Quantification of Unknown Nanoscale Biomolecules Using the Average-Weight-Difference Method
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of GluNP/Drug (INH and DOX)
2.3. Characterization of GluNP/INH
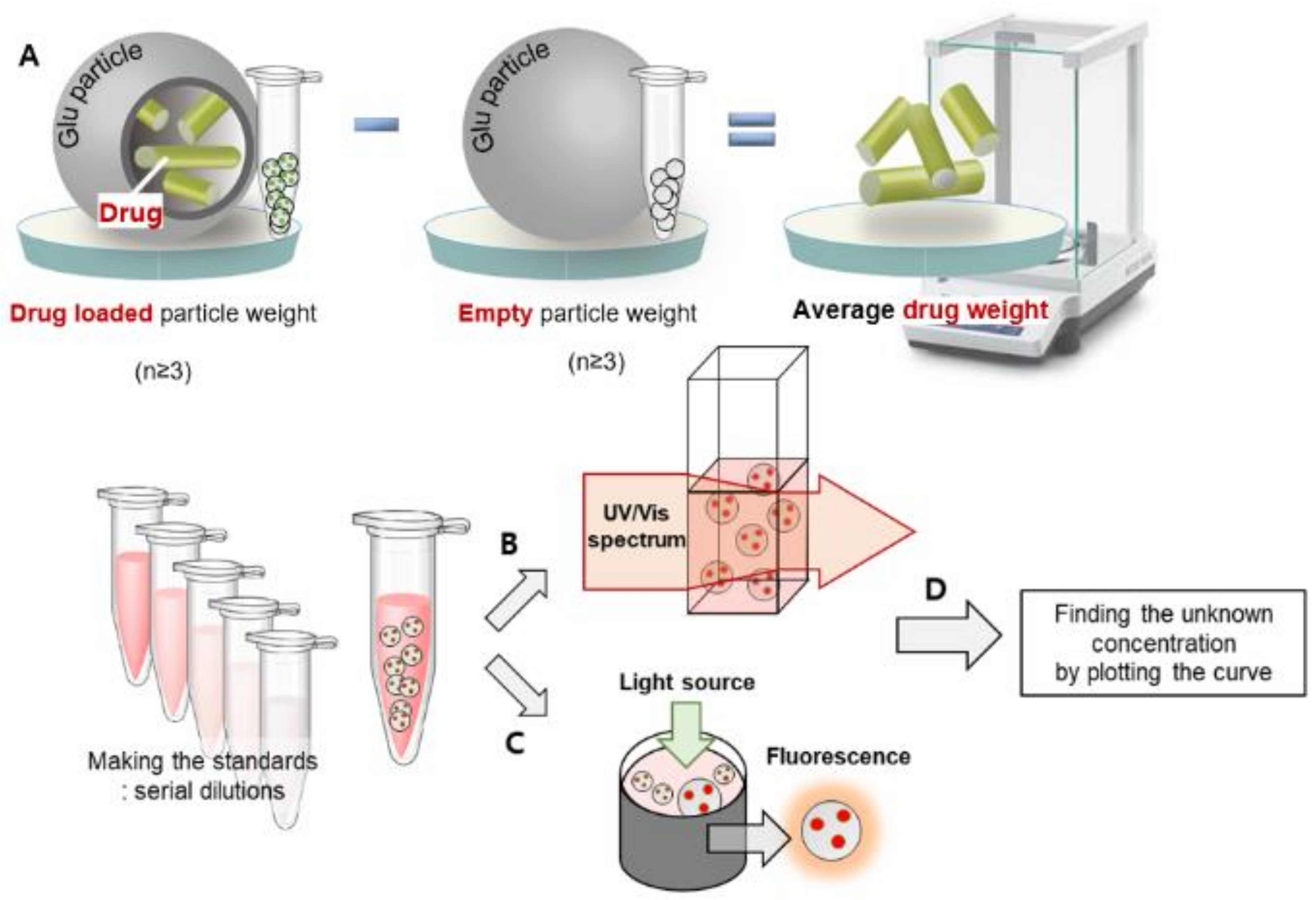
2.4. Quantification Analysis
2.4.1. Average Weight Difference
2.4.2. UV/Vis Spectroscopy
2.4.3. Fluorescence Spectroscopy
3. Results and Discussion
4. Summary
Author Contributions
Funding
Conflicts of Interest
References
- Pan, Y.; Li, X.; Kang, T.; Meng, H.; Chen, Z.; Yang, L.; Wu, Y.; Wei, Y.; Gou, M. Efficient delivery of antigen to DCs using yeast-derived microparticles. Sci. Rep. 2015, 5, 10687. [Google Scholar] [CrossRef] [PubMed]
- Verma, M.S.; Gu, F.X. 1,3-Beta-Glucans: Drug Delivery and Pharmacology. In The Complex World of Polysaccharides; InTech: Rijeka, Croatia, 2012; Volume 12, pp. 555–572. [Google Scholar]
- Soto, E.R.; Caras, A.C.; Kut, L.C.; Castle, M.K.; Ostroff, G.R. Glucan particles for macrophage targeted delivery of nanoparticles. J. Drug Deliv. 2012, 2012, 143524. [Google Scholar] [CrossRef] [PubMed]
- Shah, V.P.; Midha, K.K.; Findlay, J.W.; Hill, H.M.; Hulse, J.D.; McGilveray, I.J.; McKay, G.; Miller, K.J.; Patnaik, R.N.; Powell, M.L.; et al. Bioanalytical method validation—A revisit with a decade of progress. Pharm. Res. 2000, 17, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Belay, A.; Ture, K.; Redi, M.; Asfaw, A. Measurement of caffeine in coffee beans with UV/vis spectrometer. Food Chem. 2008, 108, 310–315. [Google Scholar] [CrossRef]
- Hwang, J.; Lee, E.; Kim, J.; Seo, Y.; Lee, K.H.; Hong, J.W.; Gilad, A.A.; Park, H.; Choi, J. Effective delivery of immunosuppressive drug molecules by silica coated iron oxide nanoparticles. Colloids Surf. B Biointerfaces 2016, 142, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Yue, Z.; Eccleston, M.E.; Swartling, J.; Slater, N.K.; Kaminski, C.F. Fluorescence intensity and lifetime imaging of free and micellar-encapsulated doxorubicin in living cells. Nanomedicine 2008, 4, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Subedi, R.K.; Kang, K.W.; Choi, H.-K. Preparation and characterization of solid lipid nanoparticles loaded with doxorubicin. Eur. J. Pharm. Sci. 2009, 37, 508–513. [Google Scholar] [CrossRef] [PubMed]
- Van Loco, J.; Elskens, M.; Croux, C.; Beernaert, H. Linearity of calibration curves: Use and misuse of the correlation coefficient. Accreditat. Qual. Assur. 2002, 7, 281–285. [Google Scholar] [CrossRef]
- Such, V.; Traveset, J.; Gonzalo, R.; Gelpi, E. Stability assays of aged pharmaceutical formulas for thiamine and pyridoxine by high performance thin-layer chromatography and derivative ultraviolet spectrometry. Anal. Chem. 1980, 52, 412–419. [Google Scholar] [CrossRef]
- Kitamura, K.; Majima, R. Determination of salicylic acid in aspirin powder by second derivative ultraviolet spectrometry. Anal. Chem. 1983, 55, 54–56. [Google Scholar] [CrossRef]
- Choi, J.; Lee, E.K.; Choo, J.; Yuh, J.; Hong, J.W. Micro 3D cell culture systems for cellular behavior studies: Culture matrices, devices, substrates, and in-situ sensing methods. Biotechnol. J. 2015, 10, 1682–1688. [Google Scholar] [CrossRef] [PubMed]
- Black, L.; Hamerstrand, G.; Nakayama, F.; Rasnik, B. Gravimetric analysis for determining the resin and rubber content of guayule. Rubber Chem. Technol. 1983, 56, 367–371. [Google Scholar] [CrossRef]
- Hwang, J.; Son, J.; Seo, Y.; Jo, Y.; Lee, K.; Lee, D.; Khan, M.S.; Chavan, S.; Park, C.; Sharma, A. Functional silica nanoparticles conjugated with beta-glucan to deliver anti-tuberculosis drug molecules. J. Ind. Eng. Chem. 2018, 58, 376–385. [Google Scholar] [CrossRef]
