Featured Application
The technique presented in the paper can be used to build an issue-based clustering system, which is essential for exploration and curation of research issues and their findings reported in scientific literature.
Abstract
A scholarly article often discusses multiple research issues. The clustering of scholarly articles based on research issues can facilitate analyses of related articles on specific issues in scientific literature. It is a task of overlapping clustering, as an article may discuss multiple issues, and hence, be clustered into multiple clusters. Clustering is challenging, as it is difficult to identify the research issues with which to cluster the articles. In this paper, we propose the use of the titles of the references cited by the articles to tackle the challenge, based on the hypothesis that such information may indicate the research issues discussed in the article. A technique referred to as ICRT (Issue-based Clustering with Reference Titles) was thus developed. ICRT works as a post-processor for various clustering systems. In experiments on those articles that domain experts have selected to annotate research issues about specific entity associations, ICRT works with various clustering systems that employ state-of-the-art similarity measures for scholarly articles. ICRT successfully improves these systems by identifying clusters of articles with the same research focuses on specific entity associations. The contribution is of technical and practical significance to the exploration of research issues reported in scientific literature (supporting the curation of entity associations found in the literature).
1. Introduction
A huge and ever-growing amount of scientific findings is reported in scholarly articles. A scholarly article often discusses multiple research issues. The clustering of scholarly articles based on their research issues can facilitate timely and comprehensive analysis of research issues (and their findings) reported in scientific literature, which is a task that is routinely conducted by scientists. For example, CTD (Comparative Toxicogenomics Database), GHR (Genetic Home Reference), and OMIM (Online Mendelian Inheritance in Human) recruit many scientists to routinely update their article databases in terms of their associations with specific biomedical entities (information about how CTD, GHR, and OMIM update their databases can be respectively found at [1,2,3]). This issue-based clustering task is a kind of overlapping clustering (i.e., partitioning articles into non-exclusive clusters), as an article may discuss multiple issues, and hence, often needs to be clustered into multiple clusters. Issue-based clustering is challenging as well, as it is difficult to identify the research issues in order to cluster the articles.
In this paper, we investigate how existing clustering techniques can be improved to conduct the issue-based clustering of scholarly articles. Figure 1 illustrates the main idea. An article may discuss multiple issues (e.g., article a1 discusses issues I1, I2, and I3 in Figure 1), and for each issue it cites different references (e.g., a1 cites different references related to I1 to I3 in Figure 1). Therefore, multiple articles (e.g., a1 and a2) can be clustered into a cluster if they cite those references that are related to the same issue (e.g., Issue I3 in Figure 1). The clustering task is thus based on citations (out-link references) in the articles. The clusters can provide an organized view of “cited issues” in scientific literature.
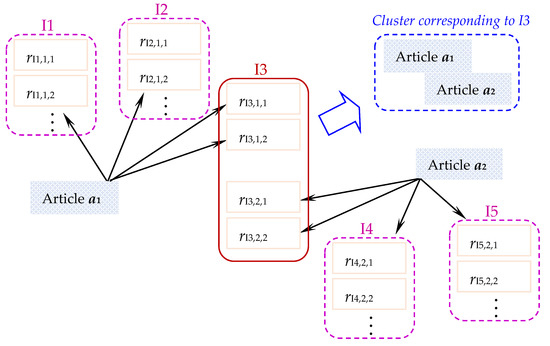
Figure 1.
Main idea of issue-based clustering of scholarly article.
More specifically, we present a technique ICRT (Issue-based Clustering with Reference Titles), which employs titles of the references cited by articles to conduct issue-based clustering. Section 2 discusses the technical contributions of ICRT when compared with previous related studies. The development of ICRT is presented in Section 3. It is based on an observation that a scholarly article often discusses several research issues, and for each issue, cites related references whose titles may indicate the subject. ICRT works as a post-processor for various clustering systems. Section 4 presents experiments to evaluate ICRT. The results show that, when collaborating with those clustering systems that employ state-of-the-art similarity measures for scholarly articles, ICRT successfully improves these systems in identifying clusters of articles having the same research focuses as judged by domain experts. The contribution is of technical and practical significance to the exploration and curation of research issues reported in scientific literature.
2. Background
Object clustering aims at grouping similar objects into the same cluster. Previous clustering techniques fell into two types: hard and soft clustering. Hard clustering aims at grouping an object into a cluster, while soft clustering (also known as fuzzy clustering) aims at assigning different degrees of membership of an object to all clusters (for a survey of fuzzy clustering techniques, the readers are referred to [4]). By setting thresholds on the degrees of membership, fuzzy clustering techniques can achieve overlapping clustering (i.e., partitioning objects into non-exclusive clusters). Fuzzy C-means (FCM) is a representative fuzzy clustering technique [4]. It worked on a similarity measure based on the distance (e.g., Euclidean distance) between objects and cluster means in a vector space [5]. For those domains in which good inter-object similarity measures have been developed, fuzzy C-medoids (FCMdd) was developed to employ these domain-specific similarity measures to improve overlapping clustering. FCMdd works to find a fixed number of representative data objects (i.e., medoids, rather than cluster means) for clusters, with each of the other data objects having a degree of membership to each cluster, so that the sum of within-cluster dissimilarities is minimized (i.e., within-cluster similarities is maximized [6,7]).
The clustering of scholarly articles is a domain in which many good inter-article similarity measures have been developed, and hence, many previous studies employed these similarity measures to cluster scholarly articles. State-of-the-art inter-article similarity measures worked on two types of contents: (1) citations among articles, and (2) the main contents of articles (e.g., titles, abstracts, and main texts). The former was based on the expectation that two articles that share certain citations may be similar to each other, while the latter was based on the expectation that two articles that have similar main contents may be similar. The technical contribution of ICRT can thus be highlighted by explaining how these previous similarity measures have weaknesses in issue-based article clustering.
2.1. Citation-Based Similarity Measures
Many similarity measures worked on citations among articles, including in-link citations and out-link citations. For an article a, in-link citations are those articles that cite a, while out-link citations are those articles that a cites. Some techniques treated the citations as undirected and unweighted links in a graph on which article clusters were produced (a survey and comparison of these techniques can be found in [8]). These techniques thus clustered articles by analyzing the inter-connections among articles, rather than similarity between articles, which is employed by ICRT and many text clustering techniques.
