Effects of Heavy Metals on Phyllosphere and Rhizosphere Microbial Community of Bothriochloa ischaemum
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Plant and Soil Sampling
2.3. Plant and Soil Chemical Properties
2.4. Determination of Phyllospheric Bacterial Community Structure
2.5. DNA Extraction, Polymerase Chain Reaction, and Denaturing Gradient Gel Electrophoresis
2.6. Denaturing Gel Gradient Electrophoresis Analysis
2.7. Statistical Analysis
3. Results
3.1. Physiological Characteristics and Heavy Metal Distribution of B. ischaemum
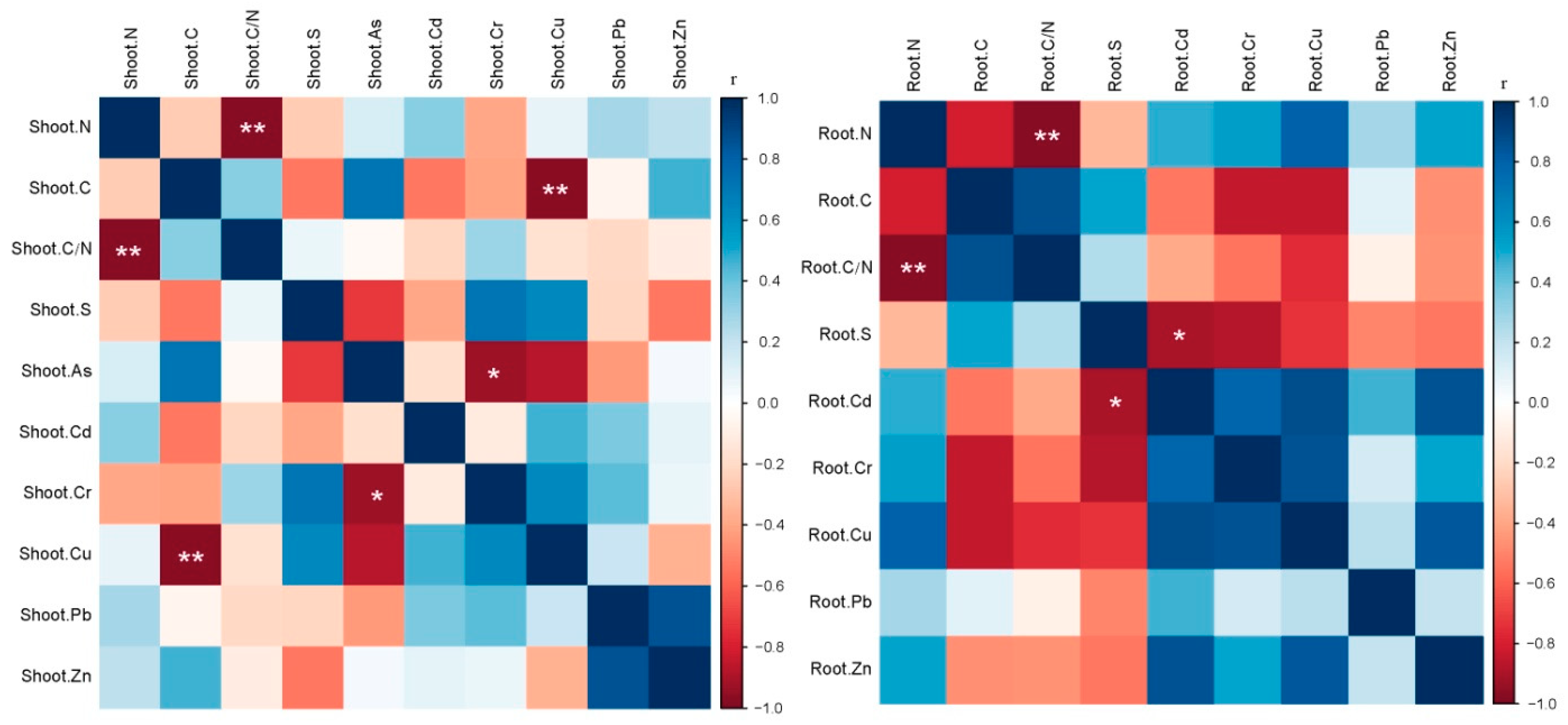
3.2. Correlations among B. ischaemum Soil Physicochemical Characteristics and Heavy Metals
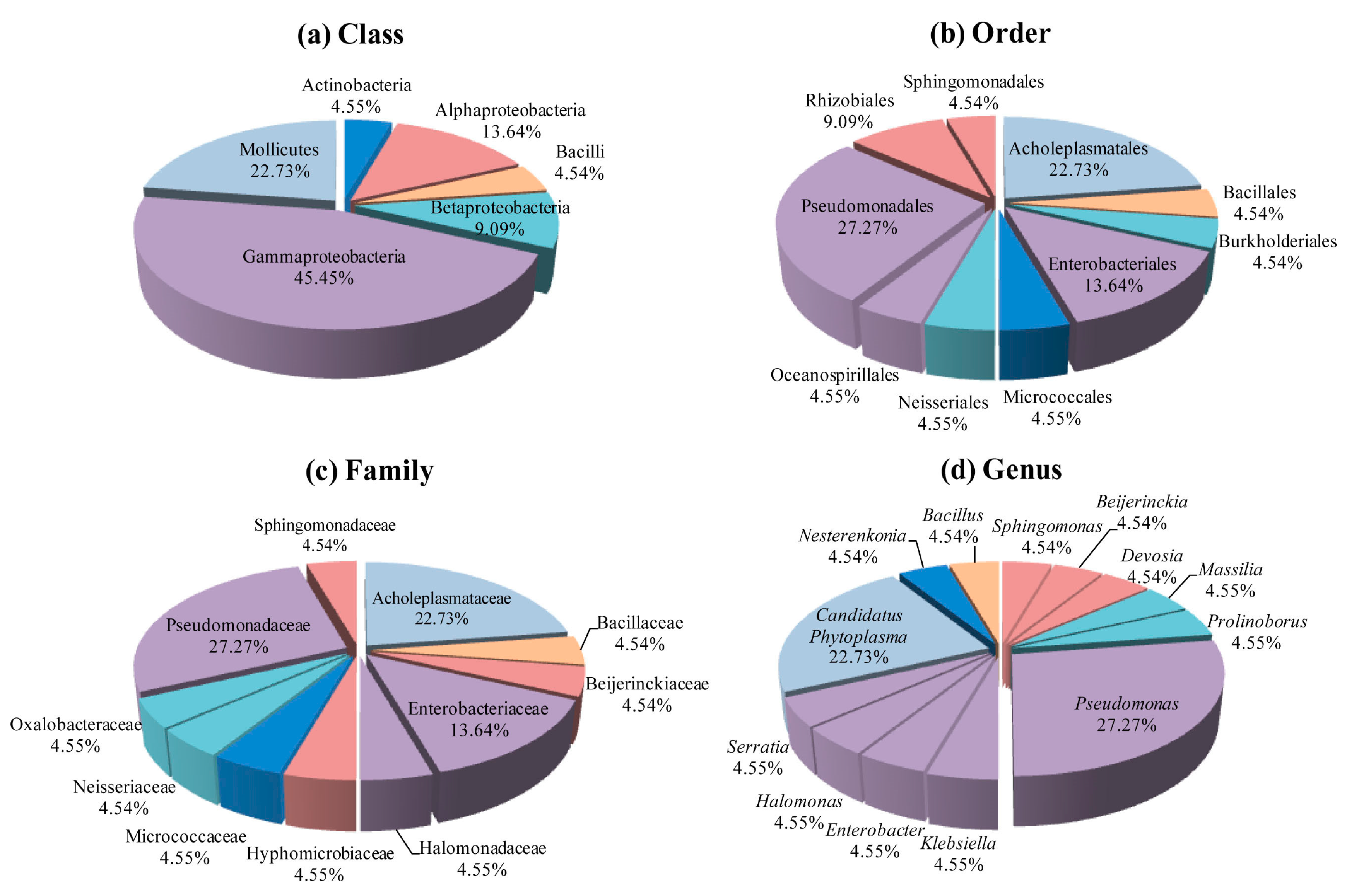
3.3. B. ischaemum Phyllosphere Bacteria Community Structure and Diversity
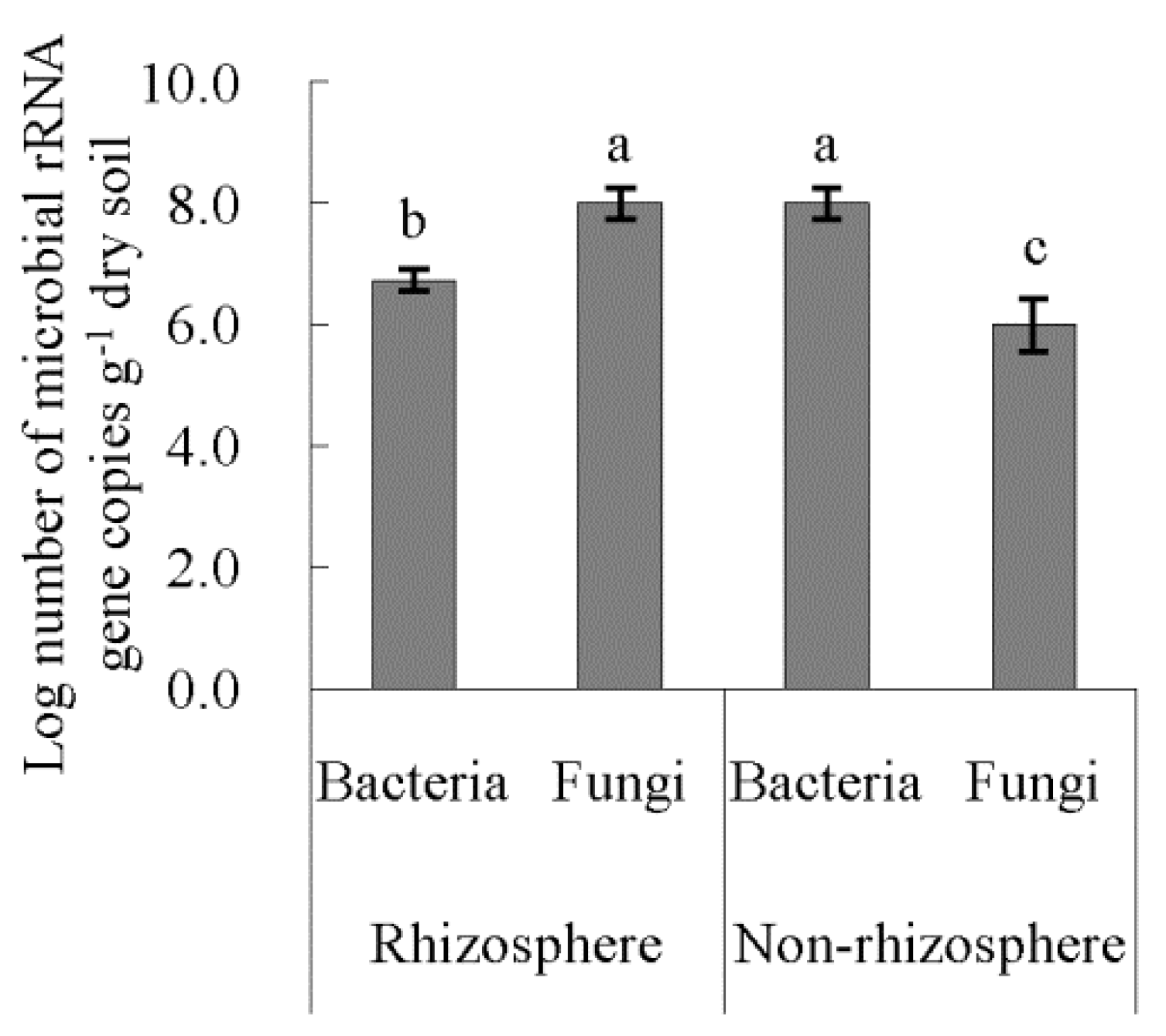
3.4. Bacterial Diversities of Rhizosphere and Non-Rhizosphere Soil and Their Driving Factors
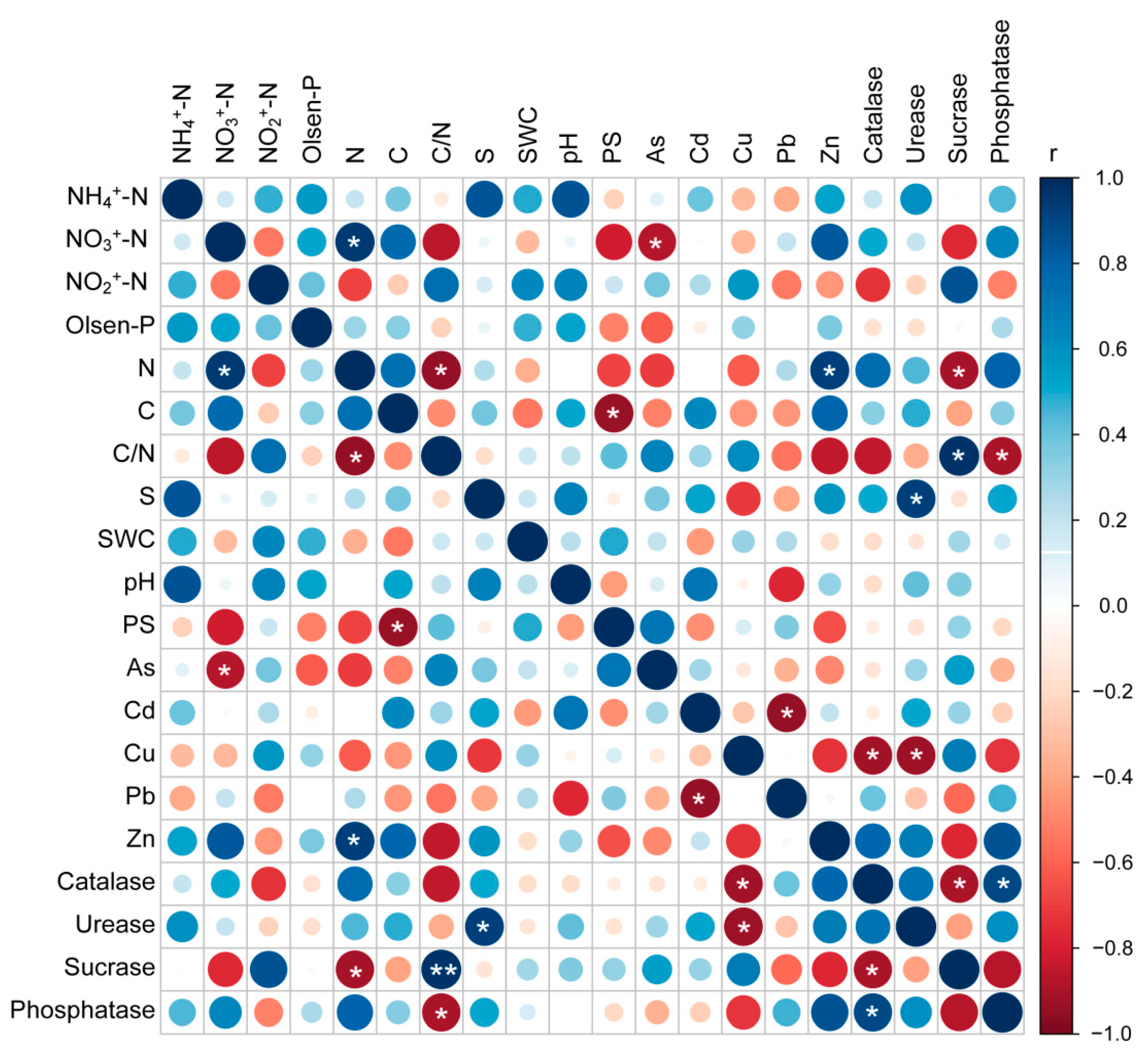
3.5. Relationship between Soil Enzyme Activities and Diversity Indices of Soil Microbial Community
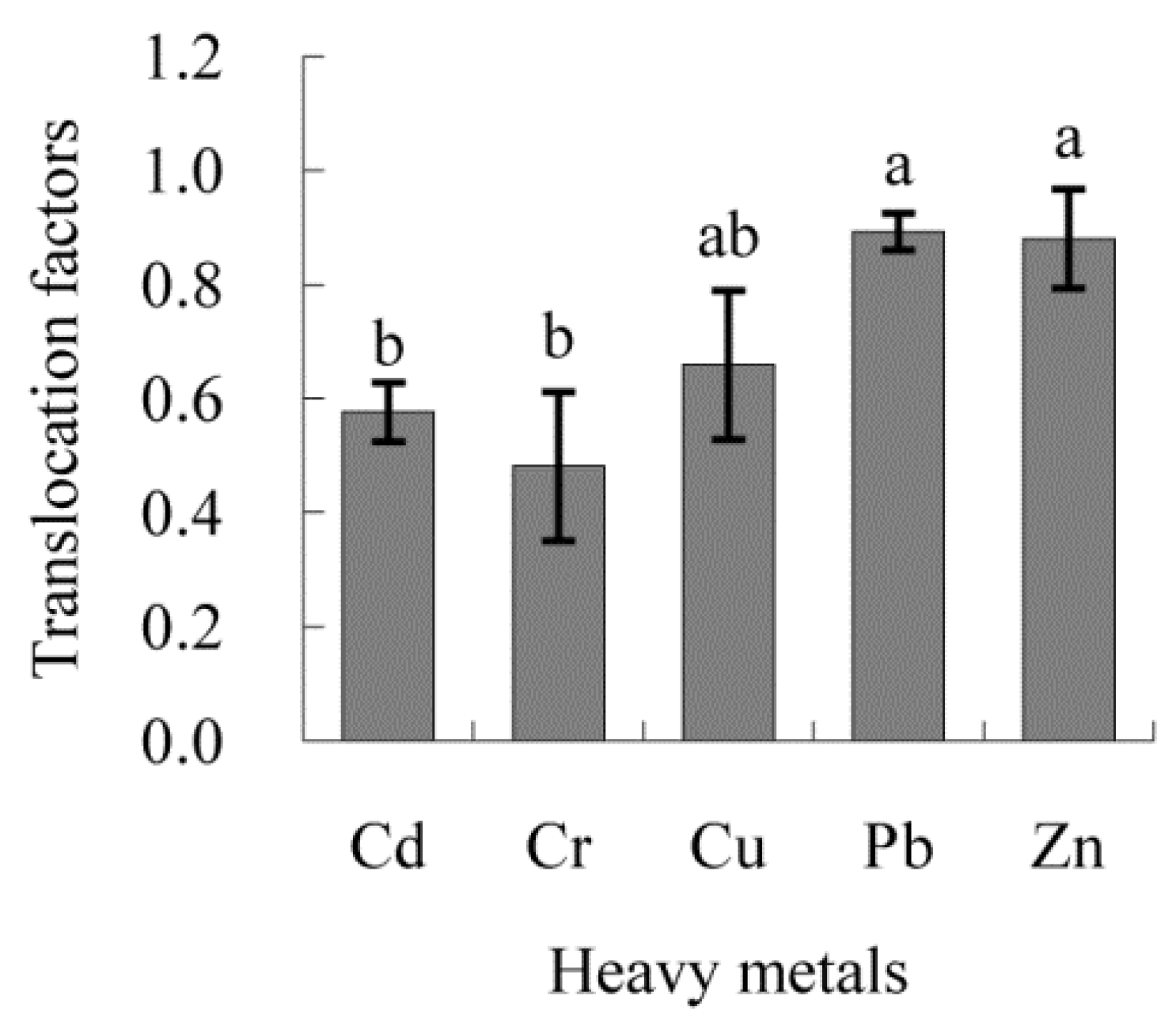
3.6. Transfer Factors of Heavy Metals in B. ischaemum and Their Driving Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wang, S.L.; Liao, W.B.; Yu, F.Q.; Liao, B.; Shu, W.S. Hyperaccumulation of lead, zinc, and cadmium in plants growing on a lead/zinc outcrop in Yunnan Province, China. Environ. Geol. 2009, 58, 471–476. [Google Scholar] [CrossRef]
- Conesa, H.M.; Faz, A.; Arnaldos, R. Initial studies for the phytostabilization of a mine tailing from the Cartagena-La Union Mining District (SE Spain). Chemosphere 2007, 66, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.H. Ecological restoration of mine degraded soils, with emphasis on metal contaminated soils. Chemosphere 2003, 50, 775–780. [Google Scholar] [CrossRef]
- Jia, T.; Cao, M.W.; Jing, J.H.; Liu, J.X.; Chai, B.F. Endophytic fungi and soil microbial community characteristics over different years of phytoremediation in a copper tailings dam of Shanxi, China. Sci. Total Environ. 2017, 574, 881–888. [Google Scholar]
- Deng, H.; Ye, Z.H.; Wong, M.H. Accumulation of lead, zinc, copper and cadmium by 12 wetland plant species thriving in metal-contaminated sites in China. Environ. Pollut. 2004, 132, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.X.; Li, C.; Jing, J.H.; Jia, T.; Liu, X.G.; Wang, X.Y.; Chai, B.F. Composition and environmental adaptation of microbial community in Shibahe copper tailing in Zhongtiao mountain in Shanxi. Environ. Sci. 2017, 38, 318–326. [Google Scholar]
