An Automatic Analysis System for High-Throughput Clostridium Difficile Toxin Activity Screening
Abstract
1. Introduction
1.1. Clinical Definition and Motivation
1.2. Related Works
2. Methods
2.1. Automated Classification System
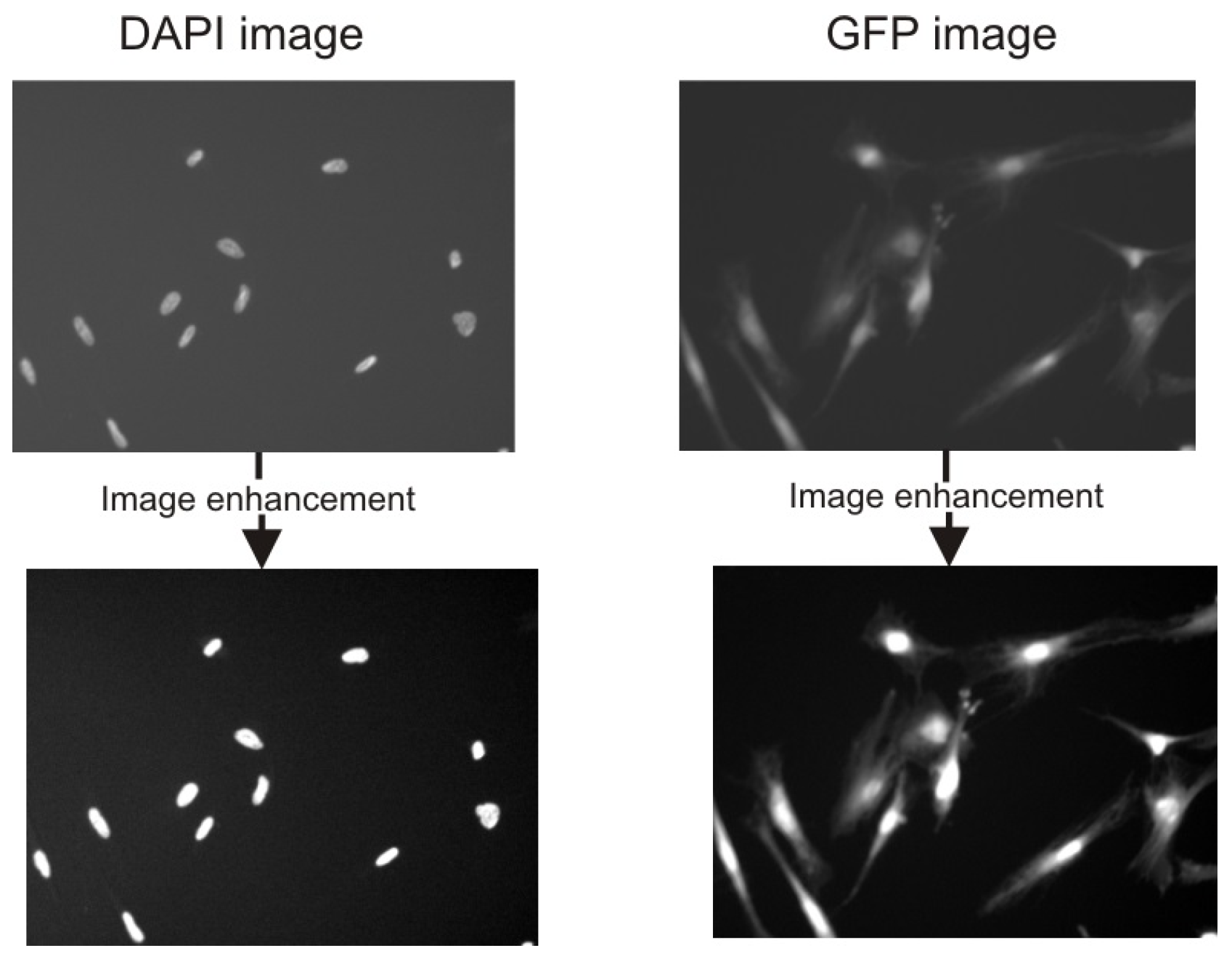
2.2. Image Preprocessing
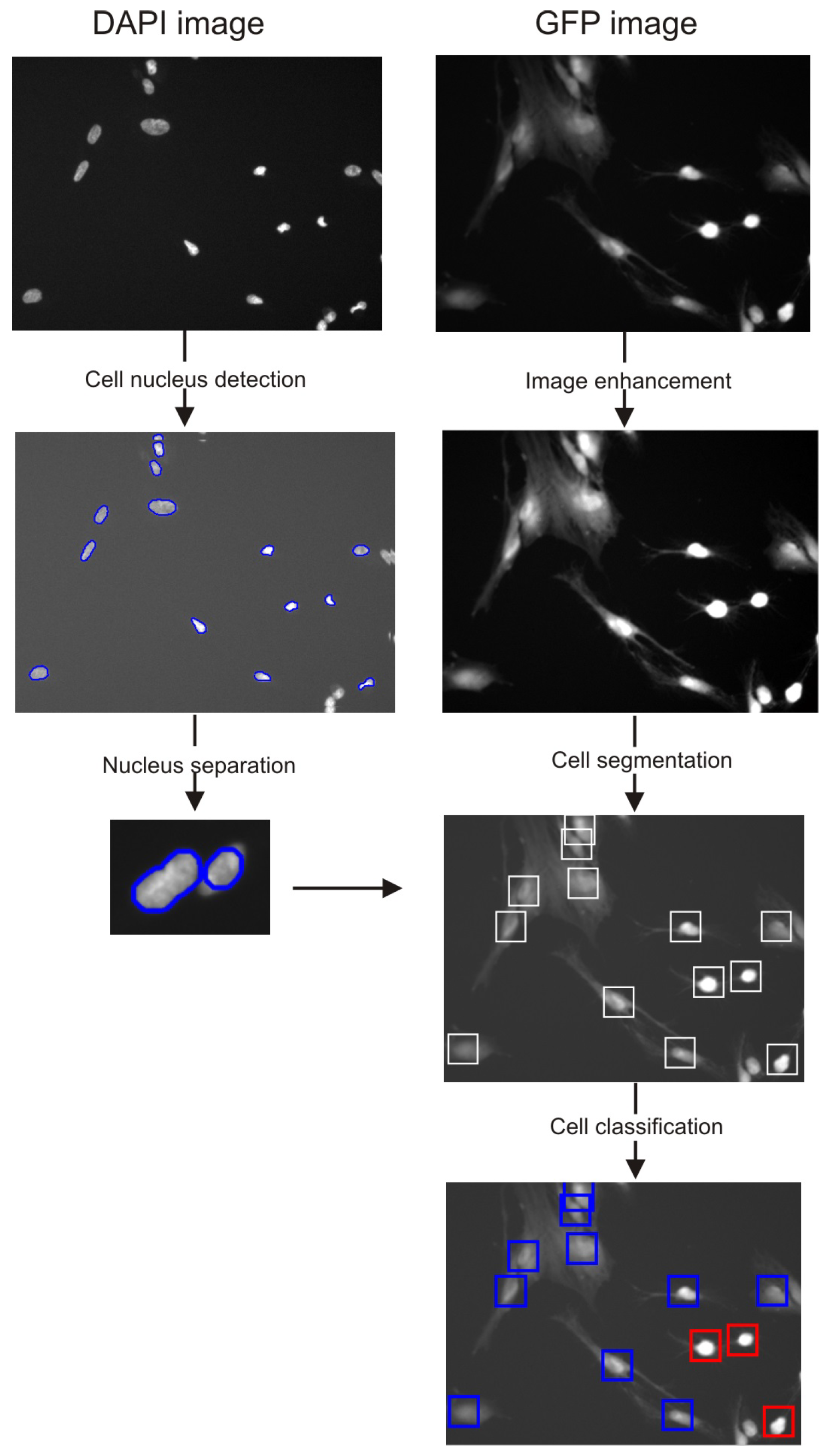
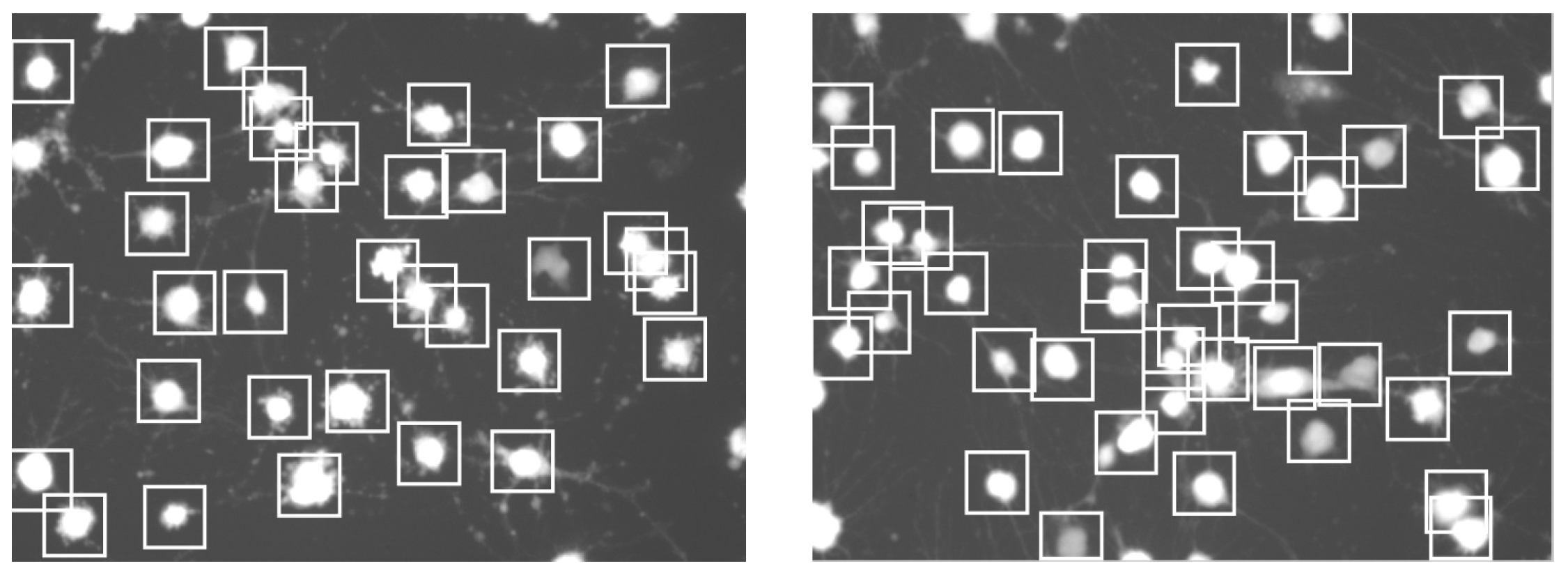
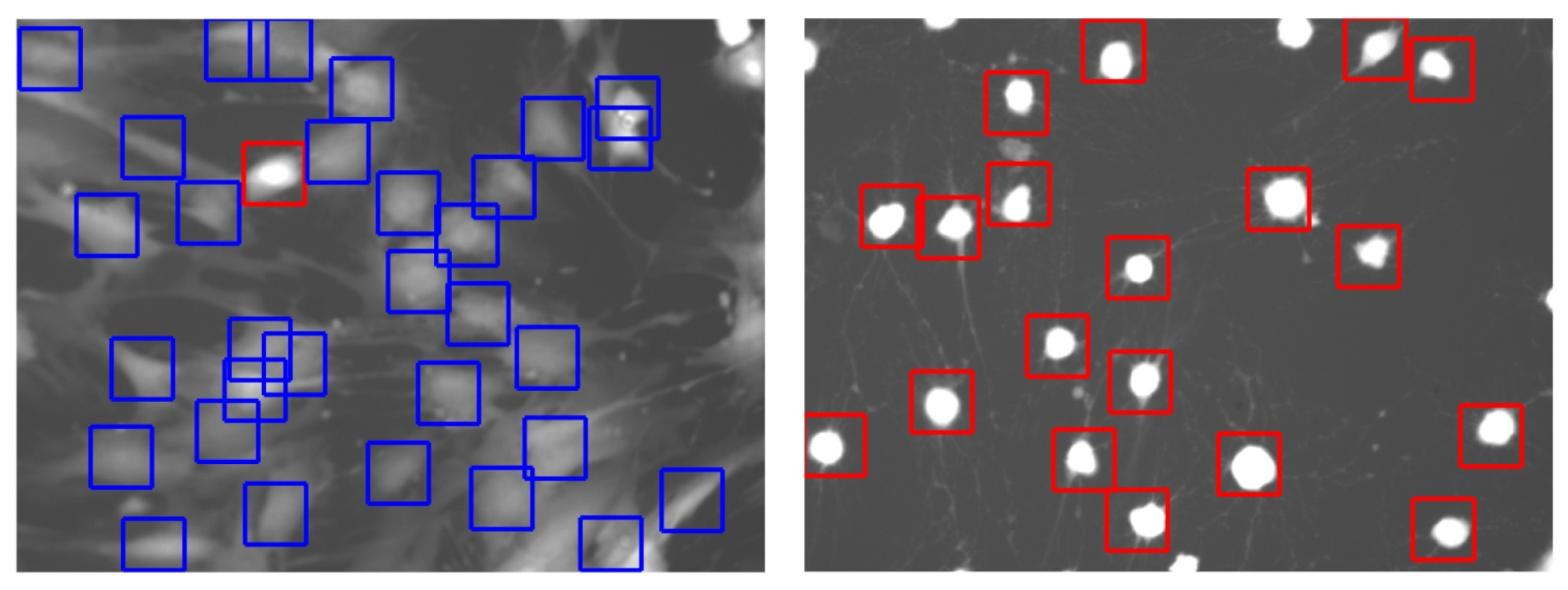
2.3. Cell Detection
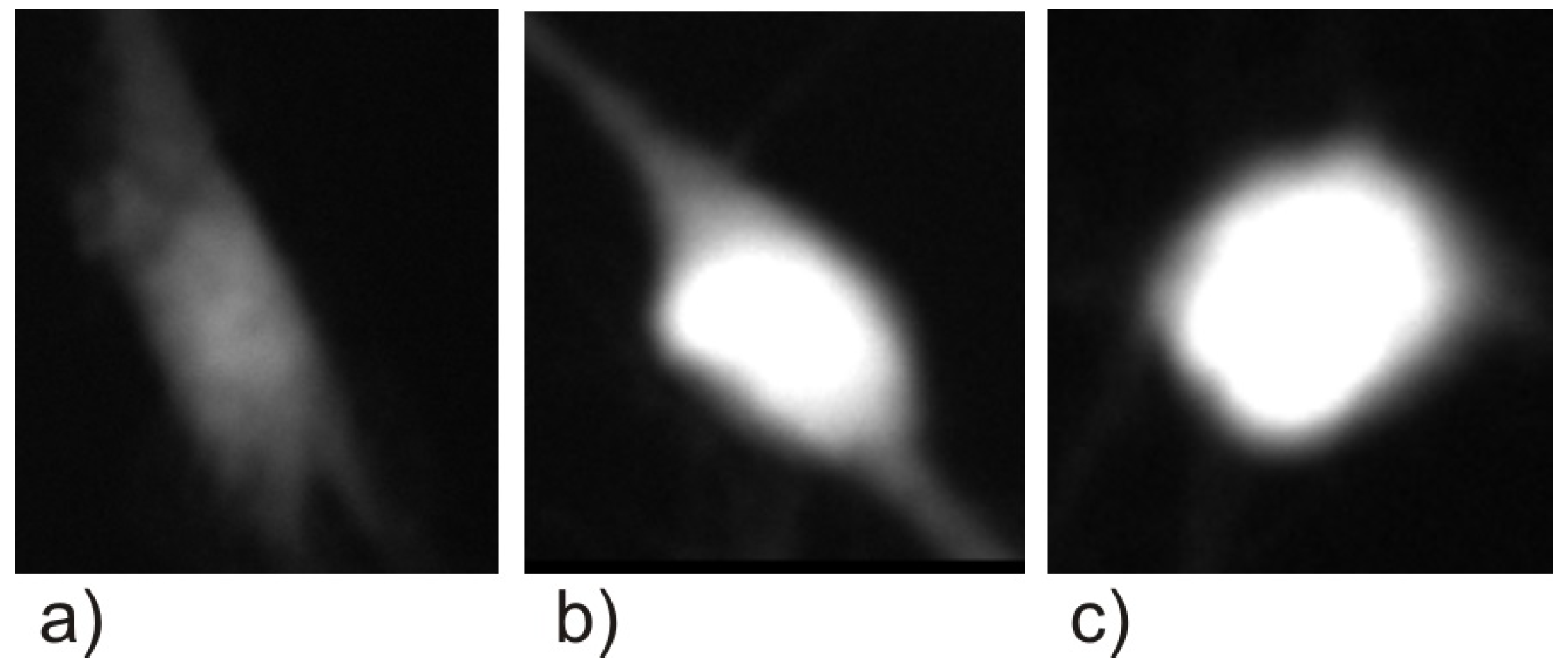
2.4. Cell Segmentation
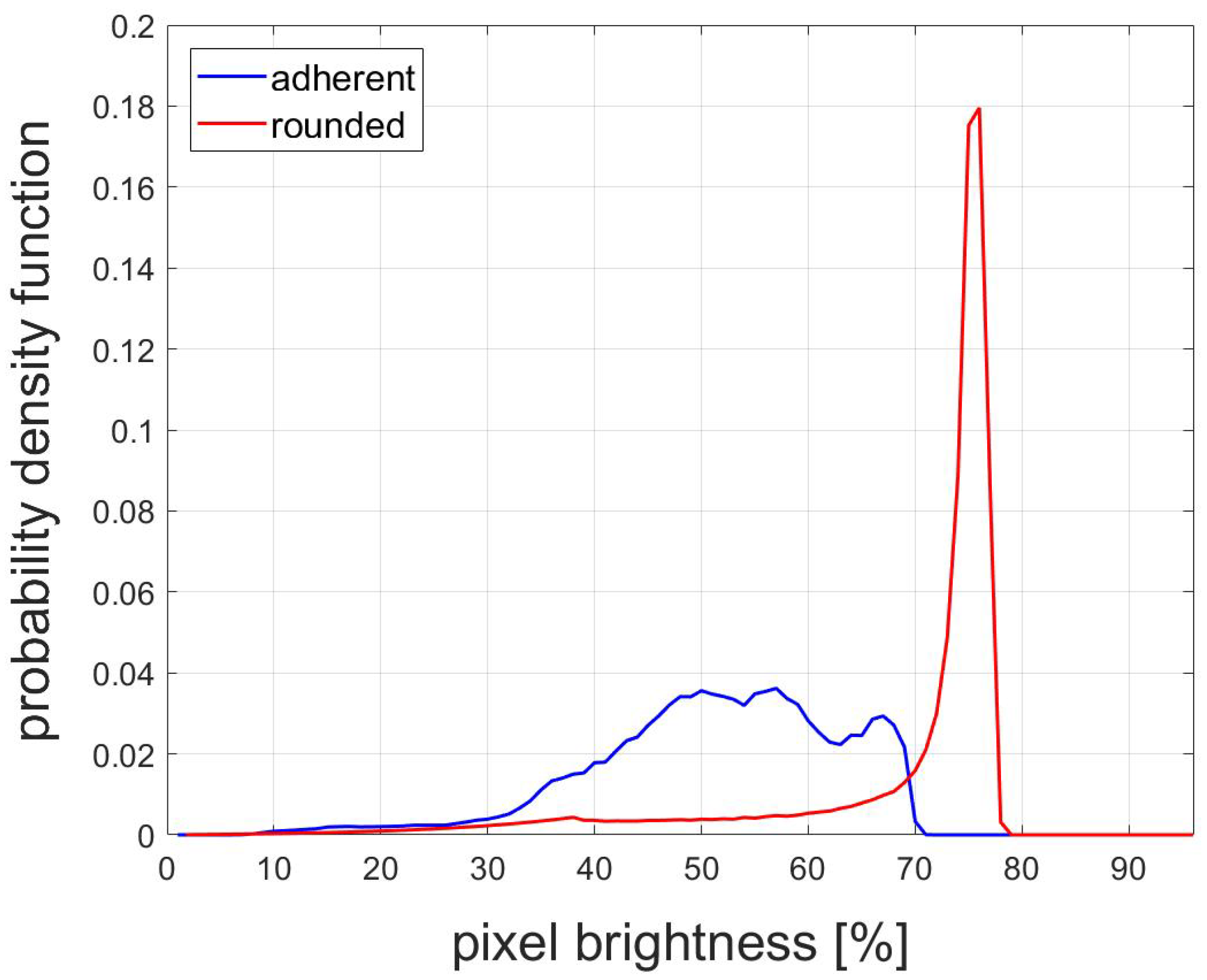
2.5. Classification
2.6. Cell-Rounding Assay
3. Results
3.1. Image Database
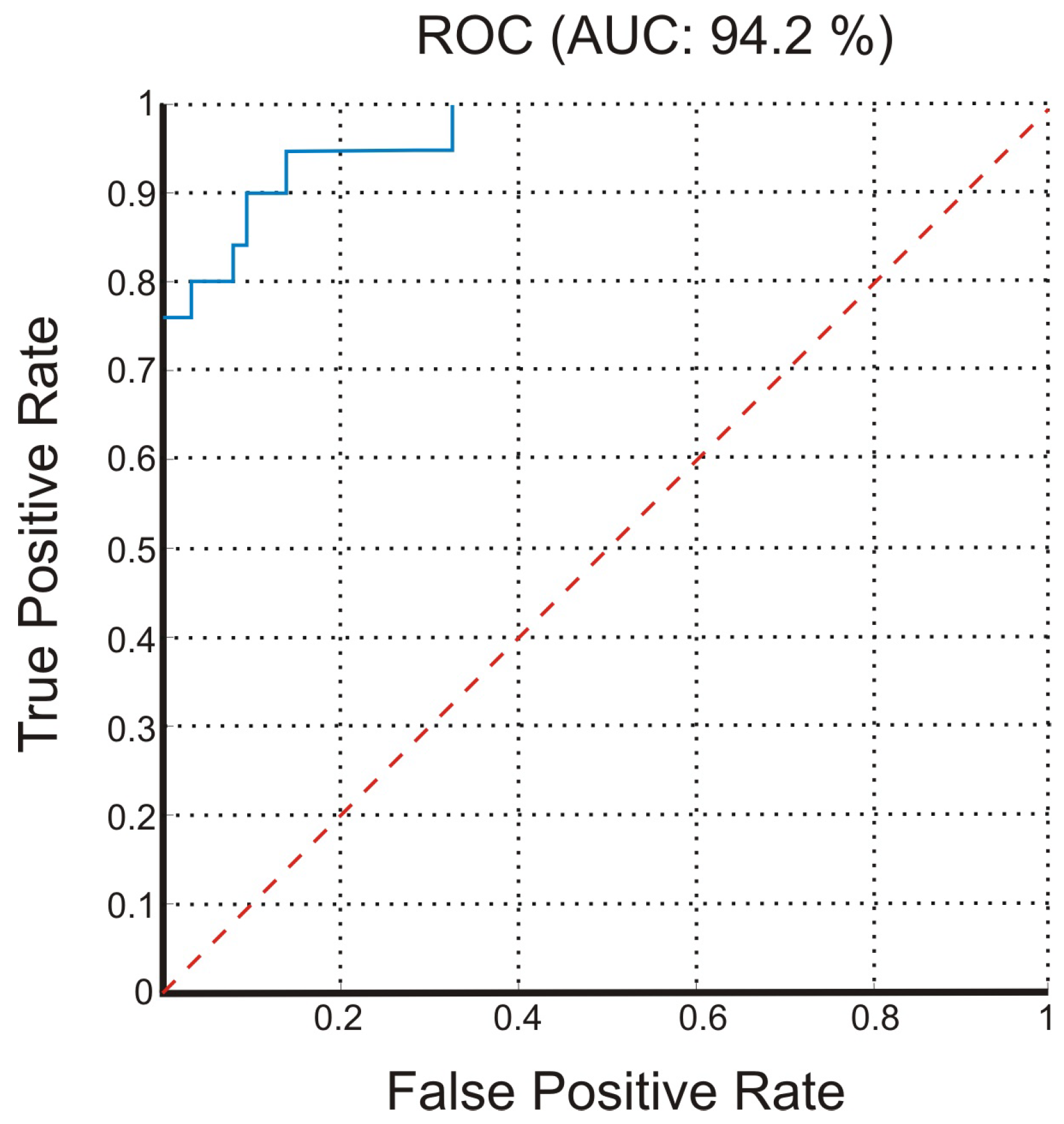
3.2. Classification Results
