Super-Resolution Lensless Imaging of Cells Using Brownian Motion
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
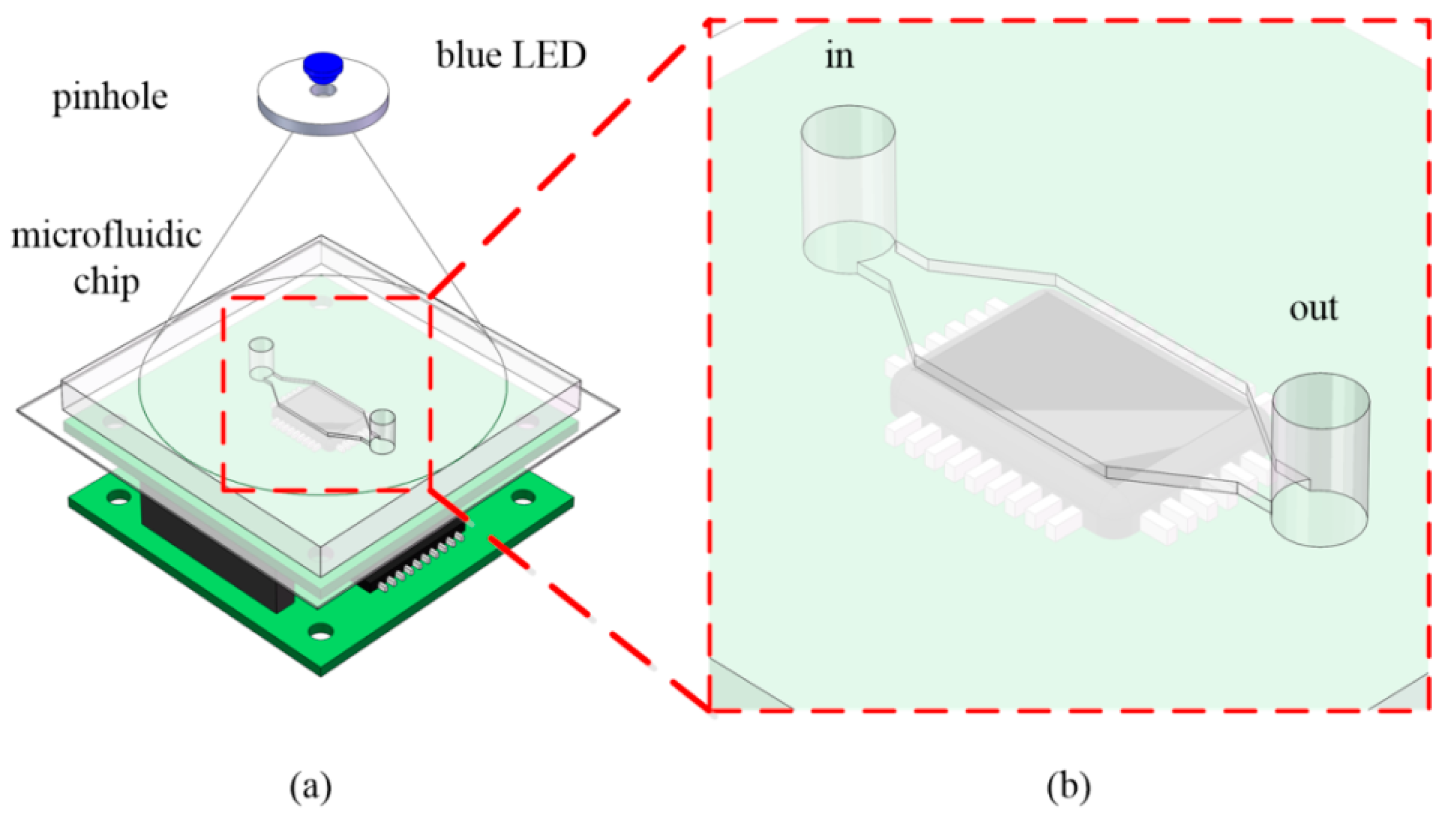
2.1. Lensless Imaging System
2.2. Shifting Parameters of Brownian Motion of Cells
2.3. Super-Resolution Reconstruction Algorithm for Cell Image in a Lensless System
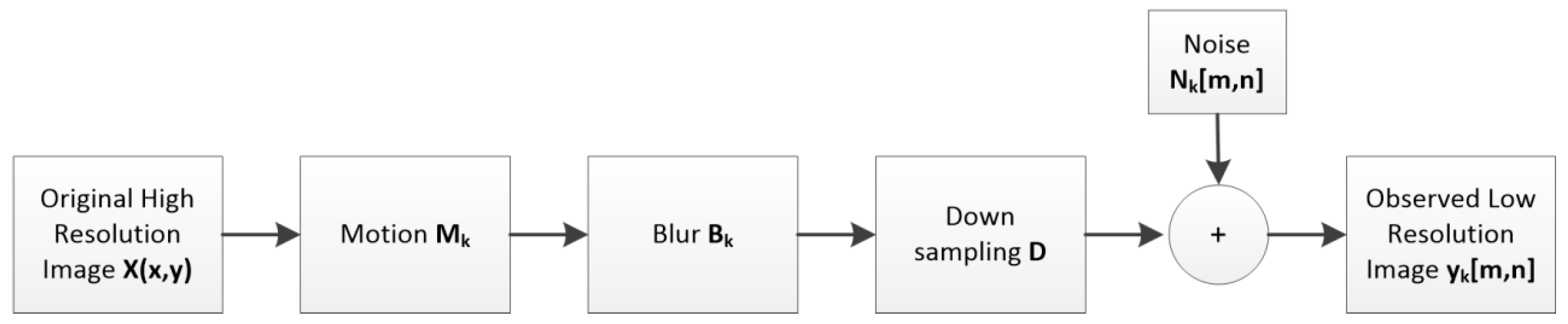
2.3.1. Motion Estimation Algorithm
2.3.2. Normalized Convolution Super-Resolution Algorithm
2.3.3. Reconstruction of the Focus Plane Image Algorithm
2.4. System Experimental Method
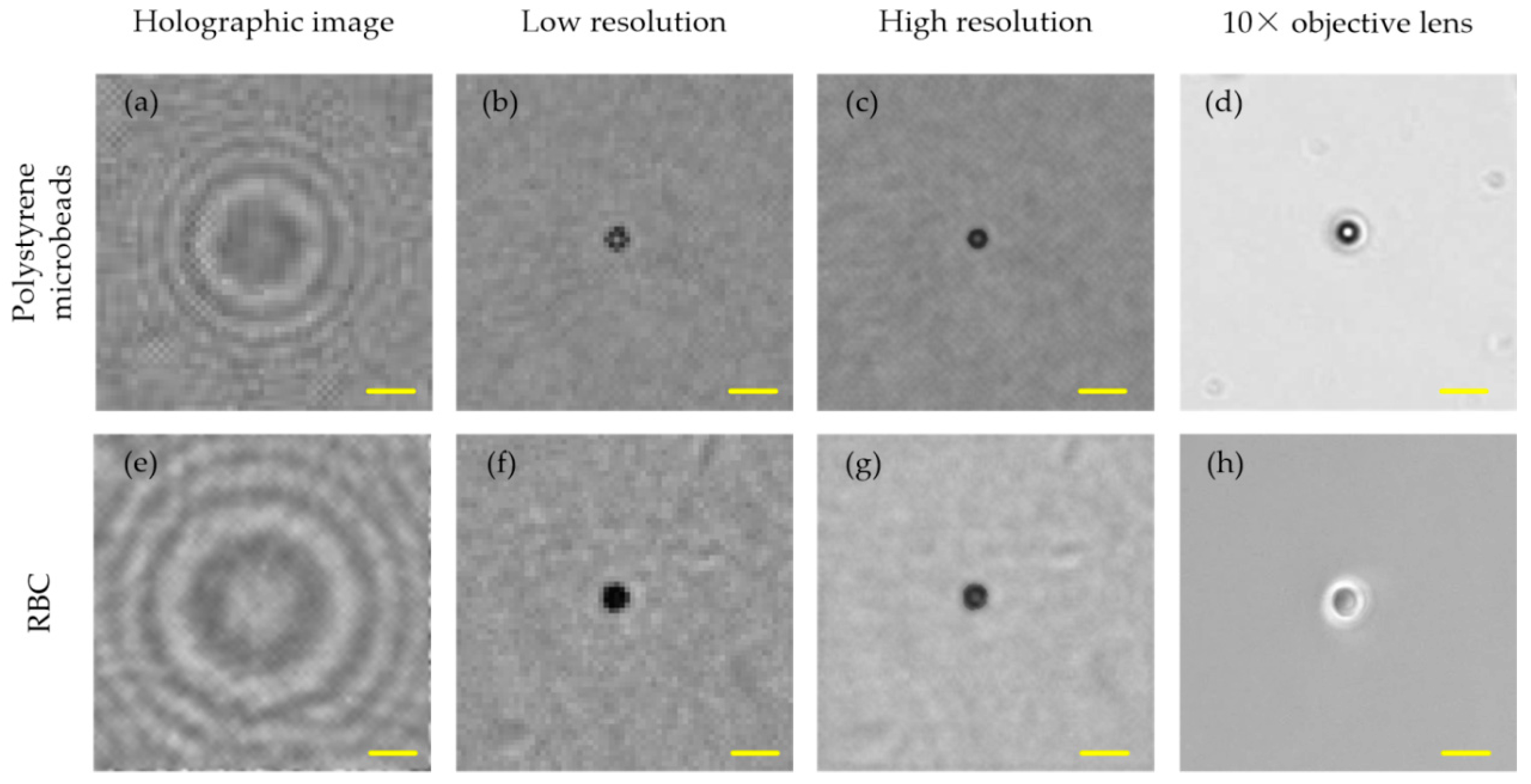
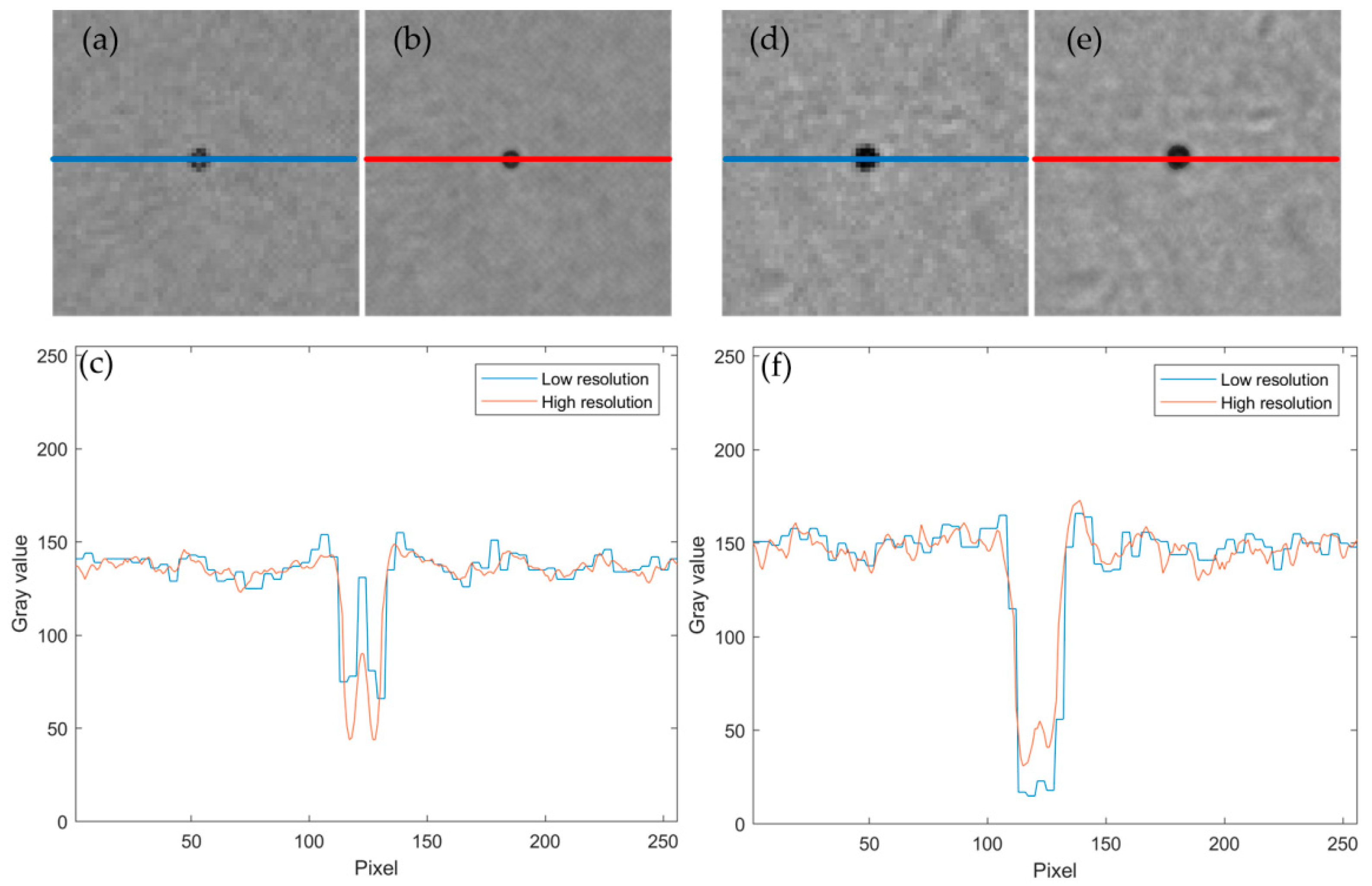
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Mudanyali, O.; Tseng, D.; Oh, C.; Isikman, S.O.; Sencan, I.; Bishara, W.; Oztoprak, C.; Seo, S.; Khademhosseini, B.; Ozcan, A. Compact, light-weight and cost-effective microscope based on lensless incoherent holography for telemedicine applications. Lab Chip 2010, 10, 1417–1428. [Google Scholar] [CrossRef]
- Mudanyali, O.; Oztoprak, C.; Tseng, D.; Erlinger, A.; Ozcan, A. Detection of waterborne parasites using field-portable and cost-effective lensfree microscopy. Lab Chip 2010, 10, 2419–2423. [Google Scholar] [CrossRef]
- Penwill, L.A.; Batten, G.E.; Castagnetti, S.; Shaw, A.M. Growth phenotype screening of Schizosaccharomyces pombe using a Lensless microscope. Biosens. Bioelectron. 2014, 54, 345–350. [Google Scholar] [CrossRef]