- He, Z.G.; Xie, X.H.; Xiao, S.M.; Liu, J.S.; Qiu, G.Z. Microbial diversity of mine water at Zhong Tiaoshan copper mine, China. J. Basic Microbiol. 2010, 47, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, A.D.; Humphreys, M.O.; Johnson, M.S. The value of heavy metal tolerance in the revegetation of metalliferous mine wastes. Environ. Manag. Mineral. Wastes 1978, 311–314. [Google Scholar] [CrossRef]
- Marques, A.P.G.C.; Rangel, A.O.S.S.; Castro, P.M.L. Remediation of heavy metal contaminated soils, phytoremediation as a potentially promising clean-up technology. Crit. Rev. Environ. Sci. Technol. 2009, 39, 622–654. [Google Scholar] [CrossRef]
- González, R.C.; González-Chávez, M.C.A. Metal accumulation in wild plants surrounding mining wastes. Environ. Pollut. 2006, 144, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Bech, J.; Poschenrieder, C.; Barceló, J.; Lansac, A. Plants from mine spoils in the south american area as potential sources of germplasm for phytoremediation technologies. Eng. Life Sci. 2002, 22, 5–11. [Google Scholar] [CrossRef]
- Mendez, M.O.; Glenn, E.P.; Maier, R.M. Phytostabilization potential of quailbush for mine tailings. J. Environ. Qual. 2007, 36, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Conesa, H.M.; Faz, A.; Arnaldos, R. Heavy metal accumulation and tolerance in plants from mine tailings of the semiarid Cartagena-La Union mining district (SE Spain). Sci. Total Environ. 2006, 366, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.J.; Li, J.H.; Jin, D.S.; Lu, J.J.; Gao, C.H.; Zhang, M. Discussion on microbial remediation technology of heavy metal contaminated soil. J. Shanxi Agric. Sci. 2018, 46, 150–154. [Google Scholar]
- Pan, X.; Achal, V.; Zhao, C.X.; Yang, J.Y.; Kumari, D. Microbial remediation of heavy metals and arsenic-contaminated environments in the arid zone of northwest China. In Twenty Years of Research and Development on Soil Pollution and Remediation China; Springer: Singapore, 2018; pp. 477–486. [Google Scholar]
- Ojuederie, O.B.; Babalola, O.O. Microbial and plant-assisted bioremediation of heavy metal polluted environments, a review. Int. J. Environ. Res. Public Health 2017, 14, 1504. [Google Scholar] [CrossRef] [PubMed]