4. Discussion and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Carroll, K.C.; Bartlett, J.G. Biology of Clostridium difficile: Implications for epidemiology and diagnosis. Annu. Rev. Microbiol. 2011, 65, 501–521. [Google Scholar] [PubMed]
- Goudarzi, M.; Seyedjavadi, S.S.; Goudarzi, H.; Mehdizadeh Aghdam, E.; Nazeri, S. Clostridium difficile Infection: Epidemiology, Pathogenesis, Risk Factors, and Therapeutic Options. Scientifica 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Magill, S.S.; Edwards, J.R.; Bamberg, W.; Beldavs, Z.G.; Dumyati, G.; Kainer, M.A.; Lynfield, R.; Maloney, M.; McAllister-Hollod, L.; Nadle, J. Multistate point-prevalence survey of health care-associated infections. N. Engl. J. Med. 2014, 370, 1198–208. [Google Scholar] [CrossRef] [PubMed]
- Hughes, D.; Karlen, A. Discovery and preclinical development of new antibiotics. Ups. J. Med. Sci. 2014, 119, 162–169. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, S. Meeting the societal need for new antibiotics: The challenges for the pharmaceutical industry. Br. J. Clin. Pharmacol. 2015, 79, 168–172. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, R.N.; Lacy, D.B. Toward a structural understanding of Clostridium difficile toxins A and B. Front. Cell. Infect. Microbiol. 2012, 2, 28. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. QuickStats: Rates of Clostridium Difficile Infection among Hospitalized Patients Aged ≥65 Years,* by Age Group—National Hospital Discharge Survey, United States, 1996–2009. Available online: https://www.cdc.gov/mmwr/preview/mmwrhtml/mm6034a7.htm (accessed on 22 August 2018).