- Huang, X.; Jiang, Y.; Liu, X.; Xu, H.; Han, Z.; Rong, H.; Yang, H.; Yan, M.; Yu, H. Machine Learning Based Single-Frame Super-Resolution Processing for Lensless Blood Cell Counting. Sensors 2016, 16, 1836. [Google Scholar] [CrossRef]
- Lee, J.; Kwak, Y.H.; Paek, S.-H.; Han, S.; Seo, S. CMOS image sensor-based ELISA detector using lens-free shadow imaging platform. Sens. Actuators B Chem. 2014, 196, 511–517. [Google Scholar] [CrossRef]
- Jin, G.; Yoo, I.H.; Pack, S.P.; Yang, J.W.; Ha, U.H.; Paek, S.H.; Seo, S. Lens-free shadow image based high-throughput continuous cell monitoring technique. Biosens. Bioelectron. 2012, 38, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Roy, M.; Jin, G.; Pan, J.-H.; Seo, D.; Hwang, Y.; Oh, S.; Lee, M.; Kim, Y.J.; Seo, S. Staining-free cell viability measurement technique using lens-free shadow imaging platform. Sens. Actuators B Chem. 2016, 224, 577–583. [Google Scholar] [CrossRef]
- Roy, M.; Jin, G.; Seo, D.; Nam, M.-H.; Seo, S. A simple and low-cost device performing blood cell counting based on lens-free shadow imaging technique. Sens. Actuators B Chem. 2014, 201, 321–328. [Google Scholar] [CrossRef]
- Scholz, G.; Xu, Q.; Schulze, T.; Boht, H.; Mattern, K.; Hartmann, J.; Dietzel, A.; Scherneck, S.; Rustenbeck, I.; Prades, J.; et al. LED-Based Tomographic Imaging for Live-Cell Monitoring of Pancreatic Islets in Microfluidic Channels. Proceedings 2017, 1, 552. [Google Scholar] [CrossRef]
