Abstract
Bacterial type IV secretion systems (T4SSs) are related to not only secretion of effector proteins and virulence factors, but also to bacterial conjugation systems that promote bacterial horizontal gene transfer. The subgroup T4BSS, with a unique mosaic architecture system, consists of nearly 30 proteins that are similar to those from other secretory systems. Despite being intensively studied, the secretion mechanism of T4BSS remains unclear. This review systematically summarizes the protein composition, coding gene set, core complex, and protein interactions of T4BSS. The interactions of proteins in the core complex of the system and the operation mechanism between each element needs to be further studied.
1. Introduction
When bacterial growth is stimulated by the external environment, some effective factors are often secreted to enhance its viability. These effectors are usually secreted into host cells or the surrounding environment using unique recognition and transmission mechanisms through a protein secretion system (a system in which the bacteria depend on a secretory pathway for transcellular membrane transport of proteins) [1]. The protein secretion systems discovered so far can be divided into nine types according to the secretion mechanism of the outer membrane (OM), namely, type I to type IX secretion systems (T1SS to T9SS). Early results indicate that the first six secretory systems are prevalent in Gram-negative bacteria and T7SS was only found in Gram-positive bacteria, although the presence of T4SS in Gram-positive bacteria was also recently reported [2]. Many reports are found in the literature on T1SS to T6SS and T8SS, whereas T7SS and T9SS are newly discovered protein secretion systems whose assembly and secretion mechanisms are still unclear.
Unlike other Gram-negative bacterial secretion systems, T4SSs typically transport macromolecular substances such as nucleoprotein particles and deoxyribonucleic acid (DNA). At the same time, it can also mediate the horizontal transfer of DNA via conjugation, thereby promoting the plasticity of the bacterial genome and the transmission of resistance genes and other virulence genes [3,4,5,6]. T4SSs can be divided into different types according to different classification standards (Table 1) [7,8,9]. Based on biochemical composition and structural characteristics, it can be divided into T4ASS (typically represented by VirB/D4 systems in Agrobacterium tumefaciens), T4BSS (typically represented as Dot/Icm systems in Legionella pneumophila), T4CSS (typically represented as GI-type T4SS-like system in Gram-positive strain Streptococcus suis) [2], and genomic island (GI) T4SS (typically represented as ICE Hin1056 in Haemophilus influenza) [10]. The most prevalent and best studied are T4ASS and T4BSS. T4ASS is comprised of proteins encoded by 12 genes, whereas T4BSS is more complex, being comprised of ~30 different proteins. Unlike T4ASS, the constituent proteins of T4BSS contain components homologous to other secretory systems (T2SS, T3SS, T4ASS, and T6SS), highlighting the special mosaic structure of T4BSS [11].

Table 1.
Classification of T4SS.
2. Gene Composition of T4BSS
A typical T4BSS consists of proteins encoded by ~30 genes, which are named dot (defect in organelle trafficking) or icm (intracellular multiplication), as shown in Figure 1. The coding genes for the Dot/Icm system are mostly found in plasmids except the genes for T4BSS in the genera Legionella, Coxiella and Rickettsiella are in the chromosome [11]. The similarity of sequence of the genes suggests that these genes may have originated from a common ancestor (for instance between the tra system in R64 and Dot/Icm system) [12]. The genes encoding the T4BSS proteins tend to form different gene clusters: (a) the dotD-dotC-dotB gene cluster; (b) the dotM/icmP-dotL/icmO gene cluster; and (c) the dotI/icmL-dotH/icmK–dotG/icmE gene cluster [11]. Compared with other T4SSs (Figure 2b) [1,7,13], only dotL (icmO), dotG (icmE), and dotO (icmB) have homology with the corresponding genes virD4, virB10 and virB4 in T4ASS in sequence level.
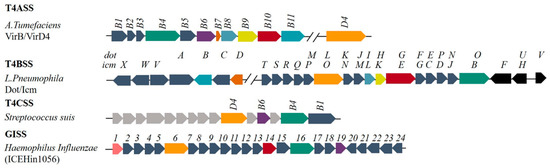
Figure 1.
Composition of different T4SSs and homology comparison. Blocks of the same color (green, purple, red, orange, deep orange, celadon, cyan and yellow) represent homologous proteins; light grey blocks in T4CSS represent proteins of unknown function.
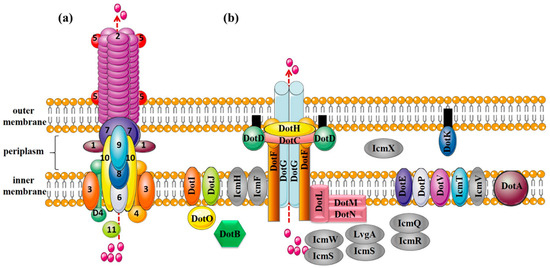
Figure 2.
Protein composition of T4ASS (the VirB/VirD4 system from Agrobacterium tumefaciens) and T4BSS (the Dot/Icm system from Legionella pneumophila). (a) T4ASS. Outer membrane components: VirB7, VirB9, and VirB10; inner membrane components: VirB3, VirB6, and VirB8; pilus components: VirB2 (major pilus component) and VirB5 (minor pilus component); lytic transglycosylase: VirB1; nucleoside triphosphatases: VirB4 and VirB11; coupling protein: VirD4; (b) T4BSS. Core complex: DotC-DotD-DotH-DotF-DotG; substrate recognition protein: DotF, DotL, DotM, DotN, IcmSW and IcmS-LvgA; other functional proteins: DotE, DotV, DotP, DotI, DotJ, DotA, DotB, IcmQ, IcmR, IcmF, IcmH, DotK, and DotO; proteins with unknown functions: IcmT, IcmV, IcmX.