- Abt, M.C.; McKenney, P.T.; Pamer, E.G. Clostridium difficile colitis: pathogenesis and host defence. Nat. Rev. Microbiol. 2016, 14, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Khanna, S.; Pardi, D.S. Clostridium difficile Infection: New Insights Into Management. Mayo Clin. Proc. 2012, 87, 1106–1117. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.; Louie, T.J.; Gerding, D.N.; Cornely, O.A.; Chasan-Taber, S.; Fitts, D.; Gelone, S.P.; Broom, C.; Davidson, D.M. For the Polymer Alternative for CDI Treatment (PACT) Investigators. Vancomycin, metronidazole, or tolevamer for Clostridium difficile infection: Results from two multinational, randomized, controlled trials. Clin. Infect. Dis. 2014, 59, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.P. Can we identify patients at high risk of recurrent Clostridium difficile infection? Clin. Microbiol. Infect. 2012, 18, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Garland, M.; Loscher, S.; Bogyo, M. Chemical Strategies To Target Bacterial Virulence. Chem. Rev. 2017, 117, 4422–4461. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, A.; Mooyottu, S.; Yin, H.; Surendran Nair, M.; Bhattaram, V.; Venkitanarayanan, K. Inhibiting Microbial Toxins Using Plant-Derived Compounds and Plant Extracts. Medicines 2015, 2, 186–211. [Google Scholar] [CrossRef] [PubMed]
- Kyne, L.; Warny, M.; Qamar, A.; Kelly, C.P. Asymptomatic Carriage of Clostridium difficile and Serum Levels of IgG Antibody against Toxin A. N. Engl. J. Med. 2000, 342, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.K.; Johnson, S.; Samore, M.H.; Bliss, D.Z.; Gerding, D.N. Primary symptomless colonisation by Clostridium difficile and decreased risk of subsequent diarrhoea. Lancet 1998, 351, 633–636. [Google Scholar] [CrossRef]
- Shen, A. Clostridium difficile toxins: Mediators of inflammation. J. Innate Immun. 2012, 4, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Bender, K.O.; Garland, M.; Ferreyra, J.A.; Hryckowian, A.J.; Child, M.A.; Puri, A.W.; Solow-Cordero, D.E.; Higginbottom, S.K.; Segal, E.; Banaei, N.; et al. A small-molecule antivirulence agent for treating Clostridium difficile infection. Sci. Transl. Med. 2015, 7. [Google Scholar] [CrossRef] [PubMed]
- Lacy, D.B. Pre-Clinical Evaluation of Clostridium Difficile Toxin Inhibitors. Available online: http://grantome.com/grant/NIH/I01-BX002943-01 (accessed on 22 August 2018).