- Heng, X.; Erickson, D.; Baugh, L.R.; Yaqoob, Z.; Sternberg, P.W.; Psaltis, D.; Yang, C. Optofluidic microscopy—A method for implementing a high resolution optical microscope on a chip. Lab Chip 2006, 6, 1274–1276. [Google Scholar] [CrossRef]
- Lee, S.A.; Leitao, R.; Zheng, G.; Yang, S.; Rodriguez, A.; Yang, C. Color capable sub-pixel resolving optofluidic microscope and its application to blood cell imaging for malaria diagnosis. PLoS ONE 2011, 6, e26127. [Google Scholar] [CrossRef]
- Lee, S.A.; Yang, C. A smartphone-based chip-scale microscope using ambient illumination. Lab Chip 2014, 14, 3056–3063. [Google Scholar] [CrossRef]
- Zheng, G.; Lee, S.A.; Antebi, Y.; Elowitz, M.B.; Yang, C. The ePetri dish, an on-chip cell imaging platform based on subpixel perspective sweeping microscopy (SPSM). Proc. Natl. Acad. Sci. USA 2011, 108, 16889–16894. [Google Scholar] [CrossRef]
- Coskun, A.F.; Sencan, I.; Su, T.W.; Ozcan, A. Wide-field lensless fluorescent microscopy using a tapered fiber-optic faceplate on a chip. Analyst 2011, 136, 3512–3518. [Google Scholar] [CrossRef]
- Coskun, A.F.; Sencan, I.; Su, T.W.; Ozcan, A. Lensfree fluorescent on-chip imaging of transgenic Caenorhabditis elegans over an ultra-wide field-of-view. PLoS ONE 2011, 6, e15955. [Google Scholar] [CrossRef]
- Greenbaum, A.; Luo, W.; Khademhosseinieh, B.; Su, T.-W.; Coskun, A.F.; Ozcan, A. Increased space-bandwidth product in pixel super-resolved lensfree on-chip microscopy. Sci. Rep. 2013, 3, 1717. [Google Scholar] [CrossRef]