3. Mosaic Structure Model of T4BSS
Components of T4BSS include nearly 30 proteins, and most of them are homologous with the components of T2SS, T3SS, T4ASS and T6SS, highlighting the greater complexity of the components of this secretion system. Similar to other secretion systems, secretion of T4BSS effector proteins and conjugal transfer of DNA are also achieved through a series of interactions between different constituent proteins, including receptor proteins that recognize substrates, ATPases that provide energy, proteins that constitute transport channels, and other auxiliary proteins. Depending on the function of the constituent proteins, they can be classified into the following four categories.
3.1. Core Complex
The core complex of T4ASS, the encoding genes which are located in the IncN plasmid pKM101, consists of three proteins, VirB7, VirB9, and VirB10, in a 1:1:1 ratio. VirB7 is in the outermost layer, surrounding the complex to form a ring; VirB9 is in the middle, forming the sides of the complex; and VirB10 is oriented toward the central cavity and inserted into the outer membrane with two α-helices per monomer [14]. The core complex of T4BSS consists of five proteins, DotC-DotD-DotH-DotF-DotG, forming the skeleton structure of the whole secretion system (Figure 2b). DotC and DotD are OM lipoproteins, and their coding genes are located in the same gene cluster. Although there are few reports on DotC, previous studies showed that the ring structure of the core complex in any mutant (ΔdotC, ΔdotD or ΔdotH) cannot be detected by transmission electron microscopy, and the expression levels of other proteins are also reduced, indicating the importance of DotC in T4BSS [15]. OM protein DotH binds to DotC and DotD to form a DotC-DotD-DotH OM complex [16,17]. The expression levels of these three proteins are related and crucial to the formation of circular channels in the core complex. The similarity between the N-terminal domain of VirB9 and residues 168–227 of DotH is as high as 49% (Table 2, Figure 3j). DotH structure was predicted by various programs (I-TASSER, Phyre2 and Quark), was speculated that DotH contains two or more separate domains (Figure 3c) [18]. OM-lipoprotein DotD consists of a disordered N-terminal domain and a globular C-terminal N0 secretin domain [19], in which a disordered N-terminal consisting of 46 amino acid residues is homologous to the mature VirB7 (a small peptide consisting of 33 amino acid residues) [20]. Both DotG and DotF are membrane proteins with a single transmembrane helix at the N-terminus. The DotG protein spans the inner membrane and outer membrane, forming the central channel of the core complex, with some homology to VirB10 at the extreme C-terminal domain sequence (Table 2) [21,22]. Because DotG has a variable region of about 600 residues that is rich in repeats, it tends to be longer than the VirB10 protein, as shown in Figure 3d,k [23]. In the absence of DotG, the outer diameter of the ring structure of the core complex decreases slightly, and the central hole increases (11.3 ± 0.1 nm in diameter) by electron microscopy analysis [15]. The inner membrane (IM) protein DotF (~30 kDa) consists of 269 amino acid residues in three domains: the cytoplasmic domain (~20 amino acids), the transmembrane domain (~50 amino acids), and the periplasmic domain (~200 amino acids) [17]. Ghosal et al. [18] speculated that the β-helices of DotG might have the same function in T4BSSs as pili in T4ASSs. Both DotF and OM protein DotD are part of the ring structure. Studies have shown that DotF is not essential for the transfer of effectors and is mainly responsible for the stabilization of the DotG channel or for triggering conformational changes [15,18]. Kubori et al. [15] knocked out the dotF gene in L. pneumophila and found one of ring structure of the core complex with a larger central pore that is similar to the ring structure of the ΔdotG mutant strain, indicating that DotF possesses the function of promoting the stability or assembly of active core complexes containing DotG.

Table 2.
Homologous proteins in T4BSS and other secretion systems.
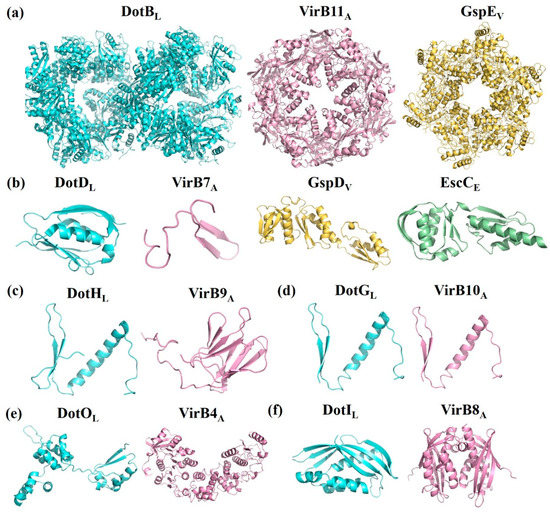
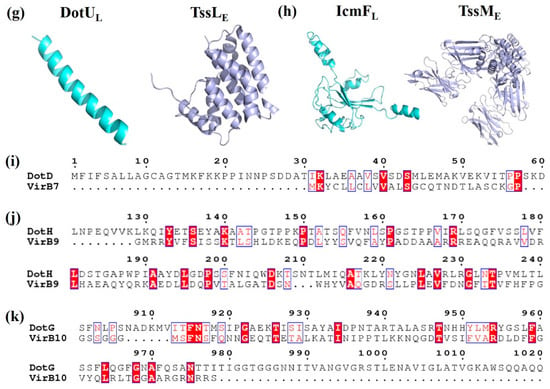
Figure 3.