- Larabee, J.L.; Bland, S.J.; Hunt, J.J.; Ballard, J.D. Intrinsic toxin-derived peptides destabilize and inactivate Clostridium difficile TcdB. MBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Letourneau, J.J.; Stroke, I.L.; Hilbert, D.W.; Sturzenbecker, L.J.; Marinelli, B.A.; Quintero, J.G.; Sabalski, J.; Ma, L.; Diller, D.J.; Stein, P.D.; et al. Identification and initial optimization of inhibitors of Clostridium difficile (C. difficile) toxin B (TcdB). Bioorg. Med. Chem. Lett. 2018, 28, 756–761. [Google Scholar] [CrossRef] [PubMed]
- Tam, J.; Beilhartz, G.L.; Auger, A.; Gupta, P.; Therien, A.G.; Melnyk, R.A. Small Molecule Inhibitors of Clostridium difficile Toxin B-Induced Cellular Damage. Chem. Biol. 2015, 22, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Dordea, A.C.; Bray, M.-A.; Allen, K.; Logan, D.J.; Fei, F.; Malhotra, R.; Gregory, M.S.; Carpenter, A.E.; Buys, E.S. An open-source computational tool to automatically quantify immunolabeled retinal ganglion cells. Exp. Eye Res. 2016, 147, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Rueden, C.T.; Hiner, M.C.; Eliceiri, K.W. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 2015, 82, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Chaumont, F. Manual Counting: Documentation. Available online: www.icy.bioimageanalysis.org/plugin/Manual_Counting. (accessed on 22 August 2018).
- Helmy, I.M.; Abdel Azim, A.M. Efficacy of ImageJ in the assessment of apoptosis. Diagn. Pathol. 2012, 7, 15. [Google Scholar] [CrossRef] [PubMed]
- Brocher, J.; Wagner, T. BioVoxxel Toolbox (ImageJ/Fiji). Available online: www.biovoxxel.de (accessed on 22 August 2018).
- Feng, Y.; Zhao, H.; Li, X.; Zhang, X.; Li, H. A multi-scale 3D Otsu thresholding algorithm for medical image segmentation. Digit. Signal Process. 2017, 60, 186–199. [Google Scholar] [CrossRef]
- Meyer, F. Topographic distance and watershed lines. Signal Process. 1994, 38, 113–125. [Google Scholar] [CrossRef]
- Zanaty, E.A.; Afifi, A. A watershed approach for improving medical image segmentation. Comput. Meth. Biomech. Biomed. Eng. 2013, 16, 1262–1272. [Google Scholar] [CrossRef] [PubMed]
- Hastie, T.; Tibshirani, R.; Friedman, J.H. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd ed.; Springer: Berlin, Germany, 2009. [Google Scholar]
- Japkowicz, N.; Shah, M. Evaluating Learning Algorithms: A Classification Perspective; Cambridge University Press: Cambridge, MA, USA, 2011. [Google Scholar]
- Bishop, C.M. Pattern Recognition and Machine Learning; Springer: Berlin, Germany, 2006. [Google Scholar]
- Devroye, L.; Gyorfi, L.; Lugosi, G. A Probabilistic Theory of Pattern Recognition; Springer: Berlin, Germany, 1996. [Google Scholar]
- Duda, R.O.; Hart, P.E.; Stork, D.G. Pattern Classification, 2nd ed.; Wiley: New York, NY, USA, 2000. [Google Scholar]
- Mitchell, T. Machine Learning; McGraw Hill: New York, NY, USA, 1997. [Google Scholar]
- Webb, A.R.; Copsey, K.D. Statistical Pattern Recognition, 3rd ed.; Wiley: New York, NY, USA, 2011. [Google Scholar]
- Al-Aidaroos, K.M.; Bakar, A.A.; Othman, Z. Medical data classification with Naïve Bayes approach. Inform. Technol. J. 2012, 11, 1166–1174. [Google Scholar] [CrossRef]
- John, G.H.; Langley, P. Estimating continuous distributions in bayesian classifiers. In Proceedings of the 11th Conference on Uncertainty in Artificial Intelligence, Montreal, QC, Canada, 18–20 August 1995. [Google Scholar]
- Khan, A.U.R.; Khan, M.; Khan, M.B. Naïve Multi-label Classification of YouTube Comments Using Comparative Opinion Mining. Procedia Comput. Sci. 2016, 82, 57–64. [Google Scholar] [CrossRef]
