRef] [PubMed]
- Jeukens, J.; Freschi, L.; Kukavica-Ibrulj, I.; Emond-Rheault, J.G.; Tucker, N.P.; Levesque, R.C. Genomics of antibiotic-resistance prediction in pseudomonas aeruginosa. Ann. N. Y. Acad. Sci. 2019, 1435, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Monk, J.M. Predicting antimicrobial resistance and associated genomic features from whole-genome sequencing. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef] [PubMed]
- Bédubourg, G.; Le Strat, Y. Evaluation and comparison of statistical methods for early temporal detection of outbreaks: A simulation-based study. PLoS ONE 2017, 12, e0181227. [Google Scholar] [CrossRef]
- Unkel, S.; Farrington, C.P.; Garthwaite, P.H.; Robertson, C.; Andrews, N. Statistical methods for the prospective detection of infectious disease outbreaks: A review. J. R. Stat. Soc. Ser. A (Stat. Soc.) 2012, 175, 49–82. [Google Scholar] [CrossRef]
- Solgi, M.; Karami, M.; Poorolajal, J. Timely detection of influenza outbreaks in iran: Evaluating the performance of the exponentially weighted moving average. J. Infect. Public Health 2018, 11, 389–392. [Google Scholar] [CrossRef] [PubMed]
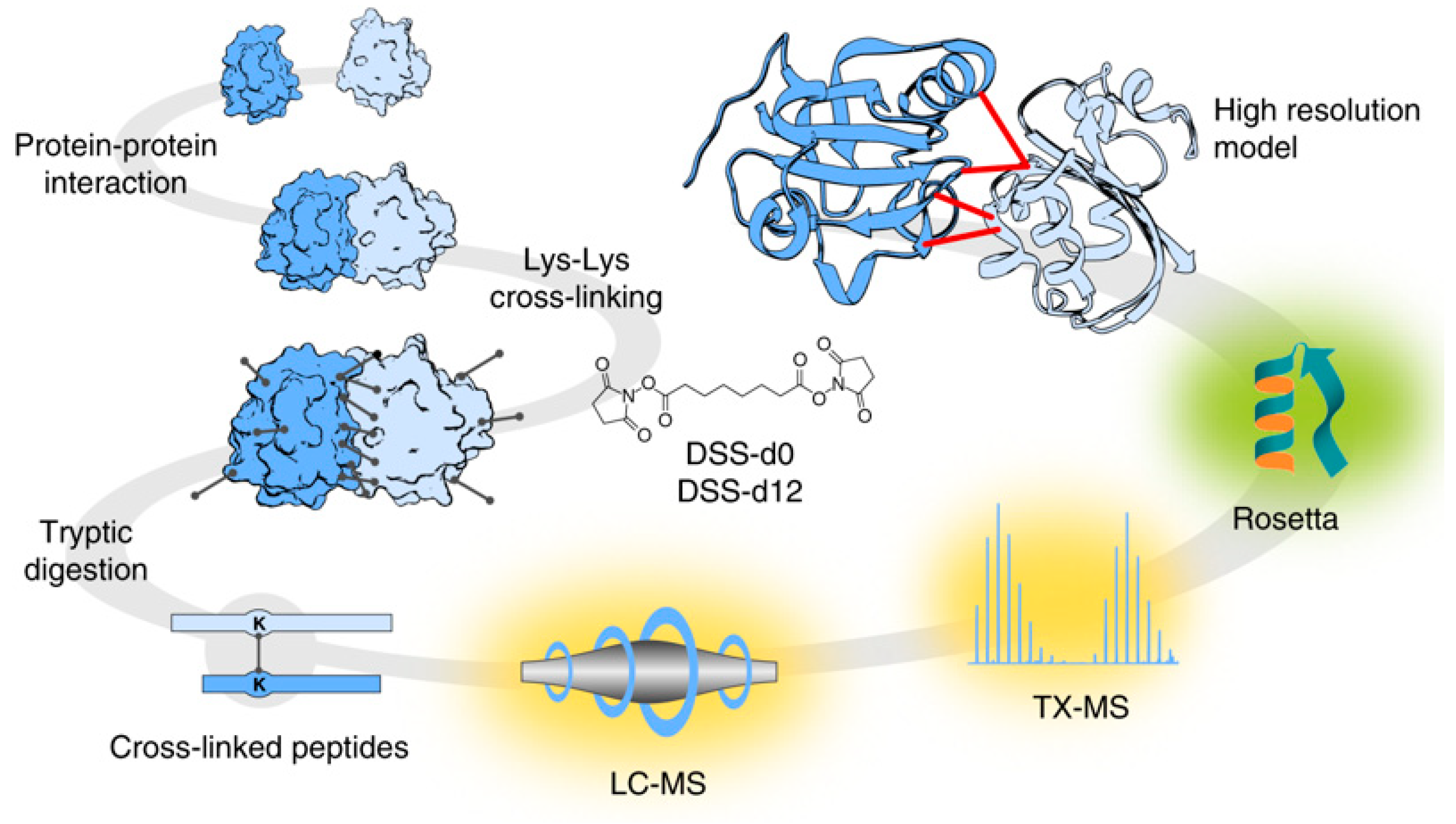

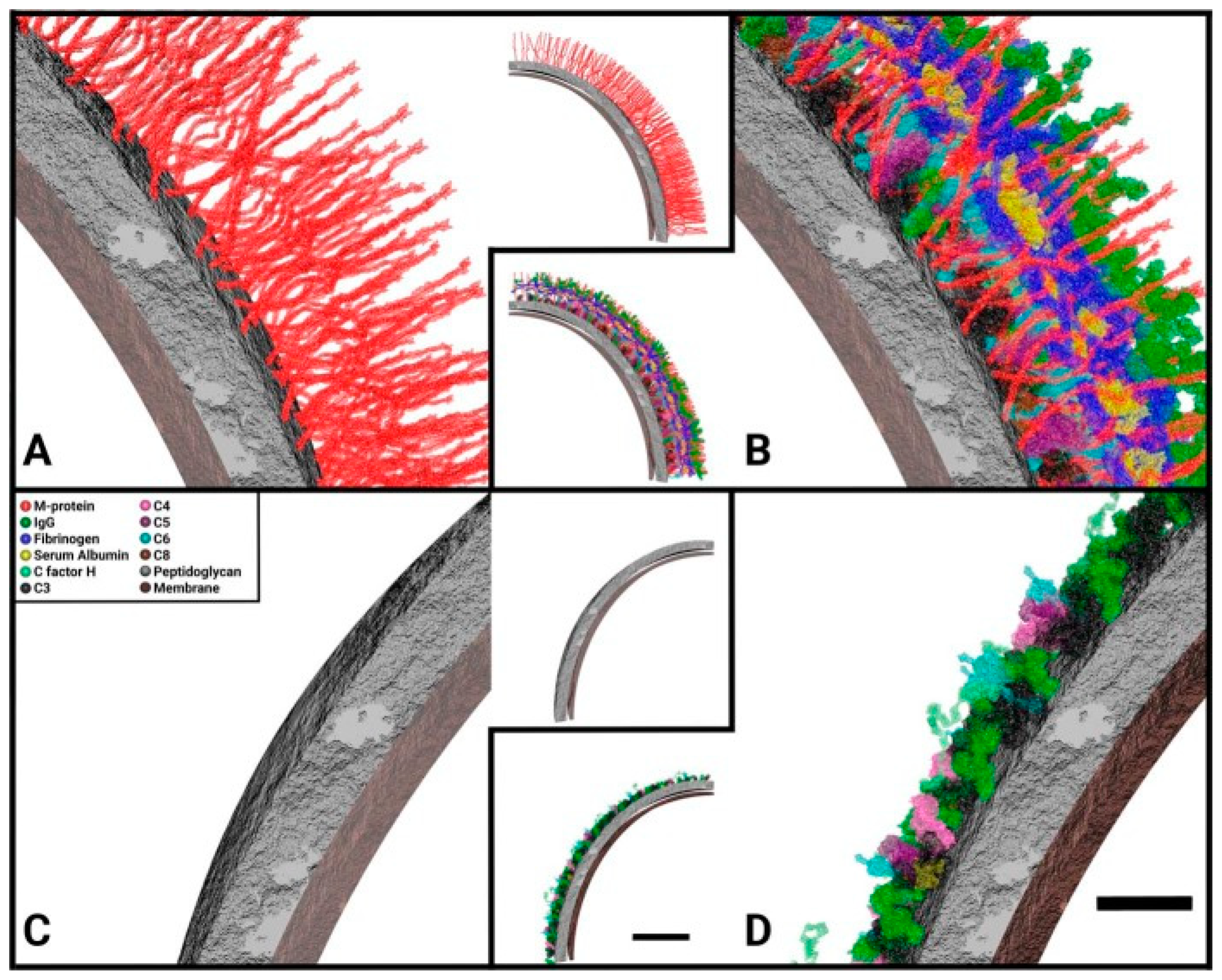
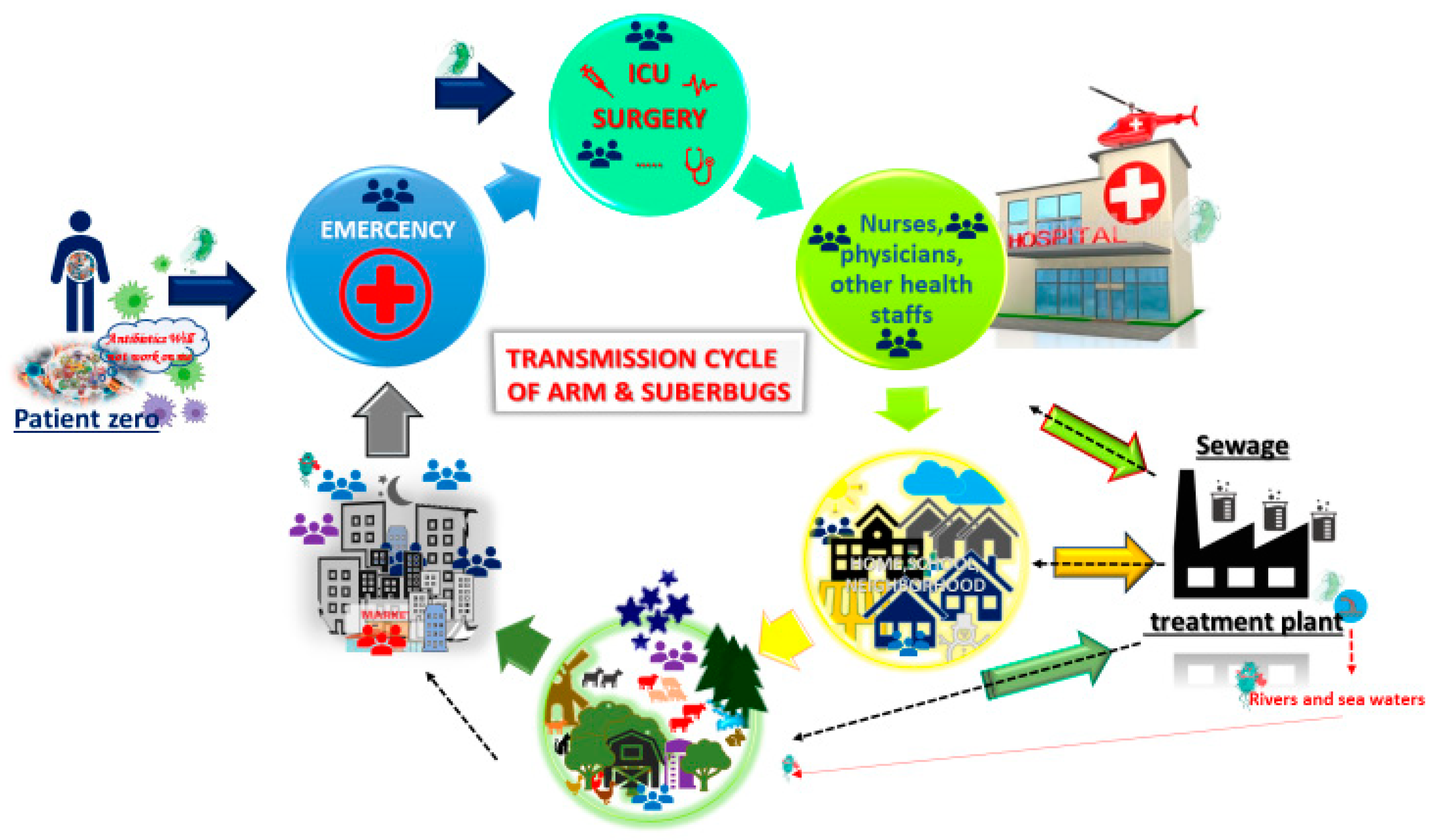
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cartelle Gestal, M.; Dedloff, M.R.; Torres-Sangiao, E. Computational Health Engineering Applied to Model Infectious Diseases and Antimicrobial Resistance Spread. Appl. Sci. 2019, 9, 2486. https://doi.org/10.3390/app9122486
Cartelle Gestal M, Dedloff MR, Torres-Sangiao E. Computational Health Engineering Applied to Model Infectious Diseases and Antimicrobial Resistance Spread. Applied Sciences. 2019; 9(12):2486. https://doi.org/10.3390/app9122486
Chicago/Turabian StyleCartelle Gestal, Mónica, Margaret R. Dedloff, and Eva Torres-Sangiao. 2019. "Computational Health Engineering Applied to Model Infectious Diseases and Antimicrobial Resistance Spread" Applied Sciences 9, no. 12: 2486. https://doi.org/10.3390/app9122486
APA StyleCartelle Gestal, M., Dedloff, M. R., & Torres-Sangiao, E. (2019). Computational Health Engineering Applied to Model Infectious Diseases and Antimicrobial Resistance Spread. Applied Sciences, 9(12), 2486. https://doi.org/10.3390/app9122486