- Lazaridou, E.; Giannopoulou, C.; Fotiadou, C.; Vakirlis, E.; Trigoni, A.; Ioannides, D. The potential role of microorganisms in the development of rosacea. J. Deutsch. Dermatol. Ges. 2011, 9, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Kaschuk, G.; Alberton, O.; Hungria, M. Three decades of soil microbial biomass studies in Brazilian ecosystems, lessons learned about soil quality and indications for improving sustainability. Soil. Biol. Biochem. 2010, 42, 1–13. [Google Scholar] [CrossRef]
- Finkenbein, P.; Kretschmer, K.; Kuka, K.; Klotz, S.; Heilmeier, H. Soil enzyme activities as bioindicators for substrate quality in revegetation of a subtropical coal mining dump. Soil. Biol. Biochem. 2013, 56, 87–89. [Google Scholar] [CrossRef]
- Dangi, S.R.; Stahl, P.D.; Wick, A.F.; Ingram, L.J.; Buyer, J.S. Soil microbial community recovery in reclaimed soils on a surface coal mine site. Soil Sci. Soc. Am. J. 2012, 76, 915–924. [Google Scholar] [CrossRef]
- Claassens, S.; Rensburg, P.J.J.V.; Maboeta, M.S.; Rensburg, L.V. Soil Microbial community function and structure in a post-mining chronosequence. Water Air Soil Pollut. 2008, 194, 315–329. [Google Scholar] [CrossRef]
- Li, J.J.; Zhou, X.M.; Yan, J.X.; Li, H.J.; He, J.Z. Effects of regenerating vegetation on soil enzyme activity and microbial structure in reclaimed soils on a surface coal mine site. Appl. Soil Ecol. 2015, 87, 56–62. [Google Scholar] [CrossRef]
- Cheng, F.; Peng, X.; Zhao, P.; Yuan, J.; Zhong, C.; Cheng, Y.; Cui, C.; Zhang, S. Soil microbial biomass, basal respiration and enzyme activity of main forest types in the Qinling Mountains. PLoS ONE 2013, 8, e67353. [Google Scholar] [CrossRef] [PubMed]
- Bakker, P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar]
- Raaijmakers, J.M.; Paulitz, T.C.; Steinberg, C.; Alabouvette, C.; Moënneloccoz, Y. The rhizosphere, a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 2009, 321, 341–361. [Google Scholar] [CrossRef]
- Klemedtsson, L.; Svensson, B.H.; Rosswall, T. Relationships between soil moisture content and nitrous oxide production during nitrification and denitrification. Biol. Fertil. Soils 1988, 6, 106–111. [Google Scholar] [CrossRef]
- Zhao, Y.; Wu, L.; Chu, L.; Yang, Y.; Li, Z.; Azeem, S.; Zhang, Z.; Fang, C.; Lin, W. Interaction of Pseudostellaria heterophylla with Fusarium oxysporum f.sp. heterophylla mediated by its root exudates in a consecutive monoculture system. Sci. Rep. 2015, 5, 8197. [Google Scholar] [CrossRef] [PubMed]