Co-citation is a representative technique that considers in-link citations to estimate inter-article similarity [9]. Two articles may be related to each other if they are co-cited by other articles. Clustering techniques based on co-citation has been developed and applied to various domains (e.g., patent analysis [10]). However, applicability of the techniques based on in-link citations is limited, as many scholarly articles have very few (or even no) in-link citations. ICRT works on out-link citations of scholarly articles, which are more publicly available, even for newly-published articles that have not yet received any in-link citations.
Bibliographic coupling (BC) is a representative technique that considers out-link citations [11], which were found to be more helpful than in-link citations in the classification [12] and clustering [13] of scholarly articles. BC was also a typical citation-based similarity measure that was integrated with content-based similarity measures by hybrid measures [12,13,14,15]. The hybrid measures did not always perform significantly better than BC [12], and they even performed significantly worse on occasion [16]. Equation (1) is a typical way to estimate BC similarity between two articles, a1 and a2 [12], where Oa1 and Oa2 are the sets of articles that a1 and a2 cite respectively. When both Oa1 and Oa2 are empty, the similarity is set to 0.
BC has a weakness in dealing with two articles that are related but cite different references, and hence, the similarity measure, DescriptiveBC, was proposed to employ titles of the references to tackle the weakness [16], as different but related references may share certain key terms in their titles. As defined in Equation (2), Jaccard index is employed to estimate similarity between titles of two references r1 and r2 (where Title(r) is the set of terms in the title of a reference r), and the DescriptiveBC similarity between two articles a1 and a2 is defined in Equation (3), where Ra is the set of references in article a.
However, these similarity measures did not aim at issue-based clustering. They did not estimate issue-based similarity; rather, they estimated the similarity between two articles based on all reference titles in the article, without considering the common case where these references are about different issues. As an article may discuss multiple issues, two articles may be in a cluster if they discuss an issue, even though they are not entirely similar to each other overall; and conversely, three articles should not be in a cluster if they discuss different issues, even though they are similar to each other overall. ICRT is thus composed of two novel components for (1) estimation of issue-based similarity, and (2) clustering of scholarly articles based on the issue-based similarity.
2.2. Content-Based Similarity Measures
Many similarity measures work on the main contents of articles, including titles, abstracts, and main texts. We are concerned with those that worked on the titles and abstracts (TA) of articles, as they are much more publicly available than main texts, making the clustering systems applicable to most scholarly articles.
The vector space model (VSM) is a typical technique to estimate content-based similarity. It represents each article as a vector of term weights, and similarity between two articles is simply the cosine similarity on their vectors. VSM was employed by many scholarly article retrieval systems (e.g., [17]). However, cosine similarity did not always perform well [18,19]. Latent Semantic Analysis (LSA) was a typical technique that employed singular value decomposition to improve the vector representation of scholarly articles [20,21]. However LSA may not perform well for scholarly articles either [19].
OK was developed based on BM25 [22], which was one of the best techniques for finding related scholarly articles [19]. It was shown to be one of the best techniques to identify related articles as well [18]. OK employs Equation (4) to estimate the similarity between two articles, a1 and a2, where k1 and b are two parameters, |a| is the number of terms in article a (i.e., length of a), avgal is the average number of terms in an article (i.e., average length of articles), TF(t,a) is the frequency of term t appearing in article a, and IDF(t) is the inverse document frequency of term t, which measures how rarely t appears in a large collection of articles.
PubMed is a popular search engine for biomedical scholarly articles. It estimates similarity between two scholarly articles by considering several kinds of information [23,24], including (1) term stemming, (2) article lengths, (3) term positions in the articles (e.g., terms in titles of the articles), (4) key terms of the articles, and (5) weights of the terms in the articles (including TF and IDF). The similarity measure used by PubMed to estimate inter-article similarity is summarized in [23]. It was found to be one of the best to cluster scholarly articles [19].
Another kind of similarity measure can be derived by topic modeling, which identifies latent topics embedded in a collection of articles. An article can be a mixture of topics, and hence, it can be represented as a topic proportion vector in which a dimension corresponds to the degree of relatedness to the corresponding topic (for a review of probabilistic topic modeling techniques, the readers are referred to [25]). Similarity between articles can be estimated based on the topic proportion vectors of the articles; hence, by treating the topics as clusters of articles, various topic modeling techniques were applied to the clustering of scholarly articles [26] and news articles [27].
However, none of these similarity measures aims at issue-based clustering, as it is quite difficult to identify research issues from the main contents of articles. ICRT recognizes the research issues by analyzing the titles of the references cited by the articles, based on the observation that, to discuss a research issue, an article often cites related references whose titles may indicate the issue. We will implement several systems that employ typical similarity measures to conduct clustering (see Section 4.2), and evaluate how these systems can be further improved to conduct issue-based clustering by employing ICRT as a post-processor for them.
3. Development of ICRT
ICRT is designed to work as a post-processor of various clustering systems that may employ various effective techniques to produce article clusters. These clustering systems provide ICRT with preliminary clusters of scholarly articles, and ICRT outputs a refined clustering result based on the preliminary clusters. Development of ICRT is thus challenging in two ways: (1) estimation of issue-based similarity by identifying the common cited issue, and (2) clustering of each article into potentially multiple clusters (i.e., overlapping clustering, as an article may discuss multiple issues).
Figure 2 illustrates the main process flow of ICRT. For each pair of test articles, issue-based similarity (IBS) is estimated to measure how the two articles are related to each other by discussing a similar research issue, and then a set of issue-indicative terms (IIT) are identified to describe the issue with certain terms (see Figure 2a). Each article thus has many IITs with other articles; hence, by clustering the IITs, IIT clusters can be constructed (see Figure 2b). For each IIT cluster, involving articles can form an article cluster, and hence, article clusters can be constructed, with each article belonging to potentially multiple clusters (or issues, see Figure 2c). Detailed design of ICRT is presented in the following subsections.
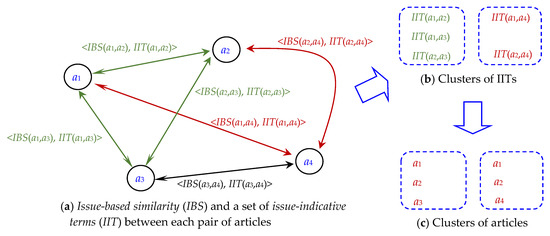
Figure 2.
Main steps of ICRT: (a) estimating IBS and identifying IIT between each pair of articles; (b) clustering the IITs, and (c) producing clusters of articles accordingly.