- Goyal, G.; Hwang, J.; Aviral, J.; Seo, Y.; Jo, Y.; Son, J.; Choi, J. Green synthesis of silver nanoparticles using β-glucan, and their incorporation into doxorubicin-loaded water-in-oil nanoemulsions for antitumor and antibacterial applications. J. Ind. Eng. Chem. 2017, 47, 179–186. [Google Scholar] [CrossRef]
- Choi, D.; Heo, J.; Park, J.H.; Jo, Y.; Jeong, H.; Chang, M.; Choi, J.; Hong, J. Nano-film coatings onto collagen hydrogels with desired drug release. J. Ind. Eng. Chem. 2016, 36, 326–333. [Google Scholar] [CrossRef]
- Sletmoen, M.; Stokke, B.T. Higher order structure of (1, 3)-β-d-glucans and its influence on their biological activities and complexation abilities. Biopolymers 2008, 89, 310–321. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Lee, K.; Gilad, A.A.; Choi, J. Synthesis of Beta-glucan Nanoparticles for the Delivery of Single Strand DNA. Biotechnol. Bioprocess Eng. 2018, 23, 144–149. [Google Scholar] [CrossRef]
- Markarian, S.; Evangelopoulos, D.; Harutyunyan, L.; Pepoyan, E.; Guzman, J.; McHugh, T.; Bhakta, S. The properties of solutions of isoniazid in water and dimethylsulfoxide. J. Solut. Chem. 2012, 41, 1462–1476. [Google Scholar] [CrossRef]
- Bogorad, M.I.; Searson, P.C. Real-time imaging and quantitative analysis of doxorubicin transport in a perfusable microvessel platform. Integr. Biol. 2016, 8, 976–984. [Google Scholar] [CrossRef]
- Mohan, P.; Rapoport, N. Doxorubicin as a molecular nanotheranostic agent: Effect of doxorubicin encapsulation in micelles or nanoemulsions on the ultrasound-mediated intracellular delivery and nuclear trafficking. Mol. Pharm. 2010, 7, 1959–1973. [Google Scholar] [CrossRef]
- Choi, J.; Tung, S.-H.; Wang, N.S.; Reipa, V. Small-angle neutron scattering measurement of silicon nanoparticle size. Nanotechnology 2008, 19, 085715. [Google Scholar] [CrossRef] [PubMed]