- Isikman, S.O.; Bishara, W.; Mavandadi, S.; Yu, F.W.; Feng, S.; Lau, R.; Ozcan, A. Lens-free optical tomographic microscope with a large imaging volume on a chip. Proc. Natl. Acad. Sci. USA 2011, 108, 7296–7301. [Google Scholar] [CrossRef]
- Isikman, S.O.; Bishara, W.; Mudanyali, O.; Sencan, I.; Su, T.W.; Tseng, D.; Yaglidere, O.; Sikora, U.; Ozcan, A. Lensfree On-Chip Microscopy and Tomography for Bio-Medical Applications. IEEE J. Sel. Top. Quantum Electron. 2011, 18, 1059–1072. [Google Scholar] [CrossRef]
- Seo, S.; Isikman, S.O.; Sencan, I.; Mudanyali, O.; Su, T.W.; Bishara, W.; Erlinger, A.; Ozcan, A. High-throughput lens-free blood analysis on a chip. Anal. Chem. 2010, 82, 4621–4627. [Google Scholar] [CrossRef]
- Su, T.W.; Erlinger, A.; Tseng, D.; Ozcan, A. Compact and light-weight automated semen analysis platform using lensfree on-chip microscopy. Anal. Chem. 2010, 82, 8307–8312. [Google Scholar] [CrossRef]
- Tseng, D.; Mudanyali, O.; Oztoprak, C.; Isikman, S.O.; Sencan, I.; Yaglidere, O.; Ozcan, A. Lensfree microscopy on a cellphone. Lab Chip 2010, 10, 1787–1792. [Google Scholar] [CrossRef]
- Zhu, H.; Yaglidere, O.; Su, T.W.; Tseng, D.; Ozcan, A. Cost-effective and compact wide-field fluorescent imaging on a cell-phone. Lab Chip 2011, 11, 315–322. [Google Scholar] [CrossRef]