3.1. Estimation of Issue-Based Similarity (IBS)
Issue-based similarity (IBS) between two articles, a1 and a2, is based on how they cite a certain number of references with similar titles. It is the fundamental basis of clustering. A larger IBS indicates that the distance (dissimilarity) between a1 and a2 is smaller. ICRT identifies the set of references in a1 that are the most similar to those references in a2. This set of references is MostSimRef(a1, a2) defined in Equation (5), where Ref(a) is the set of references in article a, and RefSim(r1, a2) measures how a reference r1 in a1 is similar to the references in a2 (see Equation (6), where Title(r) is the set of terms in reference r, and IRF(t) is the inverse reference frequency of term t defined in Equation (7) to indicate how rare t is in the references).
With MostSimRef defined above, ICRT can identify which references in a1 are the most similar to the references in a2, and vice versa. These references can collectively indicate how a1 and a2 discuss similar issues; hence, issue-based similarity (IBS) between a1 and a2 can be defined in Equation (8), where Rareness(r) measures how rarely a reference r appears in articles (see Equation (9)). The basic idea is that two articles are similar in discussing certain issues if they share similar and rare references.
3.2. Identification of the Set of Issue-Indicative Terms (IIT)
In addition to IBS defined above, each pair of articles also has an IIT, which is a set of terms to describe the issue discussed by the two articles. Therefore, each article has many IITs with other articles, and issue-based clustering can be approached by clustering the IITs. Given two articles, a1 and a2, Equation (10) defines candidate IIT terms, which need to appear in titles of similar references (IITref) defined in Equation (11). The basic idea is that a candidate term should appear in titles of references in MostSimRef and their corresponding similar references in both articles.
For a candidate term t, Equation (12) is defined to estimate its strength of being an IIT term for a1 and a2, which is a harmonic fusion of two “one-way” strengths of t defined in Equation (13). The basic idea is that a rare candidate term that appears in titles of rare and similar references in a1 and a2 can get a high strength of being an IIT term for a1 and a2.
For two articles, a1 and a2, IIT simply includes those candidate terms that have top-10 IIT strengths, as defined in Equation (14). These terms aim at indicating the issue discussed in both a1 and a2.
3.3. Clustering of IITs and Involving Articles
As an IIT between two articles contains those terms that indicate the shared issue of the two articles, issue-based clustering of articles can be done by clustering the IITs. Therefore, with IBS and IIT defined for any two articles (ref. Equations (8) and (14) defined above), ICRT conducts issue-based clustering of articles by clustering the IITs (ref. Figure 2c noted above). ICRT is designed to work as a post-processor of existing clustering systems, which provides clustering results to be refined by ICRT. The algorithm is outlined in Figure 3.

Figure 3.
The algorithm to cluster articles by clustering IITs.
Given PreClusters as the set of clusters produced by an underlying clustering system, ICRT refines PreClusters by identifying and clustering the IITs of all pairs of articles in PreClusters (see Steps 1 to 4 in Figure 3). An IIT between two articles actually corresponds to a cluster of the two articles; hence, given two articles, a1 and a2 (in a cluster in PreClusters), an initial cluster c can be constructed and represented as <cIBS(c), cIIT(c), ArticlePair(c)>, where cIBS(c) is the cluster IBS for c initially set to IBS(a1,a2); cIIT(c) is the cluster IIT for c initially set IIT(a1,a2); and ArticlePair(c) is the set of involving article pairs in c, and hence, is initially set to {<a1,a2>} (see Step 2 in Figure 2).
ICRT then ranks all clusters according to their cIBS values (see Step 3 in Figure 3), and tries to group any two clusters. Those clusters that have higher cIBS values have a higher priority to be grouped with other clusters (see Step 4 in Figure 3). Two clusters c1 and c2 can be grouped to form a new cluster if four conditions are satisfied: (1) articles in c1 and c2 are grouped in a cluster in PreClusters as well; (2) cIBS(c1) and cIBS(c2) are not less than 0.5; (3) sum of IDF values of intersecting terms in cIIT(c1) and cIIT(c2) is not less than a threshold (tuned by training data); and (4) an article pair <a1,a2> exists whose IBS is not less than 0.5, and a1 (a2) is an article in c1 (c2) but not in c2 (c1). For a new cluster c produced by grouping c1 and c2 (cIBS(c1) > cIBS(c2)), cIBS(c) is the average IBS of all article pairs in ArticlePair(c1) and ArticlePair(c2); cIIT(c) is composed of intersecting terms in cIIT(c1) and cIIT(c2); and ArticlePair(c) is the union of ArticlePair(c1) and ArticlePair(c2) (see Step 4.1.2.1 in Figure 3). Cluster c1 is then replaced with the new cluster c, and c2 is removed (see Steps 4.1.2.2 and 4.1.2.3 in Figure 3) so that c may be further grouped with other clusters.
To further refine the clustering results, ICRT continues to consider potential clusters of articles that have high IBS values (see Steps 5 to 7 in Figure 3). A pair of articles that are not grouped into a cluster in PreClusters can be a potential cluster if IBS between the two articles is not less than 0.5 (see Step 5 in Figure 3). By adding these potential clusters into the clustering results, ICRT can consider those clusters that are not identified by the underlying clustering system. ICRT re-ranks the clusters according to their corresponding cIBS values (see Step 6 in Figure 3), and triggers the same clustering procedure noted above (Step 4 and Step 7 in Figure 3 are basically the same), except that two clusters c1 and c2 can be grouped to form a new cluster if the 2nd to the 4th conditions noted above are satisfied (i.e., the 1st condition is not considered, as ICRT is now considering those clusters that are not in PreClusters).
After the above two clustering processes, the clusters are re-ranked again (according to their IBS values), and the articles in each cluster are retuned as a ranked list of article clusters (see Steps 8 and 9 in Figure 3). If the user sets a fixed number K of resulting clusters, only top-K clusters are returned.
4. Experiments
4.1. Experimental Data
Experimental data is collected from CTD (available at [28]), which recruits domain experts to curate associations that are of three types: <chemical, gene>, <chemical disease>, and <gene, disease> [29,30]. We download all the associations (downloaded in 30 January 2017), and remove those disease-related associations (i.e., associations of two types <chemical, disease> and <gene, disease>) that are not supported by ‘direct evidence’ (i.e., the diseases do not have ‘marker/mechanism’ or ‘therapeutic’ relations to the chemicals or the genes). There are thus three datasets of associations (i.e., the three types of associations) that CTD experts have confirmed based on the results reported in scientific literature.