- Elbl, J.; Záhora, J. The comparison of microbial activity in rhizosphere and non-rhizosphere soil stressed by drought. In Proceedings of the Mendel Net 2014, Brno, Czech Republic, 19–20 November 2014; pp. 234–240. [Google Scholar]
- Whipps, J.M.; Hand, P.; Pink, D.; Bending, G.D. Phyllosphere microbiology with special reference to diversity and plant genotype. J. Appl. Microbiol. 2008, 105, 1744–1755. [Google Scholar] [CrossRef] [PubMed]
- Chinnadurai, C.; Balachandar, D.; Sundaram, S.P. Characterization of 1-aminocyclopropane-1-carboxylate deaminase producing methylobacteria from phyllosphere of rice and their role in ethylene regulation. World J. Microbiol. Biotechnol. 2009, 25, 1403–1411. [Google Scholar] [CrossRef]
- Janarthine, S.R.S.; Eganathan, P. Plant growth promoting of endophytic sporosarcina aquimarina SjAM16103 isolated from the pneumatophores of Avicennia marina L. Int. J. Microbiol. 2012, 2012, 532060. [Google Scholar] [CrossRef] [PubMed]
- Balintkurti, P.; Simmons, S.J.; Blum, J.E.; Ballaré, C.L.; Stapleton, A.E. Maize leaf epiphytic bacteria diversity patterns are genetically correlated with resistance to fungal pathogen infection. Mol. Plant-Microbe Interact. 2010, 23, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Costa, D.M.D.; Samarasinghe, S.S.T.; Dias, H.R.D.; Dissanayake, D.M.N. Control of rice sheath blight by phyllosphere epiphytic microbial antagonists. Phytoparasitica 2008, 36, 52–65. [Google Scholar] [CrossRef]
- Liu, J.; Li, C.; Jing, J.; Zhao, P.; Luo, Z.; Cao, M.; Ma, Z.; Jia, T.; Chai, B. Ecological patterns and adaptability of bacterial communities in alkaline copper mine drainage. Water Res. 2018, 133, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Qiao, S.; Zhou, Y.; Liu, J.; Jing, J.; Jia, T.; Li, C.; Yang, X.; Chao, B. Characteristics of soil bacterial community structure in coniferous forests of Guandi mountains, Shanxi province. Sci. Silvae Sin. 2017, 53, 89–99. [Google Scholar]
- Li, J.; Liu, F.; Zhou, X. Effects of different reclaimed scenarios on soil microbe and enzyme activities in mining areas. Environ. Sci. 2015, 36, 1836–1841. [Google Scholar]
- Jorquera, M.A.; Maruyama, F.; Ogram, A.V.; Navarrete, O.U.; Lagos, L.M.; Inostroza, N.G.; Acuna, J.J.; Rilling, J.I.; Mora, M.l.l.M. Rhizobacterial community structures associated with native plants grown in Chilean extreme environments. Microb. Ecol. 2016, 72, 633–646. [Google Scholar] [CrossRef] [PubMed]