Crystal structure and amino acid sequences alignment of T4BSS proteins and their structural homologs. (a) ATPases of Legionella pneumophila DotBL (PDB id: 6GEB), Agrobacterium tumefaciens VirB11A, Vibrio fluvialis GspEV; (b) Outer membrane proteins of L. pneumophila DotDL (PDB id: 3ADY), A. tumefaciens VirB7A, Vibrio fluvialis GspDV, and Escherichia coli EscC (PDB id: 3GR5); (c) Periplasmic proteins of L. pneumophila DotHL, A. tumefaciens VirB9A; (d) Membrane-spanning proteins of L. pneumophila DotGL and A. tumefaciens VirB10A; (e) Inner membrane (IM)-associated proteins of L. pneumophila DotOL and A. tumefaciens VirB4A; (f) IM proteins of L. pneumophila DotIL and A. tumefaciens VirB8A; (g) Inner membrane proteins of L. pneumophila DotUL, and E. coli TssLE (PDB id: 3U66); (h) IM proteins of L. pneumophila IcmFL and E. coli TssME (PDB id: 4Y7M); (i) N-terminal domain of L. pneumophila DotDL and A. tumefaciens VirB7A; (j) Residues 168–227 of L. pneumophila DotHL and N-terminal domain of A. tumefaciens VirB9A; (k) C-terminal domain of L. pneumophila DotGL and A. tumefaciens VirB10A.
3.2. Substrate Recognition Protein
In addition to the robust central channel protein DotG, DotF also has a substrate recognition function [24], which recognizes the effector RalF secreted by the Dot/Icm system. Sutherland et al. [25] explored the ability of DotF to recognize other effectors and the results indicate that DotF can only bind to certain types of substrates, that is, DotF is not the main substrate recognition protein in the Dot/Icm system.
IM AAA+ ATPase DotL is a member of the type IV coupling protein that is functionally similar to other T4SS coupling proteins [26]. Structurally, the C-terminal cytoplasmic domain of DotL has a conserved Walker A motif and the N-terminus forms a hexameric assembly in the IM, which is related to the FtsK/SpoIIIE family of DNA translocation motors [27,28]. As key components of T4SS, coupling proteins are mainly responsible for the localisation of substrates at transmembrane receptors of the secretion system [29] and the hydrolysis of ATP to provide energy for substrate transfer [30]. DotL also forms a subcomplex with DotM, DotN, IcmS, and IcmW [31]. The C-terminal tail of DotL bound to DotN and binary complex IcmSW separately [32] and the recruitment of effectors independent of IcmSW is mediated by a secretion signal sequence rich in Glu residues and located at the C terminus, which is proved that DotM acts as a potential binding platform for the recruitment of acidic Glu-rich, IcmSW-independent effectors [33,34]. In view of the fact that DotL binds directly to DotM, it is suggested that DotM may regulate the expression of DotL. DotN is a IM/cytoplasmic protein rich in cysteine with a small molecular weight that can stabilize the structure of DotL and DotM complexes, although its specific mechanism of action remains unclear [35]. IcmS and IcmW are small molecular weight soluble cytosolic proteins, often found in IcmSW binary complexes. There is a binding site for IcmSW in the C-terminus of DotL. When this binding site is mutated, the transport capacity of IcmSW-dependent substrates such as SdeA, SidD, and VipA decreases. Conversely, there is no effect on the transport of some non-IcmSW-dependent substrates such as RalF, LnaB, and LidA [31,35]. When the expression levels of IcmS or IcmW are reduced or even eliminated, the infection ability of DotL decreases, indicating that IcmSW may be used as a targeting factor to recruit substrates to a location near DotL.
OM protein LvgA is approximately 27 kDa in size. Although the existence of the ternary complex has not been confirmed, lacking of LvgA in L. pneumophila affects the protein stability of IcmS and IcmW, indicating that LvgA is associated with IcmS and IcmW. Its coding gene lvgA is a novel kind of L. pneumophila virulence factor. Deletion of the LvgA-encoding gene causes partial defects in intracellular replication but it does not affect the expression level of DotA and IcmX [36]. LvgA is not clearly related to any of these previously described virulence factors on genetic or structural grounds. Up to now, the mechanism by which lvgA exerts its phenotype is unclear [37].
3.3. Other Functional Proteins
Both DotE and DotV are IM proteins composed of four-transmembrane helix bundles that functionally resemble TraQ in I-type conjugation systems. Similar to DotE and DotV, DotP has a cleavable signal sequence at the N-terminus in addition to two transmembrane helices. The genes encoding these three proteins are downstream of the DotF-encoding gene and are arranged in the order of dotF, dotE, dotV, and dotP [11].
DotI is also an IM protein with a molecular weight of 23 kDa. The amino acid sequence is homologous to that of the TraM protein in the plasmid R64 conjugative mobilization system, and the C-terminal protein structure is structural homologs to that of VirB8 in T4ASS (Table 2, Figure 3f). Both DotI and VirB8 form a stack of oligomeric rings in vivo [38,39] (Figure 3f). The expression levels of DotI are related to its partial homolog DotJ and the protein forms an IM hetero-complex with DotJ at a weight ratio of 1.87:1, which is closely related to the integrity of the inner proteins but does not appear to be associated with the core complex of T4BSS. Contrary to DotJ, VirB8 has an indispensable role in T4ASS. The dotI and dotJ genes, coding for DotI and DotJ, are upstream of the core complex DotF, DotG, and DotH protein-encoding genes. DotIJ forms a ring around the substrate translocation channel, and its role may be related to the IM-associated ATPase DotO [21,38]. DotI is present in all known T4BSS, whereas DotJ is currently only found in bacteria of the order Legionellales, such as genus Legionella and Rickettsiella. There is a certain similarity between the N-terminal sequences of DotJ and DotI, although DotJ does not have a periplasmic domain [39]. Similar to VirB4 of T4ASS, DotO contains Walker A and B motifs (Table 2) [11], is also an IM-associated ATPase, and both of them are the most conserved components of the T4SS superfamily (Figure 3e) [40].