- Gajendiran, M.; Choi, J.; Kim, S.J.; Kim, K.; Shin, H.; Koo, H.J.; Kim, K. Conductive biomaterials for tissue engineering applications. J. Ind. Eng. Chem. 2017, 51, 12–26. [Google Scholar] [CrossRef]



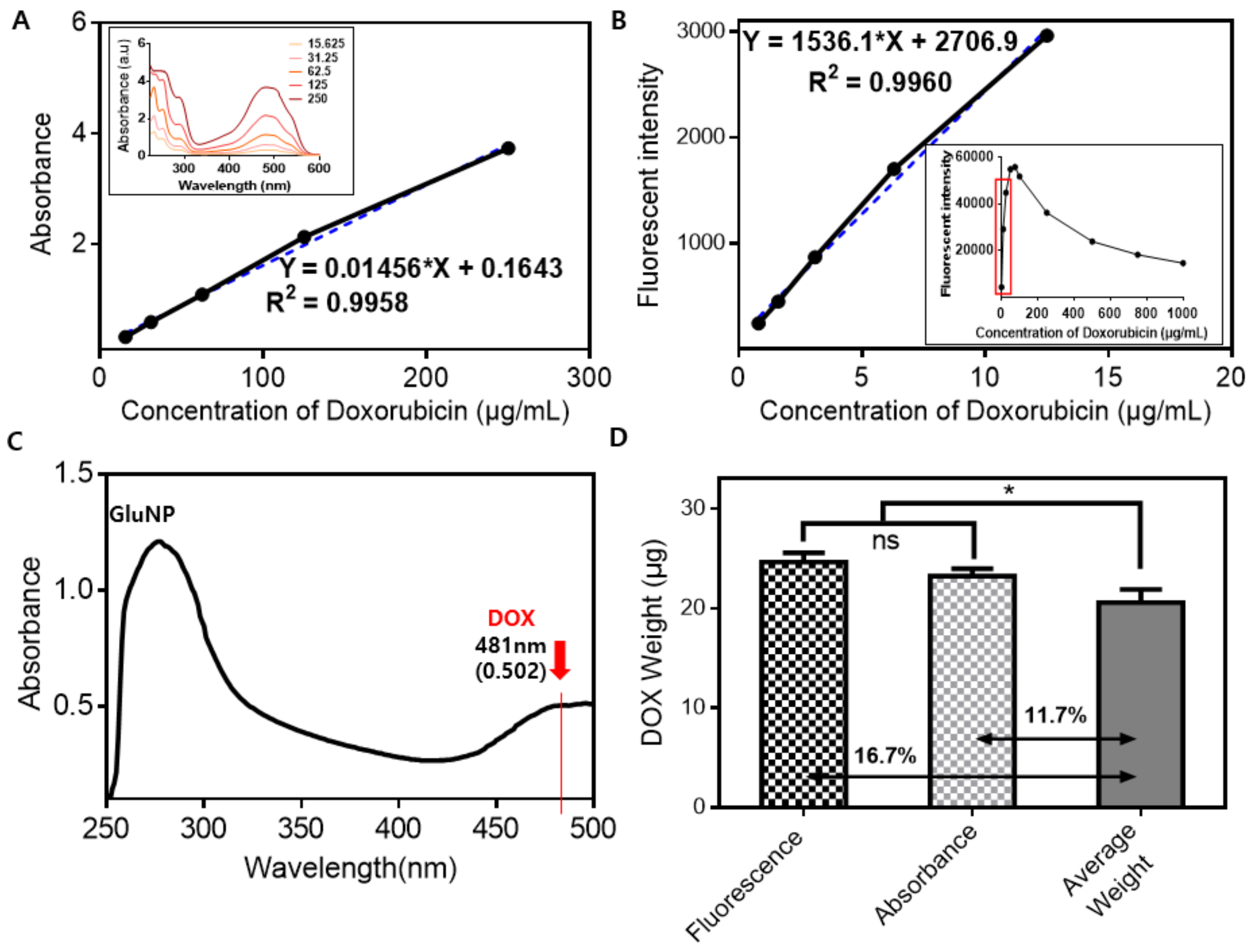
| n | Average Weight | Fluorescence (ex 490, em 590) | Absorbance (481 nm) | ||||
|---|---|---|---|---|---|---|---|
| GluNP/DOX Weight (g, ①) | GluNP Weight (g, ②) | Average Weight Difference (μg, ①-②) | Intensity | Amount of DOX (μg) | Absorption | Amount of DOX (μg) | |
| 1 | 0.7887 | 0.76695 | 21.75 | 21145.3 | 24.007 | 0.4935 | 22.610 |
| 2 | 0.7948 | 0.77567 | 19.13 | 22458.7 | 25.717 | 0.5147 | 24.066 |
| 3 | 0.8087 | 0.78792 | 20.78 | 21232.2 | 24.120 | 0.4990 | 22.988 |
| Average | 0.7974 | 0.776847 | 20.55 | 21612.1 | 24.615 | 0.5024 | 23.221 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, K.; Choi, Y.; Kim, K.; Koo, H.-J.; Choi, J. Quantification of Unknown Nanoscale Biomolecules Using the Average-Weight-Difference Method. Appl. Sci. 2019, 9, 130. https://doi.org/10.3390/app9010130
Lee K, Choi Y, Kim K, Koo H-J, Choi J. Quantification of Unknown Nanoscale Biomolecules Using the Average-Weight-Difference Method. Applied Sciences. 2019; 9(1):130. https://doi.org/10.3390/app9010130
Chicago/Turabian StyleLee, Kyungwoo, Yonghyun Choi, Kyobum Kim, Hyung-Jun Koo, and Jonghoon Choi. 2019. "Quantification of Unknown Nanoscale Biomolecules Using the Average-Weight-Difference Method" Applied Sciences 9, no. 1: 130. https://doi.org/10.3390/app9010130
APA StyleLee, K., Choi, Y., Kim, K., Koo, H.-J., & Choi, J. (2019). Quantification of Unknown Nanoscale Biomolecules Using the Average-Weight-Difference Method. Applied Sciences, 9(1), 130. https://doi.org/10.3390/app9010130