- Bassam, B.J.; Caetanoanollés, G.; Gresshoff, P.M. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal. Biochem. 1991, 196, 80–83. [Google Scholar] [CrossRef]
- Yang, Y.; Song, Y.; Scheller, H.V.; Ghosh, A.; Ban, Y.; Chen, H.; Tang, M. Community structure of arbuscular mycorrhizal fungi associated with Robinia pseudoacacia in uncontaminated and heavy metal contaminated soils. Soil Biol. Biochem. 2015, 86, 146–158. [Google Scholar] [CrossRef]
- Niklas, K.J.; Cobb, E.D. N, P, and C stoichiometry of Eranthis hyemalis (Ranunculaceae) and the allometry of plant growth. Am. J. Bot. 2005, 92, 1256–1263. [Google Scholar] [CrossRef] [PubMed]
- Nies, D.H.; Silver, S. Plasmid-determined inducible efflux is responsible for resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus. J. Bacteriol. 1989, 171, 896–900. [Google Scholar] [CrossRef] [PubMed]
- Nishizono, H.; Ichikawa, H.; Suziki, S.; Suzuki, S.; Ishii, F. The role of the root cell wall in the heavy metal tolerance of Athyrium yokoscense. Plant Soil 1987, 101, 15–20. [Google Scholar] [CrossRef]
- Salt, D.E.; Kramer, U. Mechanisms of metal hyperaccumulation in plants. In Phytoremediation of Toxic Metals: Using Plants to Clean up the Environment; Raskin, I., Ensley, B.D., Eds.; John Wiley & Sons: New York, NY, USA, 2000; pp. 231–246. [Google Scholar]
- Gabbrielli, R.; Mattioni, C.; Vergnano, O. Accumulation mechanisms and heavy metal tolerance of a nickel hyperaccumulator. J. Plant Nutr. 1991, 14, 1067–1080. [Google Scholar] [CrossRef]
- Bringel, F.; Couée, I. Pivotal roles of phyllosphere microorganisms at the interface between plant functioning and atmospheric trace gas dynamics. Front. Microbiol. 2015, 6, 486. [Google Scholar] [CrossRef] [PubMed]
- Trouvelot, S.; Heloir, M.C.; Poinssot, B.; Gauthier, A.; Paris, F.; Guillier, C.; Combier, M.; Trda, L.; Daire, X.; Adrian, M. Carbohydrates in plant immunity and plant protection, roles and potential application as foliar sprays. Front. Plant Sci. 2014, 5, 592. [Google Scholar] [CrossRef] [PubMed]
- Brandl, M.T.; Lindow, S.E. Contribution of indole-3-acetic acid production to the epiphytic fitness of erwinia herbicola. Appl. Environ. Microbiol. 1998, 64, 3256–3263. [Google Scholar] [PubMed]
- Teng, Y.; Huang, C.Y. Ecological effect of heavy metals on soil microbes and research advances on the mechanisms of bioremediation. Soil Environ. Sci. 2002, 11, 85–89. [Google Scholar]