- Ontivero-Ortega, M.; Lage-Castellanos, A.; Valente, G.; Goebel, R.; Valdes-Sosa, M. Fast Gaussian Naïve Bayes for searchlight classification analysis. Neuroimage 2017, 163, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Sheikhian, H.; Delavar, M.R.; Stein, A. Predictive Modelling of Seismic Hazard Applying Naïve Bayes and Granular Computing Classifiers. Procedia Environ. Sci. 2015, 26, 49–52. [Google Scholar] [CrossRef]
- Vangelis, M.; Ion, A.; Geogios, P. Spam Filtering with Naïve Bayes—Which Naïve Bayes? In Proceedings of the 3rd Conference on Email and Anti-Spam, Mountain View, CA, USA, 27–28 July 2006.
- Hajian-Tilaki, K. Sample size estimation in diagnostic test studies of biomedical informatics. J. Biomed. Inform. 2014, 48, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, T. An Introduction to ROC Analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Feng, D.; Cortese, G.; Baumgartner, R. A comparison of confidence/credible interval methods for the area under the ROC curve for continuous diagnostic tests with small sample size. Stat. Methods Med. Res. 2015, 26, 2603–2621. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Chung, B.D.; Lee, J.S. Incorporating receiver operating characteristics into naive Bayes for unbalanced data classification. Computing 2017, 99, 203–218. [Google Scholar] [CrossRef]
- Airola, A.; Pahikkala, T.; Waegeman, W.; De Baets, B.; Salakoski, T. An experimental comparison of cross-validation techniques for estimating the area under the ROC curve. Comput. Stat. Data Anal. 2011, 55, 1828–1844. [Google Scholar] [CrossRef]
- Bamber, D. The area above the ordinal dominance graph and the area below the receiver operating characteristic graph. J. Math. Psychol. 1975, 12, 387–415. [Google Scholar] [CrossRef]
- Cortes, C.; Mohri, M. AUC optimization vs. error rate minimization. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 2004. [Google Scholar]




| Software | Classification Algorithm | SN (%) | SP (%) | ACC (%) |
|---|---|---|---|---|
| ImageJ | Circularity | 67 | 71 | 69 |
| BioVoxxel Toolbox | Shape analysis | 82 | 84 | 82 |
| Described algorithm | Preprocessing, image enhancement, shape analysis, pixel brightness | 93 | 91 | 92.6 |
| Adherent (Predicted) | Rounded (Predicted) | Total | |
|---|---|---|---|
| Adherent (Actual) | 2552 () | 213 () | 2765 |
| Rounded (Actual) | 164 () | 2167 () | 2331 |
| Sensitivity: 93% | Specificity: 91% | Total: 5096 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garland, M.; Jaworek-Korjakowska, J.; Libal, U.; Bogyo, M.; Sieńczyk, M. An Automatic Analysis System for High-Throughput Clostridium Difficile Toxin Activity Screening. Appl. Sci. 2018, 8, 1512. https://doi.org/10.3390/app8091512
Garland M, Jaworek-Korjakowska J, Libal U, Bogyo M, Sieńczyk M. An Automatic Analysis System for High-Throughput Clostridium Difficile Toxin Activity Screening. Applied Sciences. 2018; 8(9):1512. https://doi.org/10.3390/app8091512
Chicago/Turabian StyleGarland, Megan, Joanna Jaworek-Korjakowska, Urszula Libal, Matthew Bogyo, and Marcin Sieńczyk. 2018. "An Automatic Analysis System for High-Throughput Clostridium Difficile Toxin Activity Screening" Applied Sciences 8, no. 9: 1512. https://doi.org/10.3390/app8091512
APA StyleGarland, M., Jaworek-Korjakowska, J., Libal, U., Bogyo, M., & Sieńczyk, M. (2018). An Automatic Analysis System for High-Throughput Clostridium Difficile Toxin Activity Screening. Applied Sciences, 8(9), 1512. https://doi.org/10.3390/app8091512