DotA is an IM protein consisting of eight membrane-spanning domains, two large periplasmic domains (approximately 503 and 73 amino acids), and a small C-terminal cytoplasmic domain (122 amino acids) [41]. Matthews et al. reported that DotA and the periplasmic IcmX are in a T4BSS-dependent manner [42]. The periplasmic domain of DotA is located at the top of the central channel, which can stabilize the channel wall or be released [41]. DotA is only found in bacteria of the genus Legionella, forming a cyclic oligomer in the extracellular domain and binding to a protein of unknown function with 46 kDa [11]. When the bacterial dotA gene is deleted, the mutant strain is unable to infect the host cell through the T4BSS secreting effector protein [43].
DotB is an ATPase mostly present in the cytoplasm, which is similar to ATPases of T2SS and T4SS, all of which form hexameric rings. DotB is essential to the normal function of T4BSS. Sexton et al. [44] studied the structure and function of DotB by constructing a series of dotB allele mutants, and found that DotB is composed of an N-terminal domain interacting with cell membranes, an ATPase domain, and a C-terminal domain of unknown function. It is involved in the secretion of effector proteins by T4BSS, and the ΔdotB mutant failed to secrete T4BSS substrates and productively infect host cells [43]. The IcmQ and IcmR proteins located in the in bacterial cytoplasm are essential for the activity of the Dot/Icm system [45]. IcmQ has a small N-terminal domain and a large C-terminal domain. The purified IcmQ is prone to aggregation, and addition of IcmR inhibits this aggregation, indicating that IcmQ and IcmR have a relationship similar to a substrate and a molecular chaperone. The behavior of IcmR regulates IcmQ is similar to that observed in some chaperones [46,47]. The crystal structure of IcmR-IcmQ complex shows an amphipathic four-helix bundle (two helices each from IcmR and IcmQ). IcmQ acts on the lipid vesicles that forms and causes them to rupture. The mechanism of action consists of insertion of the N-terminal domain into vesicles of the phospholipid bilayer, and destruction of the membrane structure by the C-terminal domain with targeting of the membrane by electrostatic action. The protein also has an NAD+ binding site that allows IcmR-IcmQ to bind to the membrane, thereby interacting with or modifying the T4BSS substrate [48].
The IM proteins IcmF and IcmH are present in the T4BSS of Legionella and Legionella-related genera, which are responsible for intracellular growth, immediate cytotoxicity or salt sensitivity [49,50]. These proteins are essential for the normal function of the Dot/Icm system. When any of them is lost, the amount of core complexes decreases, especially the levels of DotG and DotH, but a partially functional Icm/Dot complex is probably present in the bacteria [50]. Therefore, IcmF and IcmH work together to maintain the stability of the core complex and recruit DotC-H subcomplexes to the poles to initiate assembly because they are the polar localization factors [18]. Lipoprotein DotK has many homologous proteins in other bacteria, is conserved in Legionella and Coxiella species, and is responsible for anchoring the Dot/Icm system to the peptidoglycan layer [51]. When the DotK coding gene is knocked out, Legionella shows partial growth defects into the protozoan host, but it has no effect on the growth on macrophages. [23,36].
IcmX is a periplasmic protein with a molecular weight of 50 kDa. IcmX is indispensable for the activity of the Dot/Icm system and is conserved in the Legionellaceae family [42]. Even though T4BSS still assembles in the absence of IcmX, the intracellular growth of the ΔicmX mutant is severely affected [18,42].
3.4. Proteins of Unknown Function
In previous studies [11,42], many proteins with unknown functions are described in T4BSS. IcmT is a small molecular weight IM protein that is essential for the function of T4BSS and the coding gene of IcmT contains significant sequence homology to a gene coding for conjugation-related proteins in the IncI plasmid R64 [12,52]. IcmV is also an IM protein with unknown function that is not conserved in all T4BSS families. Similar to IcmW coding gene icmW, the coding gene of IcmV has an overlapping regulatory region, which probably serves as binding sites for regulatory proteins [53].
4. Comparison of T4BSS with Other Secretion Systems
In the currently studied Gram-negative bacterial T1SS to T6SS, T1SS, T3SS, T4SS, and T6SS are one-step Sec-independent secretion systems [54], that is, the target protein is directly secreted from the cell to the extracellular environment, not into the periplasmic space. In contrast, T2SS and T5SS are two-step Sec-dependent secretion systems [55,56,57]: The effectors are transported by substrate recognition proteins to the transmembrane channel, which is then translocated across the IM to the periplasmic space, forming its periplasmic intermediate form. Subsequently, the periplasmic intermediate form is transported to the extracellular space through a channel protein across the OM (Figure 4).
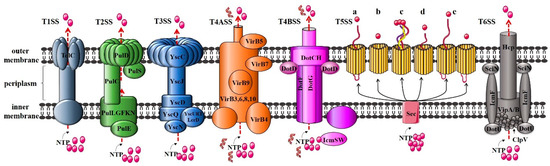
Figure 4.
Protein components of T1SS to T6SS. T1SS: haemolysin A (HlyA) secretion system in Escherichia coli; T2SS: pullulanase secretion system in Klebsiella oxytoca; T3SS: yop secretion system in Yersinia; T4ASS: VirB/VirD4 secretion system in Agrobacterium tumefaciens; T4BSS: Dot/Icm system in Legionella pneumophila; T5SS: Type Va (autotransporter system); Type Vb (two-partner secretion systems); Type Vc (trimeric autotransporters); Type Vd (encoding a patatin-like passenger domain fused to a C-terminal β-barrel domain that resembles TpsB proteins); Type Ve (inverse autotransporters and two-partner inverse autotransporters); and T6SS: secretion system in Vibrio cholera.