- Li, X.; Cheng, L.; Deng, Y.; Zhang, H. Advances in anaerobic biodegradation of hydrocarbon. Chin. J. Appl. Environ. Biol. 2008, 14, 283–289. [Google Scholar]
- Jing, J.; Liu, J.; Cui, L.I.; Jia, T.; Wang, X.; Chai, B. The structural characteristics of a soil bacterial community in a dam of copper mine tailings in Zhongtiao mountain, Shanxi. Chin. J. Appl. Environ. Biol. 2017, 23, 527–534. [Google Scholar]
- Buyer, J.S.; Zuberer, D.A.; Nichols, K.A.; Franzluebbers, A.J. Soil microbial community function, structure, and glomalin in response to tall fescue endophyte infection. Plant Soil 2011, 339, 401–412. [Google Scholar] [CrossRef]
- Tripathi, B.M.; Stegen, J.C.; Kim, M.; Dong, K.; Adams, J.M.; Lee, Y.K. Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. ISME J. 2018, 12, 1072–1083. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Meng, D.; Li, J.; Yin, H.; Liu, H.; Liu, X.; Cheng, C.; Xiao, Y.; Liu, Z.; Yan, M. Response of soil microbial communities and microbial interactions to long-term heavy metal contamination. Environ. Pollut. 2017, 231, 908–917. [Google Scholar] [CrossRef] [PubMed]
- Wichern, J.; Wichern, F.; Joergensen, R.G. Impact of salinity on soil microbial communities and the decomposition of maize in acidic soils. Geoderma 2006, 137, 100–108. [Google Scholar] [CrossRef]
- Liu, H.M.; Zhang, H.F.; Huangfu, C.H.; Jie, L.I.; Zhou, G.F.; Yang, D.L. Effects of different long-term nitrogen addition on soil microbial diversity of Stipa baicalensis steppe in Inner Mongolia, China. J. Agro-Environ. Sci. 2017, 36, 709–717. [Google Scholar]
- Wang, Y.; Shi, J.; Wang, H.; Lin, Q.; Chen, X.; Chen, Y. The influence of soil heavy metals pollution on soil microbial biomass, enzyme activity, and community composition near a copper smelter. Ecotoxicol. Environ. Saf. 2007, 67, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Hunter, P.J.; Hand, P.; Pink, D.; Whipps, J.M. Both leaf properties and microbe-microbe interactions influence within-species variation in bacterial population diversity and structure in the lettuce (Lactuca Species) phyllosphere. Appl. Environ. Microbiol. 2010, 76, 8117–8125. [Google Scholar] [CrossRef] [PubMed]
- Dohrmann, A.B.; Tebbe, C.C. Effect of elevated tropospheric ozone on the structure of bacterial communities inhabiting the rhizosphere of herbaceous plants native to Germany. Appl. Environ. Microbiol. 2005, 71, 7750–7758. [Google Scholar] [CrossRef] [PubMed]