- Zheng, G.; Lee, S.A.; Yang, S.; Yang, C. Sub-pixel resolving optofluidic microscope for on-chip cell imaging. Lab Chip 2010, 10, 3125–3129. [Google Scholar] [CrossRef]
- Isikman, S.O.; Bishara, W.; Sikora, U.; Yaglidere, O.; Yeah, J.; Ozcan, A. Field-portable lensfree tomographic microscope. Lab Chip 2011, 11, 2222–2230. [Google Scholar] [CrossRef]
- Bishara, W.; Sikora, U.; Mudanyali, O.; Su, T.W.; Yaglidere, O.; Luckhart, S.; Ozcan, A. Holographic pixel super-resolution in portable lensless on-chip microscopy using a fiber-optic array. Lab Chip 2011, 11, 1276–1279. [Google Scholar] [CrossRef]
- Lee, K.; Kim, H.D.; Kim, K.; Kim, Y.; Hillman, T.R.; Min, B.; Park, Y. Synthetic Fourier transform light scattering. Opt Express 2013, 21, 22453–22463. [Google Scholar] [CrossRef]
- Knutsson, H.; Westin, C.-F. Normalized and differential convolution. In Proceedings of the IEEE Computer Society Conference on ComputerVision and Pattern Recognition, New York, NY, USA, 15–17 June 1993; pp. 515–523. [Google Scholar]
- Pham, T.Q.; van Vliet, L.J.; Schutte, K. Robust Fusion of Irregularly Sampled Data Using Adaptive Normalized Convolution. EURASIP J. Adv. Signal Process. 2006, 2006, 083268. [Google Scholar] [CrossRef]
- Eckstein, E.C. Fractional Brownian Motion and Particle Motions in Blood Flow. In Proceedings of the International Conference of the IEEE Engineering in Medicine & Biology Society, Orlando, FL, USA, 31 October–3 November 1991. [Google Scholar]