For an association <e1, e2> between two entities e1 and e2, the CTD experts have selected scholarly articles that report conclusive findings on the association. Therefore, articles curated for an association actually focus on the same research issue (i.e., the association), and hence, they should be grouped into a cluster. An association thus corresponds to a cluster. A system that can achieve such issue-based clustering is essential for researchers (e.g., the CTD experts and research professionals) to validate and curate related conclusive findings reported in scientific literature.
Therefore, all articles that are curated for the associations in the three datasets are collected. As we are investigating how references in articles can be used to improve issue-based clustering, those articles whose references cannot be retrieved from an archive PubMed Central are removed (PubMed Central is available at [31]). Also note that as we are investigating how articles are grouped into clusters, those clusters (associations) that do not have multiple articles are removed as well. Clusters that are composed of the same set of articles are treated as a cluster (i.e., duplicate clusters are removed). Finally there are three datasets for experiments: (1) <chemical, gene>: 13307 associations (clusters) with 8450 articles; (2) <chemical disease>: 2451 associations (clusters) with 5224 articles; and (3) <gene, disease>: 571 associations (clusters) with 1150 articles. The three data sets are available as supplementary materials (see Tables S1–S3, respectively). Each data set is employed as training data to tune the clustering systems, while the other two sets are employed as test data to test the systems, and the process repeats three times so that each data set is employed as training data exactly one time. Average performance on each data set is then reported.
Table 1 presents a statistical analysis on the three datasets, which shows that the clustering task on the three datasets is quite different from many previous clustering tasks. Most associations (clusters) have very few articles (having only two to three articles, although many associations have more than three articles as well (ref. “# clusters having X articles” in Table 1). Therefore, the clustering task needs to partition the articles into clusters about specific research issues with possibly very few articles in a cluster, rather than diverse research fields with many articles in a cluster (e.g., tissue engineering, genetic algorithm, … etc. [26]). Moreover, many articles belong to multiple clusters (associations), especially in the first dataset <chemical, gene> in which about 50% of the articles belong to three or more clusters (ref. “# articles belonging to X clusters” in Table 1). This situation justifies the need of overlapping clustering, as an article often discusses multiple issues, and hence, should be put into multiple clusters in practice. Finally, most articles were published after 2006 (inclusive), although many articles were published before then as well, especially in the second dataset <chemical, disease> (ref. “# articles published in year X” in Table 1). This indicates that the clustering task is essential for researchers to check related articles that were published over diverse periods of time.

Table 1.
Statistical characteristics of the three datasets in the experiment.
4.2. Underlying Systems for Overlapping Clustering
ICRT works as a post-processor for a clustering system so that performance of the system in issue-based clustering can be improved. We thus implement several types of systems that employ typical article similarity measures to conduct overlapping clustering (i.e., partitioning articles into non-exclusive clusters, as an article may discuss multiple issues), and investigate how they can be improved by collaborating with ICRT. As summarized in Table 2, these underlying clustering systems work on two kinds of contents: (1) references (citations) in each article, and (2) title and abstract of each article, which are publicly available and employed by many systems for the clustering of scientific articles (e.g., [26]).

Table 2.
Systems for overlapping clustering with various inter-article similarity measures.
Two clustering systems that work on references employ bibliographic coupling (BC, see Equation (1)) and DescriptiveBC (see Equation (3)) as their similarity measures. As noted in Section 2.1, BC is a typical measure that performed well in the retrieval and clustering of scholarly articles [13,16]. DescriptiveBC employed titles of the references to improve BC [16].
The clustering systems that work on titles and abstracts (TA) of articles employ several kinds of inter-article similarity measures, including (1) PubMed-based similarity (PMS), (2) OK, and (3) those derived by topic modeling. The textual data is reprocessed by removing stop words (by a stop word list available at [32]) and mapping synonymous words to the same ID (by the MetaMap tool available at [33]). As noted in Section 2.2, PubMed recommends related articles for a given article, based on an article similarity measure that has considered several kinds of indicators [23,24]. The similarity measure was one of the best to cluster scholarly articles [19]. Following [19], for each article a1, a ranked list of top-50 similar articles recommended by PubMed is retrieved. Equation (15) is then employed to estimate how an article a2 is similar to a1 based on the rank of a2 in the ranked list (i.e., RankSim(a1, a2), see Equation (15)). PMS between two articles a1 and a2 is simply the average of RankSim(a1, a2) and RankSim(a2, a1), as defined in Equation (16).
OK was shown to be one of the best techniques to identify related articles as well [18]. It employs Equation (4) to estimate article similarity. Equation (4) has two parameters, k1 and b. Following the suggestion of [18], they are set to 8 and 1.0, respectively, and normalization is conducted so that OK similarity is in the range of [0, 1]. We test two versions that cluster articles based on OK similarity on titles and abstracts (OK-TA) and reference titles (OK-RT), respectively. Many previous studies have found that titles of articles are more topic-indicative than their abstracts, and hence, following [23,24], words from titles are added twice to give them higher weights in computing inter-article similarity.
To employ these state-of-the-art similarity measures to conduct overlapping clustering, fuzzy C-medoids clustering (FCMdd) is employed. As noted in Section 1, FCMdd works to find representative data objects (i.e., medoids) for clusters, so that sum of within-cluster dissimilarities is minimized. To investigate whether ICRT can further improve well-tuned underlying clustering systems, the number of clusters produced by FCMdd is set to the number of target associations that have been identified by CTD experts (ref. Section 4.1). FCMdd is suitable for our experimental purpose, as it can conduct overlapping clustering on relational data (i.e., articles and similarities between them) so that state-of-the-art inter-article similarity measures (e.g., BC, DescriptiveBC, PMS, and OK noted above) can be used for article clustering, while many other clustering techniques only worked on distance measures defined on a vector space (e.g., fuzzy c means [5]).
In the results of fuzzy clustering produced by FCMdd, each article has different degrees of relatedness to the clusters. Therefore, to determine the final clusters of articles, thresholding is conducted to tune a relatedness threshold in the hope to achieve the best performance in the evaluation criteria to be defined in Section 4.3 (i.e., BCubedF1, see Equation (21)). As noted in Section 4.1, training data is used to tune the best thresholds for the systems.