- Schmalenberger, A.; Tebbe, C.C. Genetic profiling of noncultivated bacteria from the rhizospheres of sugar beet (Beta vulgaris) reveal field and annual variability but no effect of a transgenic herbicide resistance. Can. J. Microbiol. 2003, 49, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kögel-Knabner, I. The macromolecular organic composition of plant and microbial residues as inputs to soil organic matter. Soil Biol. Biochem. 2002, 34, 139–162. [Google Scholar] [CrossRef]
- Shi, X.; Chang, X. Microbial responses to heavy metal pollution in soils. Ecol. Environ. 2003, 12, 498–499. [Google Scholar]
- Fließbach, A.; Martens, R.; Reber, H.H. Soil microbial biomass and microbial activity in soils treated with heavy metal contaminated sewage sludge. Soil Biol. Biochem. 1994, 26, 1201–1205. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, Y.; Mai, Y.E.; Dou, L.; Li, X.Y.; Mo, L.P. Bioavailability of heavy metal and transfer factors in a regional soil-to-crops system. Environ. Sci. Technol. 2017, 40, 256–266. [Google Scholar]
- Zhao, Y.; Ma, Z.J.; Zhang, X.; Xue, X.; Li, M.Y.; Cheng, Y.W.; Zha, T.G. Influencing factors of heavy metal mobility and evaluating methods of heavy metal bioavailability in soil-plant system. J. Chin. Inst. Water Resour. Hydropower Res. 2015, 13, 177–183. [Google Scholar]
- Olaniran, A.O.; Adhika, B.; Balakrishna, P. Bioavailability of heavy metals in soil, impact on microbial biodegradation of organic compounds and possible improvement strategies. Int. J. Mol. Sci. 2013, 14, 10197–10228. [Google Scholar] [CrossRef] [PubMed]


| S | H′ | dMax | En | D | ||
|---|---|---|---|---|---|---|
| Bacteria | Non-rhizosphere | 35.80 ± 3.834 a | 3.550 ± 0.105 a | 4.102 ± 0.402 a | 0.994 ± 0.007 a | 0.971 ± 0.003 a |
| Rhizosphere | 39.40 ± 1.673 a | 3.633 ± 0.079 a | 4.506 ± 0.164 a | 0.989 ± 0.010 a | 0.973 ± 0.002 a | |
| Fungi | Non-rhizosphere | 2.677 ± 0.248 b | 15.00 ± 3.808 b | 1.814 ± 0.434 b | 0.998 ± 0.001 ab | 0.929 ± 0.017 b |
| Rhizosphere | 2.710 ± 0.227 b | 15.40 ± 3.362 b | 1.858 ± 0.391 b | 0.998 ± 0.001 b | 0.932 ± 0.016 b |
| Microbial Diversity | Catalase | Urease | Sucrase | Phosphatase | ||
|---|---|---|---|---|---|---|
| Rhizosphere | Bacteria | H′ | 0.376 | 0.594 | 0.028 | 0.333 |
| S | 0.375 | 0.534 | 0.001 | 0.375 | ||
| dMax | 0.405 | 0.579 | −0.030 | 0.424 | ||
| En | 0.376 | 0.656 | 0.052 | 0.289 | ||
| D | 0.346 | 0.573 | 0.061 | 0.299 | ||