- Lucchese, L.; Cortelazzo, G.M. A Noise-Robust Frequency Domain Technique for Estimating Planar Roto-Translations. IEEE Trans. Signal Process. 2000, 48, 1769–1786. [Google Scholar] [CrossRef]
- Bigot, J.; Gamboa, F.; Vimond, M. Estimation of Translation, Rotation, and Scaling between Noisy Images Using the Fourier–Mellin Transform. SIAM J. Imaging Sci. 2009, 2, 614–645. [Google Scholar] [CrossRef]
- Keren, D.; Peleg, S.; Brada, R. Image sequence enhancement using sub-pixel displacement. In Proceedings of the Conference on Computer Vision & Pattern Recognition, Ann Arbor, MI, USA, 5–9 June 1988. [Google Scholar]
- Fang, Y.; Yu, N.; Jiang, Y.; Dang, C. High-Precision Lens-Less Flow Cytometer on a Chip. Micromachines 2018, 9, 227. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, Y.; Yu, N.; Jiang, Y. Super-Resolution Lensless Imaging of Cells Using Brownian Motion. Appl. Sci. 2019, 9, 2080. https://doi.org/10.3390/app9102080
Fang Y, Yu N, Jiang Y. Super-Resolution Lensless Imaging of Cells Using Brownian Motion. Applied Sciences. 2019; 9(10):2080. https://doi.org/10.3390/app9102080
Chicago/Turabian StyleFang, Yuan, Ningmei Yu, and Yuquan Jiang. 2019. "Super-Resolution Lensless Imaging of Cells Using Brownian Motion" Applied Sciences 9, no. 10: 2080. https://doi.org/10.3390/app9102080
APA StyleFang, Y., Yu, N., & Jiang, Y. (2019). Super-Resolution Lensless Imaging of Cells Using Brownian Motion. Applied Sciences, 9(10), 2080. https://doi.org/10.3390/app9102080