T1SS, also known as ATP-binding cassette (ABC) secretion system, is found in all bacterial genomes that have been fully sequenced. It is mainly used for translocating virulence factors for extracellular transport [58]. The composition of the T1SS secretion system is very simple, mainly including three functional proteins: ABC translocating enzyme that provides energy for effector secretion in the IM, membrane fusion protein (MFP, IM protein) for movement across the periplasmic space, and OM protein (OMP) [59], corresponding to HlyB, HlyD, and TolC in a typical α-haemolysin secretion system (E. coli), respectively. Taking Klebsiella oxytoca as an example, the main components of T2SS include a series of IM proteins referred to as PulLGFKN, that are intracellular secretory proteins transported across the membrane using energy provided by the ATPase PulE. The molecular chaperone PulC that binds to secreted proteins in the periplasmic space, the channel protein PulD inserted into the OM, and the substrate recognition protein PulS [60]. Similar to T2SS, T3SS also includes chaperones, translocators, and effector proteins. Unlike other secretory systems, the secretion of effector proteins by T3SS is dependent on the 5′ end of the mRNA, the N-terminus of the secreted protein, and the binding of secretory proteins to corresponding chaperone proteins [61]. T5SS just consists of a secreted moiety and a putative membrane translocator domain and is the simplest secretory device. It is proposed that these virulence factors are autonomous secretion systems [62,63]. Currently, T5SSs are classified into five classes (Types Va–Ve) based on their domain architecture [64,65]. The genes for T6SS are often arranged together to form a gene cluster, which encode 13 to 25 proteins that form core and auxiliary components [66].
T4BSS is obviously different from other secretory systems in its special mosaic structure and different constituent proteins, which are homologous with different secretory system components. IcmS and IcmW of T4BSS perform the same function as the coupling proteins of T3SS. Loss of these proteins affects intracellular growth but has no significant effect on cytotoxicity [67]. The function of the amorphous N-terminal of the core complex protein DotD is similar to that of VirB7 in T4ASS (Figure 3i) [68]. In addition, the spherical structure domain of the amorphous N-terminal of DotD shares also homology with the N0/T3S domain of the OM proteins GspD (T2SS) and EscC (T3SS) [19] (Table 2, Figure 3b). The C-terminus of DotG is similar to VirB10 of T4ASS in the amino acid sequence level (Figure 3d). VirB10 is adjacent to the central channel, thus indicating that DotG is likely to be involved in the formation of a circular OM core complex [69]. The ATPase DotB is closely related to the ATPase PilT in the type IV pilus system, and it shares higher homology with GspE of T2SS than VirB11 of T4ASS [70] (Table 2, Figure 3a). It is especially important that DotU/IcmH and IcmF are highly homologous to the constituent proteins TssL and TssM of T6SS [49,71] (Table 2, Figure 3g,h). These results once again confirm the unique mosaic structure of the protein composition in T4BSS.
5. Prospects
Unlike other bacterial secretion systems, T4BSS can not only transport effectors but also can horizontally transfer genetic elements such as pathogenicity genes, resistance genes and genes for degradation of organic pollutants that are difficult to degrade. Compared with T4ASS, T4BSS with its mosaic structure has a wider host range and is more complex. If the assembly model and secretion mechanism of T4BSS can be fully clarified, it will be beneficial to the prevention or control of some diseases and the solution of environmental pollution problems. Both medical disease treatment and environmental engineering protection have a breakthrough significance. Many reports on the Dot/Icm system of L. pneumophila are found in the literature, and the core complex composed of DotC-DotD-DotH-DotG-DotF is intensively studied. However, the structure and function of many proteins in the system other than the core complex are still unknown, and urgently need to be further studied. In addition to solving the molecular structure and function of all participating proteins, how these proteins communicate with each other, how the signal transduction is induced, and the proteins cooperate with each other to jointly complete the secretion of effector proteins and DNA, should be addressed in future studies.
Author Contributions
Conceptualization, S.L. and S.W.; investigation, D.W. and D.D.; writing—original draft preparation, S.W.; writing—review & editing, S.L. and W.Y.
Funding
This research was funded by National Natural Science Foundation of China, grant number 31670512 and 31300438, Natural Science Basic Research Plan in Shaanxi Province of China, program number 2018JM3039 and China Postdoctoral Science Foundation funded project, number 2014M552456.