As noted in Section 2.2, another typical kind of content-based similarity measure can be derived based on topic modeling, which identifies latent topics in a collection of articles. The topic modeling systems are implemented with a toolkit that is available at [34]. To investigate whether ICRT can further improve well-tuned systems based on topic modeling, the number of topics is set to the number of target associations that have been identified by CTD experts (ref. Section 4.1). As an article can be a mixture of the topics, it can be represented as a topic proportion vector in which a dimension corresponds to the degree of relatedness to the corresponding topic (cluster). Therefore, thresholding on the degrees of relatedness is conducted to determine the final clusters as well, and as for OK noted above, there are two versions, TM-TA and TM-RT, that respectively apply topic modeling to the two different textual contents (i.e., titles and abstracts, as well as reference titles). Similar strategies of producing clusters with topic modeling were employed by previous studies as well (e.g., [26,27]); however, we revise them so that the system can cluster an article into multiple clusters (rather than a single cluster) with a well-tuned threshold. Another way to conduct overlapping clustering by topic modeling is to estimate cosine similarities based on the topic proportion vectors of articles, and then conduct fuzzy clustering with the similarities. There are thus two more versions TM-TA-FC and TM-RT-FC, which are respectively variants of TM-TA and TM-RT, by invoking fuzzy clustering (FC) to cluster articles.
4.3. Evaluation Criteria
As we are evaluating how systems perform in overlapping clustering in which articles are partitioned into non-exclusive clusters, many external evaluation metrics become inappropriate in measuring multiple assignments for a single item [35]. Therefore, precision and recall were often employed and revised to evaluate the quality of overlapping clustering (e.g., [35,36]). Among many typical evaluation criteria, the BCubed measure was extended to satisfy the most essential constraints in evaluating overlapping clustering [35]. BCubed computes precision and recall on each article, and then accordingly computes precision and recall of the whole result of clustering. More specifically, given a pair of articles <a1, a2>, Equations (17) and (18) respectively defines pair-based precision (Ppair) and recall (Rpair), where C(a) and G(a) are respectively the sets of identified clusters and target clusters in which article a appears. Therefore, Ppair and Rpair collectively measure the quality of clustering a1 and a2 in the same clusters.
The quality of the whole clustering result can then be measured based on the pair-based precision and recall. By estimating the average precision and recall values on individual articles, BCubed precision (BCubedP) and recall (BCubedR) are respectively defined in Equations (19) and (20), where A is the set of test articles. Equation (21) defines BCubed F1 (BCubedF1), which integrates BCubedP and BCubedR in a harmonic way.
4.4. Results
Figure 4, Figure 5 and Figure 6 respectively show test results on three datasets: chemical-gene, chemical-disease, and gene-disease datasets defined above (ref. Section 4.1). ICRT successfully works as a post-processor to improve all the underling systems in all the datasets. It is interesting to note that, with the help of ICRT, all the systems achieve similar performance on individual datasets, indicating that ICRT can work with various clustering systems to conduct issue-based clustering. Among the three datasets, these systems achieve worse performance on the chemical-gene dataset (compare BCubedF1 values in Figure 4, Figure 5 and Figure 6). As noted in Section 4.1, in this dataset, many articles belong to more clusters (about 50% of the articles belong to three or more clusters, ref. “# articles belonging to X clusters” of the first dataset in Table 1), making it more difficult to identify appropriate clusters. Overlapping clustering of the articles is thus an essential and challenging task.
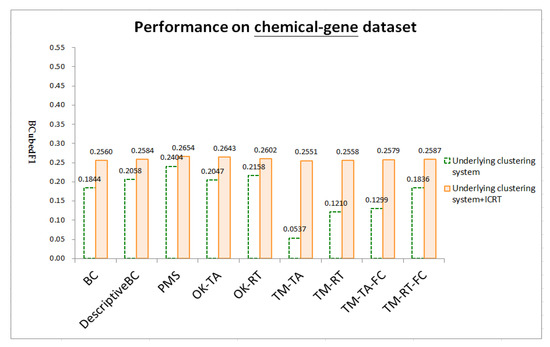
Figure 4.
Test results on the chemical-gene dataset.
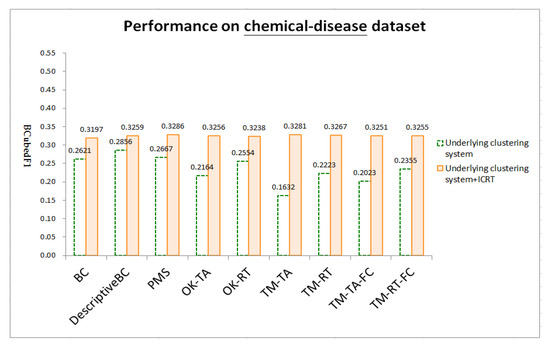
Figure 5.
Test results on the chemical-disease dataset.
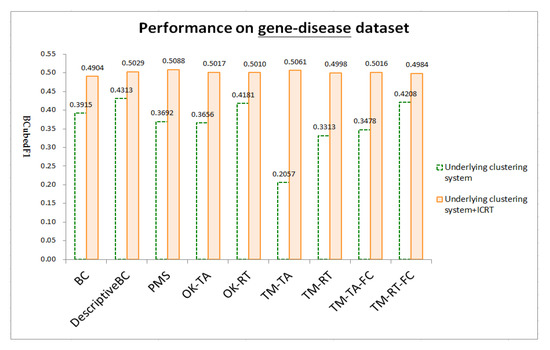
Figure 6.
Test results on the gene-disease dataset.
To further investigate whether the performance difference between an underlying system U and the enhance system U+ICRT is statistically significant, Equations (22)–(24) are defined to respectively estimate precision (Particle), recall (Rarticle), and F1 (F1article) on each individual article, where Ppair and Rpair have been defined above (ref. Equations (17) and (18)). Note that Rarticle is always computable, becasue all experimental articles belong to certain target clusters. However, Particle for an article a may be incomputable, because the system may not put a into any cluster. In this case, Particle is set to 1.0 [37,38]. Moreover, when both Particle and Rarticle for an article are 0, F1article is set to 0 as well [38], becasue in that case the clustering result for the article is totally incorrect. A paired t-test with 99% confidence level is then conducted, and the results show that all the performance differences are statistically significant, reconfirming the contribution of ICRT to the identification of article clusters corresponding to the research issues (i.e., associations between specific entities) curated by CTD experts.