| Fungi | H′ | 0.679 | 0.362 | −0.773 | 0.924 * | |
| S | 0.724 | 0.394 | −0.814 | 0.942 * | ||
| dMax | 0.717 | 0.396 | −0.807 | 0.938 * | ||
| En | −0.738 | −0.218 | 0.913 * | −0.890 * | ||
| D | 0.631 | 0.325 | −0.729 | 0.903 * | ||
| Non-rhizosphere | Bacteria | H′ | 0.053 | 0.682 | 0.311 | −0.170 |
| S | 0.219 | 0.793 | 0.183 | 0.002 | ||
| dMax | 0.232 | 0.799 | 0.170 | 0.011 | ||
| En | −0.852 | −0.649 | 0.644 | −0.854 | ||
| D | 0.078 | 0.705 | 0.287 | -0.137 | ||
| Fungi | H′ | 0.736 | 0.442 | −0.886 * | 0.758 | |
| S | 0.708 | 0.384 | −0.884 * | 0.739 | ||
| dMax | 0.716 | 0.398 | −0.887 * | 0.746 | ||
| En | −0.790 | −0.662 | 0.667 | -0.448 | ||
| D | 0.760 | 0.502 | −0.881 * | 0.772 | ||
| Log Number of Microbial rRNA Gene Copies g−1 Dry Soil | TF-Cd | TF-Cr | TF-Cu | TF-Pb | TF-Zn | |
|---|---|---|---|---|---|---|
| Non-rhizosphere | Bacteria | −0.130 | −0.069 | −0.013 | −0.718 | −0.155 |
| Fungi | 0.057 | 0.062 | 0.002 | −0.632 | −0.411 | |
| B/F | −0.341 | −0.262 | −0.048 | 0.292 | 0.673 | |
| Rhizosphere | Bacteria | −0.783 | −0.818 | −0.670 | 0.741 | 0.215 |
| Fungi | −0.130 | −0.069 | −0.013 | −0.718 | −0.155 | |
| B/F | −0.423 | −0.517 | −0.515 | 0.895 * | −0.004 | |
| Catalase | Urease | Sucrase | Phosphatase | |
|---|---|---|---|---|
| TF-Cd | 0.254 | −0.174 | −0.632 | 0.371 |
| TF-Cr | 0.169 | −0.340 | −0.583 | 0.321 |
| TF-Cu | −0.139 | −0.738 | −0.297 | −0.005 |
| TF-Pb | −0.644 | −0.283 | 0.679 | −0.900 * |
| TF-Zn | −0.775 | −0.905 * | 0.614 | −0.636 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, T.; Guo, T.; Cao, M.; Chai, B. Effects of Heavy Metals on Phyllosphere and Rhizosphere Microbial Community of Bothriochloa ischaemum. Appl. Sci. 2018, 8, 1419. https://doi.org/10.3390/app8091419
Jia T, Guo T, Cao M, Chai B. Effects of Heavy Metals on Phyllosphere and Rhizosphere Microbial Community of Bothriochloa ischaemum. Applied Sciences. 2018; 8(9):1419. https://doi.org/10.3390/app8091419
Chicago/Turabian StyleJia, Tong, Tingyan Guo, Miaowen Cao, and Baofeng Chai. 2018. "Effects of Heavy Metals on Phyllosphere and Rhizosphere Microbial Community of Bothriochloa ischaemum" Applied Sciences 8, no. 9: 1419. https://doi.org/10.3390/app8091419
APA StyleJia, T., Guo, T., Cao, M., & Chai, B. (2018). Effects of Heavy Metals on Phyllosphere and Rhizosphere Microbial Community of Bothriochloa ischaemum. Applied Sciences, 8(9), 1419. https://doi.org/10.3390/app8091419