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Green, E.R.; Mecsas, J. Bacterial Secretion Systems: An Overview. Microbiol. Spectr. 2016, 4, 19. [Google Scholar] [CrossRef]
- Zhang, W.; Rong, C.; Chen, C.; Gao, G.F. Type-IVC Secretion System: A Novel Subclass of Type IV Secretion System (T4SS) Common Existing in Gram-Positive Genus Streptococcus. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Fronzes, R.; Christie, P.J.; Waksman, G. The structural biology of type IV secretion systems. Nat. Rev. Microbiol. 2009, 7, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Waksman, G.; Fronzes, R. Molecular architecture of bacterial type IV secretion systems. Trends Biochem. Sci. 2010, 35, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Wallden, K.; Rivera-Calzada, A.; Waksman, G. Type IV secretion systems: Versatility and diversity in function. Cell. Microbiol. 2010, 12, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Cascales, E.; Christie, P.J. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 2003, 1, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Julin, D. Plasmid Incompatibility. Mol. Life Sci. 2014. [Google Scholar] [CrossRef]
- Darbari, V.C.; Waksman, G. Structural Biology of Bacterial Type IV Secretion Systems. Ann. Rev. Biochem. 2015, 84, 603–629. [Google Scholar] [CrossRef] [PubMed]
- Guglielmini, J.; Neron, B.; Abby, S.S.; Pilar Garcillan-Barcia, M.; de la Cruz, F.; Rocha, E.P.C. Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res. 2014, 42, 5715–5727. [Google Scholar] [CrossRef] [PubMed]
- Juhas, M.; Crook, D.W.; Dimopoulou, I.D.; Lunter, G.; Harding, R.M.; Ferguson, D.J.P.; Hood, D.W. Novel type IV secretion system involved in propagation of genomic islands. J. Bacteriol. 2007, 189, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Nagai, H.; Kubori, T. Type IVB secretion systems of Legionella and other Gram-negative bacteria. Front. Microbiol. 2011, 2. [Google Scholar] [CrossRef] [PubMed]
- Komano, T.; Yoshida, T.; Narahara, K.; Furuya, N. The transfer region of Incl1 plasmid R64: Similarities between R64 tra and Legionella icm/dot genes. Mol. Microbiol. 2000, 35, 1348–1359. [Google Scholar] [CrossRef] [PubMed]
- Juhas, M.; Crook, D.W.; Hood, D.W. Type IV secretion systems: Tools of bacterial horizontal gene transfer and virulence. Cell. Microbiol. 2008, 10, 2377–2386. [Google Scholar] [CrossRef] [PubMed]
- Franco, I.S.; Shuman, H.A.; Charpentier, X. The perplexing functions and surprising origins of Legionella pneumophila type IV secretion effectors. Cell. Microbiol. 2009, 11, 1435–1443. [Google Scholar] [CrossRef] [PubMed]
- Kubori, T.; Koike, M.; Bui, X.T.; Higaki, S.; Aizawa, S.-I.; Nagai, H. Native structure of a type IV secretion system core complex essential for Legionella pathogenesis. Proc. Natl. Acad. Sci. USA 2014, 111, 11804–11809. [Google Scholar] [CrossRef] [PubMed]
- Ghosal, D.; Chang, Y.-W.; Jeong, K.C.; Vogel, J.P.; Jensen, G.J. In situ structure of the Legionella Dot/Icm type IV secretion system by electron cryotomography. EMBO Rep. 2017, 18, 726–732. [Google Scholar] [CrossRef] [PubMed]
- Vincent, C.D.; Friedman, J.R.; Jeong, K.C.; Buford, E.C.; Miller, J.L.; Vogel, J.P. Identification of the core transmembrane complex of the Legionella Dot/Icm type IV secretion system. Mol. Microbiol. 2006, 62, 1278–1291. [Google Scholar] [CrossRef] [PubMed]
- Ghosal, D.; Chang, Y.; Jeong, K.; Vogel, J.; Jensen, G. Molecular architecture of the Legionella Dot/Icm type IV secretion system. bioRxiv 2018. [Google Scholar] [CrossRef]
- Nakano, N.; Kubori, T.; Kinoshita, M.; Imada, K.; Nagai, H. Crystal Structure of Legionella DotD: Insights into the Relationship between Type IVB and Type II/III Secretion Systems. PLoS Pathog. 2010, 6. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Martinez, C.E.; Christie, P.J. Biological Diversity of Prokaryotic Type IV Secretion Systems. Microbiol. Mol. Biol. Rev. 2009, 73, 775–808. [Google Scholar] [CrossRef] [PubMed]
- Kubori, T.; Nagai, H. The Type IVB secretion system: An enigmatic chimera. Curr. Opin. Microbiol. 2016, 29, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Christie, P.J.; Gomez Valero, L.; Buchrieser, C. Biological Diversity and Evolution of Type IV Secretion Systems. Curr. Top. Microbiol. Immunol. 2017, 413, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Segal, G.; Purcell, M.; Shuman, H.A. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 1998, 95, 1669–1674. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.Q.; Isberg, R.R. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. USA 2004, 101, 841–846. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, M.C.; Binder, K.A.; Cualing, P.Y.; Vogel, J.P. Reassessing the Role of DotF in the Legionella pneumophila Type IV Secretion System. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Buscher, B.A.; Conover, G.M.; Miller, J.L.; Vogel, S.A.; Meyers, S.N.; Isberg, R.R.; Vogel, J.P. The DotL protein, a member of the TraG-coupling protein family, is essential for viability of Legionella pneumophila strain Lp02. J. Bacteriol. 2005, 187, 2927–2938. [Google Scholar] [CrossRef] [PubMed]
- Aussel, L.; Barre, F.X.; Aroyo, M.; Stasiak, A.; Stasiak, A.Z.; Sherratt, D. FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell 2002, 108, 195–205. [Google Scholar] [CrossRef]