It is also interesting to note that, several clustering systems that work on titles and abstracts (i.e., those that have ‘TA’ in their names, including OK-TA, TM-TA, TM-TA-FC) perform worse than their counterparts that work on reference titles (i.e., those that have ‘RT’ in their names, including OK-RT, TM-RT, TM-RT-FC) on all the datasets. Moreover, BC and DescriptiveBC, which work on article references, have overall better performance among the underlying systems. Therefore, article references are helpful for issue-based clustering. ICRT significantly improves these systems with a more effective method to employ titles of the references. PMS, which employs several well-tuned indicators to estimate article similarity on titles and abstracts (ref. similarity measure of PubMed noted in Section 2.2), can be further improved by ICRT as well, indicating that reference titles can provide helpful information that is different from the information extracted from titles and abstracts of articles.
4.5. A Case Study
To further analyze the contribution of ICRT, a case study is conducted on an article with PubMed ID 17785587. As illustrated in Figure 7, based on the curation by CTD experts, the article belongs to two target clusters that respectively correspond to two issues about two associations: (1) Target cluster I: <TERT, DC>, and (2) Target cluster II: <DKC1, DC>, where TERT is “telomerase reverse transcriptase,” DC is “dyskeratosis congenita,” and DKC1 is “dyskerin pseudouridine synthase 1”.
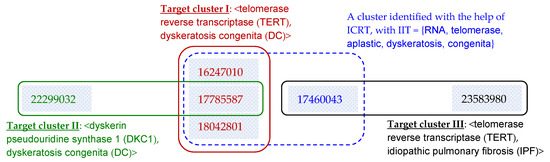
Figure 7.
A case study to analyze how ICRT successfully improves clustering systems that work on various inter-article similarity measures: Without ICRT, all the underlying systems make more errors (see the explanation in the main text).
With the help of ICRT, all systems correctly identify Target cluster II, but not Target cluster I, as they make an error in identifying a large cluster with an additional article (ID = 17460043, see the cluster marked with a dash line box in Figure 7). This article focuses on the association <TERT, IPF>, where IPF is “Idiopathic Pulmonary Fibrosis” (see Target cluster III in Figure 7). That is, both Target clusters I and III are about a gene TERT, but they are about different issues of entity associations (i.e., associations between TERT and two different diseases DC and IPF, respectively). Detailed analysis shows that the error is made (i.e., grouping article 17460043 into Target cluster I) for two reasons: (1) when discussing the target issue <TERT, IPF>, article 17460043 also discusses <TERT, DC> and cites references about the association, and (2) the article also notes the correlation of DC and IPF, and cites references about the correlation. That is, the article discusses correlations among TERT, DC and IPF, although it manly focuses on TERT and IPF only.
It is interesting to note that, without the help of ICRT, all the underlying systems make more errors in clustering the articles. Table 3 summarizes the errors made by the systems. OK-TA identifies none of the article pairs in Target clusters I (ref. the 1st row of Table 3), and all the other underlying systems make the same error noted above (i.e., grouping article 17460043 with certain articles in Target cluster I, ref. the 2nd row of Table 3). Moreover, both BC and DescriptiveBC make an additional error on article 23583980 (ref. the 3rd row of Table 3), and all the other underlying systems make an additional error on article 22299032 (ref. the 4th row of Table 3). With the help of ICRT, these errors are avoided.

Table 3.
Errors made by the systems in the case study (see the main text for explanation).
More specifically, for article 23583980, BC and DescriptiveBC find that it is quite similar to article 17460043, because they cite several references in common. The similarity estimation is good, as both articles belong to Target cluster III in Figure 7; however, it also leads the clustering systems to make an error of grouping them with articles in Target cluster I, because article 17460043 is similar to articles in Target cluster I. With the help of ICRT, the error is avoided by excluding article 23583980 from the identified cluster (i.e., the cluster marked with a dash line box in Figure 7), because IIT of 17460043 and 23583980 is quite different from IIT of the identified cluster.
For article 22299032, all the other systems make at least one of two errors: (1) without grouping it into Target cluster II, and (2) grouping it into Target cluster I (recall that the article belongs to Target cluster II but not Target cluster I, see Figure 7). The first error is made by those systems (e.g., PMS and OK-TA) that think the two articles in Target cluster II (i.e., 22299032 and 17785587) are not similar enough. With the help of ICRT, the error is avoided, as many references in the two articles are similar, meaning that issue-based similarity (IBS, ref. Equation (8)) between the two articles is high. Conversely, the second error (grouping article 22299032 into Target cluster I) is made by those systems that think article 22299032 is quite similar to article 17785587, which is also similar to articles in Target cluster I, leading the systems to group these articles together. With the help of ICRT, the error is avoided by noting that IIT of articles 22299032 and 17785587 is quite different from IIT of the identified cluster (i.e., the cluster in a dash line box in Figure 7).
5. Conclusions and Future Work
We have presented a novel technique ICRT to conduct issue-based clustering of scholarly articles, which is essential for the analysis and exploration of highly related research findings in scientific literature. The issue-based article clustering is challenging, as it is difficult to identify the research issues to cluster the articles, and ICRT tackles the challenge by employing the titles of the references cited by scholarly articles, based on the idea that an article often discusses critical issues by citing related references whose titles may indicate the issues. ICRT works as a post-processor to improve various kinds of clustering systems in issue-based clustering. Experimental results show that ICRT successfully improves several systems in identifying clusters of articles with the same research focuses on specific entity associations judged by domain experts.
The results are of technical significance. The issue-based clustering task calls for effective overlapping clustering techniques, as target clusters may overlap extremely with each other in practice (ref. the clusters about entity associations curated by CTD in the experiment). ICRT is the first technique to identify main research issues in each scholarly article, and based on the identified issues, conduct overlapping clustering for the articles.
The contribution is of practical significance as well, as the clustering of scholarly articles based on research issues can support timely and comprehensive analysis of highly related articles in scientific literature. Scientists often strive to routinely analyze specific issues published in literature (e.g., many scientific evidence databases, such as CTD, GHR, and OMIM, are devoting much effort to routinely updating their databases with focuses on specific research issues, ref. Section 1). It’s quite hard for these scientists to conduct timely and comprehensive analyses of highly related articles that focus on the same research issues, because (1) the analysis of even a research issue needs to be based on careful survey of articles in the ever-growing body of scientific literature, and moreover, (2) it is hard to know how research issues are published in the literature. ICRT should thus be integrated with existing article search engines and clustering systems so that the integrated system can conduct article retrieval and issue-based clustering to support the analysis task of the scientists.
It is interesting to extend ICRT to provide additional information about the identified clusters, including (1) descriptive terms to label each cluster, and (2) representative articles for each cluster to indicate related articles of the cluster. Both kinds of information are helpful for scientists to comprehend and navigate through the space of main research issues in scientific literature.
As ICRT identifies clusters by clustering IITs, descriptive labels for a cluster c can be selected from IITs of article pairs in c. IITstrength defined above (ref. Equation (12)) can be the basis by which to select the descriptive labels. An intelligent way to determine which and how many terms to select is thus an interesting future extension of ICRT. This extension is of technical significance as well, as previous techniques often select cluster labels for a cluster c from main texts of articles in c, by preferring those terms that are frequent in c and/or rare in all clusters [39,40] and those terms identified by topic modeling [25]. Instead of selecting terms from the main texts, terms should be selected from IITs, as the experimental results have shown that the IITs are more effective in issue-based clustering of scholarly articles. Although these previous techniques can be used to select terms from titles of references as well, selecting terms from IITs is still a better way, as the experimental results have shown that ICRT improves clustering systems that work on titles of references (ref. those systems with ‘RT’ in their names in Figure 4, Figure 5 and Figure 6).
Another kind of helpful information to annotate each cluster is representative articles for the cluster, including both citing and cited articles. For a cluster c, all articles belonging to c are candidates of citing articles, as c is constructed based on the references cited by the articles. Conversely, all references cited by the articles in c are candidates of cited articles. An effective way to determine which and how many articles to select is thus an interesting future extension of ICRT. It aims at selecting those articles that can indicate the research issue corresponding to c, rather than those articles that are similar to other articles, which is a typical goal of previous article selection strategies. As each cluster c has a cluster IIT (i.e., cIIT(c), ref. Section 3.3), we suggest preferring those citing articles whose IITs with other articles are similar to cIIT(c), as these articles should be more related to the corresponding research issue of c. To select cited articles for c, those cited articles whose titles are similar to cIIT(c) should be preferred. Articles selected to annotate c can facilitate the navigation of the research findings related to the corresponding issue of c.
Supplementary Materials
The following are available online at http://www.mdpi.com/2076-3417/8/12/2591/s1, Table S1: Gold standard clusters based on chemical-gene associations; Table S2: Gold standard clusters based on chemical-disease associations; and Table S3: Gold standard clusters based on gene-disease associations. Each row in the tables provides information about a gold standard cluster (i.e., an association curated by CTD experts): first entity, second entity, and IDs of articles that belong to the cluster. Each article has two IDs: PubMed ID and PubMed Central ID, with which readers can access the article on PubMed or PubMed Central.
Author Contributions
Conceptualization, R.-L.L.; Data curation, C.-K.H.; Formal analysis, R.-L.L. and C.-K.H.; Funding acquisition, R.-L.L.; Investigation, R.-L.L. and C.-K.H.; Methodology, R.-L.L. and C.-K.H.; Project administration, R.-L.L.; Resources, R.-L.L.; Software, C.-K.H.; Supervision, R.-L.L.; Validation, R.-L.L. and C.-K.H.; Visualization, R.-L.L. and C.-K.H.; Writing—original draft, R.-L.L.; Writing—review & editing, R.-L.L. and C.-K.H.
Funding
The APC was funded by Tzu Chi University, Taiwan.
Acknowledgments
The authors are grateful to Shu-Yu Tung and Yun-Ling Lu for collecting the experimental data.
Conflicts of Interest
The authors declare no conflict of interest.
References
- FAQ—When Is Data Updated? Available online: http://ctdbase.org/help/faq/;jsessionid=92111C8A6B218E4B2513C3B0BEE7E63F?p=6422623 (accessed on 11 December 2018).
- Expert Reviewers—Genetics Home Reference—NIH. Available online: https://ghr.nlm.nih.gov/about/expert-reviewers (accessed on 11 December 2018).
- About OMIM. Available online: https://www.omim.org/about (accessed on 11 December 2018).
- Peters, G.; Crespo, F.; Lingras, P.; Weber, R. Soft clustering—Fuzzy and rough approaches and their extensions and derivatives. Int. J. Approx. Reason. 2013, 54, 307–322. [Google Scholar] [CrossRef]
- Bezdek, J.C.; Ehrlich, R.; Full, W. FCM: The Fuzzy c-means Clustering Algorithm. Comput. Geosci. 1984, 10, 191–203. [Google Scholar] [CrossRef]
- Sisodia, D.S.; Verma, S.; Vyas, O.P. A Subtractive Relational Fuzzy C-Medoids Clustering Approach to Cluster Web User Sessions from Web Server Logs. Int. J. Appl. Eng. Res. 2017, 12, 1142–1150. [Google Scholar]
- Krishnapuram, R.; Joshi, A.; Yi, L. A Fuzzy Relative of the k-Medoids Algorithm with Application to Web Document and Snippet Clustering. In Proceedings of the IEEE International Conference on Fuzzy Systems, Seoul, Korea, 22–25 August 1999; pp. 1281–1286. [Google Scholar]
- Šubelj, L.; van Eck, N.J.; Waltman, L. Clustering Scientific Publications Based on Citation Relations: A Systematic Comparison of Different Methods. PLoS ONE 2016, 11, e0154404. [Google Scholar] [CrossRef] [PubMed]
- Small, H.G. Co-citation in the scientific literature: A new measure of relationship between two documents. J. Am. Soc. Inf. Sci. 1973, 24, 265–269. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, Y.; Liu, R.; Zhang, J. Knowledge-transfer analysis based on co-citation clustering. Scientometrics 2013, 3, 859–869. [Google Scholar] [CrossRef]