- Massey, T.H.; Mercogliano, C.P.; Yates, J.; Sherratt, D.J.; Lowe, J. Double-stranded DNA translocation: Structure and mechanism of hexameric FtsK. Mol. Cell 2006, 23, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Atmakuri, K.; Cascales, E.; Christie, P.J. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol. Microbiol. 2004, 54, 1199–1211. [Google Scholar] [CrossRef] [PubMed]
- Tato, I.; Zunzunegui, S.; de la Cruz, F.; Cabezon, E. TrwB, the coupling protein involved in DNA transport during bacterial conjugation, is a DNA-dependent ATPase. Proc. Natl. Acad. Sci. USA 2005, 102, 8156–8161. [Google Scholar] [CrossRef] [PubMed]
- Vincent, C.D.; Friedman, J.R.; Jeong, K.C.; Sutherland, M.C.; Vogel, J.P. Identification of the DotL coupling protein subcomplex of the Legionella Dot/Icm type IV secretion system. Mol. Microbiol. 2012, 85, 378–391. [Google Scholar] [CrossRef] [PubMed]
- Kwak, M.-J.; Kim, J.D.; Kim, H.; Kim, C.; Bowman, J.W.; Kim, S.; Joo, K.; Lee, J.; Jin, K.S.; Kim, Y.-G.; et al. Architecture of the type IV coupling protein complex of Legionella pneumophila. Nat. Microbiol. 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Lifshitz, Z.; Burstein, D.; Peeri, M.; Zusman, T.; Schwartz, K.; Shuman, H.A.; Pupko, T.; Segal, G. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc. Natl. Acad. Sci. USA 2013, 110, E707–E715. [Google Scholar] [CrossRef] [PubMed]
- Meir, A.; Chetrit, D.; Liu, L.; Roy, C.R.; Waksman, G. Legionella DotM structure reveals a role in effector recruiting to the Type 4B secretion system. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, M.C.; Thuy Linh, N.; Tseng, V.; Vogel, J.P. The Legionella IcmSW Complex Directly Interacts with DotL to Mediate Translocation of Adaptor-Dependent Substrates. PLoS Pathog. 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Segal, G.; Shuman, H.A. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect. Immun. 1999, 67, 2117–2124. [Google Scholar] [PubMed]
- Edelstein, P.H.; Hu, B.F.; Higa, F.; Edelstein, M.A.C. lvgA, a novel Legionella pneumophila virulence factor. Infect. Immun. 2003, 71, 2394–2403. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, J.J.; Phan, I.Q.H.; Scheib, H.; Subramanian, S.; Edwards, T.E.; Lehman, S.S.; Piitulainen, H.; Rahman, M.S.; Rennoll-Bankert, K.E.; Staker, B.L.; et al. Structural Insight into How Bacteria Prevent Interference between Multiple Divergent Type IV Secretion Systems. Mbio 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, T.; Kubori, T.; Bui, X.T.; Hyakutake, A.; Uchida, Y.; Imada, K.; Nagai, H. Molecular and structural analysis of Legionella DotI gives insights into an inner membrane complex essential for type IV secretion. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Pena, A.; Matilla, I.; Martin-Benito, J.; Valpuesta, J.M.; Carrascosa, J.L.; de la Cruz, F.; Cabezon, E.; Arechaga, I. The Hexameric Structure of a Conjugative VirB4 Protein ATPase Provides New Insights for a Functional and Phylogenetic Relationship with DNA Translocases. J. Biol. Chem. 2012, 287. [Google Scholar] [CrossRef] [PubMed]
- Roy, C.R.; Isberg, R.R. Topology of Legionella pneumophila DotA: An inner membrane protein required for replication in macrophages. Infect. Immun. 1997, 65, 571–578. [Google Scholar] [PubMed]
- Matthews, M.; Roy, C.R. Identification and subcellular localization of the Legionella pneumophila IcmX protein: A factor essential for establishment of a replicative organelle in eukaryotic host cells. Infect. Immun. 2000, 68, 3971–3982. [Google Scholar] [CrossRef] [PubMed]
- Beare, P.A.; Larson, C.L.; Gilk, S.D.; Heinzen, R.A. Two Systems for Targeted Gene Deletion in Coxiella burnetii. Appl. Environ. Microbiol. 2012, 78, 4580–4589. [Google Scholar] [CrossRef] [PubMed]
- Sexton, J.A.; Yeo, H.J.; Vogel, J.P. Genetic analysis of the Legionella pneumophila DotB ATPase reveals a role in type IV secretion system protein export. Mol. Microbiol. 2005, 57, 70–84. [Google Scholar] [CrossRef] [PubMed]
- Coers, J.; Monahan, C.; Roy, C.R. Modulation of phagosome biogenesis by Legionella pneumophila creates an organelle permissive for intracellular growth. Nat. Cell Biol. 1999, 1, 451–453. [Google Scholar] [CrossRef] [PubMed]
- Dumenil, G.; Isberg, R.R. The Legionella pneumophila IcmR protein exhibits chaperone activity for IcmQ by preventing its participation in high-molecular-weight complexes. Mol. Microbiol. 2001, 40, 1113–1127. [Google Scholar] [CrossRef] [PubMed]
- Raychaudhury, S.; Farelli, J.D.; Montminy, T.P.; Matthews, M.; Menetret, J.-F.; Dumenil, G.; Roy, C.R.; Head, J.F.; Isberg, R.R.; Akey, C.W. Structure and Function of Interacting IcmR-IcmQ Domains from a Type IVb Secretion System in Legionella pneumophila. Structure 2009, 17, 590–601. [Google Scholar] [CrossRef] [PubMed]
- Farelli, J.D.; Gumbart, J.C.; Akey, I.V.; Hempstead, A.; Amyot, W.; Head, J.F.; McKnight, C.J.; Isberg, R.R.; Akey, C.W. IcmQ in the Type 4b Secretion System Contains an NAD(+) Binding Domain. Structure 2013, 21, 1361–1373. [Google Scholar] [CrossRef] [PubMed]
- Sexton, J.A.; Miller, J.L.; Yoneda, A.; Kehl-Fie, T.E.; Vogel, J.P. Legionella pneumophila DotU and IcmF are required for stability of the Dot/Icm complex. Infect. Immun. 2004, 72, 5983–5992. [Google Scholar] [CrossRef] [PubMed]