- Kessler, M.M. Bibliographic coupling between scientific papers. Am. Doc. 1963, 14, 10–25. [Google Scholar] [CrossRef]
- Couto, T.; Cristo, M.; Gonc¸alves, M.A.; Calado, P.; Nivio Ziviani, N.; Moura, E.; Ribeiro-Neto, B. A Comparative Study of Citations and Links in Document Classification. In Proceedings of the 6th ACM/IEEE-CS Joint Conference on Digital Libraries, Chapel Hill, NC, USA, 11–15 June 2006; pp. 75–84. [Google Scholar]
- Boyack, K.W.; Klavans, R. Co-citation analysis, bibliographic coupling, and direct citation: Which citation approach represents the research front most accurately? J. Am. Soc. Inf. Sci. Technol. 2010, 61, 2389–2404. [Google Scholar] [CrossRef]
- Liu, R.-L. Passage-based Bibliographic Coupling: An Inter-Article Similarity Measure for Biomedical Articles. PLoS ONE 2015, 10, e0139245. [Google Scholar] [CrossRef]
- Janssens, F.; Glänzel, W.; De Moor, B. A hybrid mapping of information science. Scientometrics 2008, 75, 607–631. [Google Scholar] [CrossRef]
- Liu, R.-L. A New Bibliographic Coupling Measure with Descriptive Capability. Scientometrics 2017, 110, 915–935. [Google Scholar] [CrossRef]
- Tian, G.; Jing, L. Recommending scientific articles using bi-relational graph-based iterative RWR. In Proceedings of the 7th ACM Conference on Recommender Systems, Hong Kong, China, 12–16 October 2013; pp. 399–402. [Google Scholar]
- Whissell, J.S.; Clarke, C.L.A. Effective Measures for Inter-Document Similarity. In Proceedings of the 22nd ACM International Conference on Conference on Information & Knowledge Management, San Francisco, CA, USA, 17 October–1 November 2013; pp. 1361–1370. [Google Scholar]
- Boyack, K.W.; Newman, D.; Duhon, R.J.; Klavans, R.; Patek, M.; Biberstine, J.R. Clustering More than Two Million Biomedical Publications: Comparing the Accuracies of Nine Text-Based Similarity Approaches. PLoS ONE 2011, 6, e18029. [Google Scholar] [CrossRef] [PubMed]
- Glenisson, P.; Glanzel, W.; Janssens, F.; De Moor, B. Combining full text and bibliometric information in mapping scientific disciplines. Inf. Process. Manag. 2005, 41, 1548–1572. [Google Scholar] [CrossRef]
- Landauer, T.K.; Laham, D.; Derr, M. From paragraph to graph: Latent semantic analysis for information visualization. Proc. Natl. Acad. Sci. USA 2004, 101 (Suppl. 1), 5214–5219. [Google Scholar] [CrossRef] [PubMed]
- Robertson, S.E.; Walker, S.; Beaulieu, M. Okapi at TREC-7: Automatic ad hoc, filtering, VLC and interactive. In Proceedings of the 7th Text REtrieval Conference (TREC 7), Gaithersburg, MD, USA, 1 July 1998; pp. 253–264. [Google Scholar]
- PubMed Help—PubMed Help—NCBI Bookshelf. Available online: https://www.ncbi.nlm.nih.gov/books/NBK3827/#pubmedhelp.Computation_of_Similar_Articl (accessed on 11 December 2018).
- Lin, J.; Wilbur, W.J. PubMed related articles: A probabilistic topic-based model for content similarity. BMC Bioinform. 2007, 8, 423. [Google Scholar] [CrossRef] [PubMed]
- Blei, D.M. Probabilistic topic models. Commun. ACM 2012, 55, 77–84. [Google Scholar] [CrossRef]
- Yau, C.-K.; Porter, A.L.; Newman, N.C.; Suominen, A. Clustering scientific documents with topic modeling. Scientometrics 2014, 100, 767–786. [Google Scholar] [CrossRef]
- Xie, P.; Xing, E.P. Integrating Document Clustering and Topic Modeling. In Proceedings of the Twenty-Ninth Conference on Uncertainty in Artificial Intelligence, Bellevue, WA, USA, 11–15 August 2013; pp. 694–703. [Google Scholar]
- The Comparative Toxicogenomics Database | CTD. Available online: http://ctdbase.org/ (accessed on 11 December 2018).
- Davis, A.P.; Grondin, C.J.; Johnson, R.J.; Sciaky, D.; King, B.L.; McMorran, R. The Comparative Toxicogenomics Database: Update 2017. Nucleic Acids Res. 2017, 45, D972–D978. [Google Scholar] [CrossRef] [PubMed]
- Wiegers, T.C.; Davis, A.P.; Cohen, K.B.; Hirschman, L.; Mattingly, C.J. Text mining and manual curation of chemical-gene-disease networks for the Comparative Toxicogenomics Database (CTD). BMC Bioinform. 2009, 10, 326. [Google Scholar] [CrossRef]
- Home—PMC—NCBI. Available online: https://www.ncbi.nlm.nih.gov/pmc/ (accessed on 11 December 2018).
- [Table, Stopwords]—PubMed Help—NCBI Bookshelf. Available online: https://www.ncbi.nlm.nih.gov/books/NBK3827/table/pubmedhelp.T.stopwords/ (accessed on 13 March 2018).
- MetaMap—A Tool For Recognizing UMLS Concepts in Text. Available online: https://metamap.nlm.nih.gov/ (accessed on 13 March 2018).
- GitHub—Senderle/Topic-Modeling-Tool: A Point-and-Click Tool for Creating and Analyzing Topic Models Produced by MALLET. Available online: https://github.com/senderle/topic-modeling-tool (accessed on 13 March 2018).
- Amigo, E.; Gonzalo, J.; Artiles, J.; Verdejo, F. A comparison of extrinsic clustering evaluation metrics based on formal constraints. Inf. Retr. 2009, 12, 461–486. [Google Scholar] [CrossRef]
- Banerjee, A.; Krumpelman, C.; Ghosh, J.; Basu, S.; Mooney, R.J. Model based overlapping clustering. In Proceedings of the International Conference on Knowledge Discovery and Data Mining, Chicago, IL, USA, 18 August 2005; pp. 532–537. [Google Scholar]
- Lewis, D.D.; Schapire, R.E.; Callan, P.; Papka, R. Training Algorithms for Linear Text Classifiers. In Proceedings of the 19th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval, Zurich, Switzerland, 18–22 August 1996; pp. 298–306. [Google Scholar]
- Liu, R.-L. Context-based Term Frequency Assessment for Text Classification. J. Am. Soc. Inf. Sci. Technol. 2010, 61, 300–309. [Google Scholar]
- Tong, T.; Dinakarpandian, D.; Lee, Y. Literature Clustering using Citation Semantics. In Proceedings of the 42nd Hawaii International Conference on System Sciences, Big Island, HI, USA, 5–8 January 2009. [Google Scholar]
- Janssens, F.; Zhang, L.; De Moor, B.; Glänzel, W. Hybrid clustering for validation and improvement of subject-classification schemes. Inf. Process. Manag. 2009, 45, 683–702. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).