- Zusman, T.; Feldman, M.; Halperin, E.; Segal, G. Characterization of the icmH and icmF genes required for Legionella pneumophila intracellular growth, genes that are present in many bacteria associated with eukaryotic cells. Infect. Immun. 2004, 72, 3398–3409. [Google Scholar] [CrossRef] [PubMed]
- Morozova, I.; Qu, X.Y.; Shi, S.D.; Asamani, G.; Greenberg, J.E.; Shuman, H.A.; Russo, J.J. Comparative sequence analysis of the icm/dot genes in Legionella. Plasmid 2004, 51, 127–147. [Google Scholar] [CrossRef] [PubMed]
- Solomon, J.M.; Rupper, A.; Cardelli, J.A.; Isberg, R.R. Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect. Immun. 2000, 68, 2939–2947. [Google Scholar] [CrossRef] [PubMed]
- Gal-Mor, O.; Zusman, T.; Segal, G. Analysis of DNA regulatory elements required for expression of the Legionella pneumophila icm and dot virulence genes. J. Bacteriol. 2002, 184, 3823–3833. [Google Scholar] [CrossRef] [PubMed]
- Hodges, K.; Hecht, G. Interspecies communication in the gut, from bacterial delivery to host-cell response. J. Physiol. Lond. 2012, 590, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Henderson, I.R.; Navarro-Garcia, F.; Desvaux, M.; Fernandez, R.C.; Ala’Aldeen, D. Type V protein secretion pathway: The autotransporter story. Microbiol. Mol. Biol. Rev. 2004, 68, 692–744. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kendall, D.A. Sec-dependent protein export and the involvement of the molecular chaperone SecB. Cell Stress Chaperones 2000, 5, 267–275. [Google Scholar] [CrossRef]
- Van Wely, K.H.M.; Swaving, J.; Freudl, R.; Driessen, A.J.M. Translocation of proteins across the cell envelope of Gram-positive bacteria. Fems Microbiol. Rev. 2001, 25, 437–454. [Google Scholar] [CrossRef] [PubMed]
- Costa, T.R.D.; Felisberto-Rodrigues, C.; Meir, A.; Prevost, M.S.; Redzej, A.; Trokter, M.; Waksman, G. Secretion systems in Gram-negative bacteria: Structural and mechanistic insights. Nat. Rev. Microbiol. 2015, 13, 343–359. [Google Scholar] [CrossRef] [PubMed]
- Omori, K.; Idei, A. Gram-negative bacterial ATP-binding cassette protein exporter family and diverse secretory proteins. J. Biosci. Bioeng. 2003, 95, 1–12. [Google Scholar] [CrossRef]
- Michel, G.P.F.; Voulhoux, R. Type II Secretory System (T2SS) in Gram-negative Bacteria: A Molecular Nanomachine for Secretion of Sec and Tat-Dependent Extracellular Proteins. In Bacterial Secreted Proteins: Secretory Mechanisms and Role in Pathogenesis; Wooldridge, K., Ed.; Caister Academic Press: Norfolk, UK, 2009; pp. 67–92. [Google Scholar] [CrossRef]
- Gauthier, A.; Puente, J.L.; Finlay, B.B. Secretin of the enteropathogenic Escherichia coli type III secretion system requires components of the type III apparatus for assembly and localization. Infect. Immun. 2003, 71, 3310–3319. [Google Scholar] [CrossRef] [PubMed]
- Pohlner, J.; Halter, R.; Beyreuther, K.; Meyer, T.F. Gene structure and extracellular secretion of neisseria-gonorrhoeae iga protease. Nature 1987, 325, 458–462. [Google Scholar] [CrossRef] [PubMed]
- Jose, J.; Jahnig, F.; Meyer, T.F. Common structural features of iga1 protease-like outer-membrane protein autotransporters. Mol. Microbiol. 1995, 18, 378–380. [Google Scholar] [CrossRef] [PubMed]
- Casasanta, M.A.; Yoo, C.C.; Smith, H.B.; Duncan, A.J.; Cochrane, K.; Varano, A.C.; Allen-Vercoe, E.; Slade, D.J. A chemical and biological toolbox for Type Vd secretion: Characterization of the phospholipase A1 autotransporter FplA from Fusobacterium nucleatum. J. Biol. Chem. 2017, 292, 20240–20254. [Google Scholar] [CrossRef] [PubMed]
- Albenne, C.; Ieva, R. Job contenders: Roles of the beta-barrel assembly machinery and the translocation and assembly module in autotransporter secretion. Mol. Microbiol. 2017, 106, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Silverman, J.M.; Austin, L.S.; Hsu, F.; Hicks, K.G.; Hood, R.D.; Mougous, J.D. Separate inputs modulate phosphorylation-dependent and -independent type VI secretion activation. Mol. Microbiol. 2011, 82, 1277–1290. [Google Scholar] [CrossRef] [PubMed]
- Coers, J.; Kagan, J.C.; Matthews, M.; Nagai, H.; Zuckman, D.M.; Roy, C.R. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol. Microbiol. 2000, 38, 719–736. [Google Scholar] [CrossRef] [PubMed]
- Souza, D.P.; Andrade, M.O.; Alvarez-Martinez, C.E.; Arantes, G.M.; Farah, C.S.; Salinas, R.K. A Component of the Xanthomonadaceae Type IV Secretion System Combines a VirB7 Motif with a N0 Domain Found in Outer Membrane Transport Proteins. PLoS Pathog. 2011, 7. [Google Scholar] [CrossRef] [PubMed]
- Chandran, V.; Fronzes, R.; Duquerroy, S.; Cronin, N.; Navaza, J.; Waksman, G. Structure of the outer membrane complex of a type IV secretion system. Nature 2009, 462, 1011–1015. [Google Scholar] [CrossRef] [PubMed]
- Sexton, J.A.; Pinkner, J.S.; Roth, R.; Heuser, J.E.; Hultgren, S.J.; Vogel, J.P. The Legionella pneumophila PilT homologue DotB exhibits ATPase activity that is critical for intracellular growth. J. Bacteriol. 2004, 186, 1658–1666. [Google Scholar] [CrossRef] [PubMed]
- Cascales, E. The type VI secretion toolkit. EMBO Rep. 2008, 9, 735–741. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).