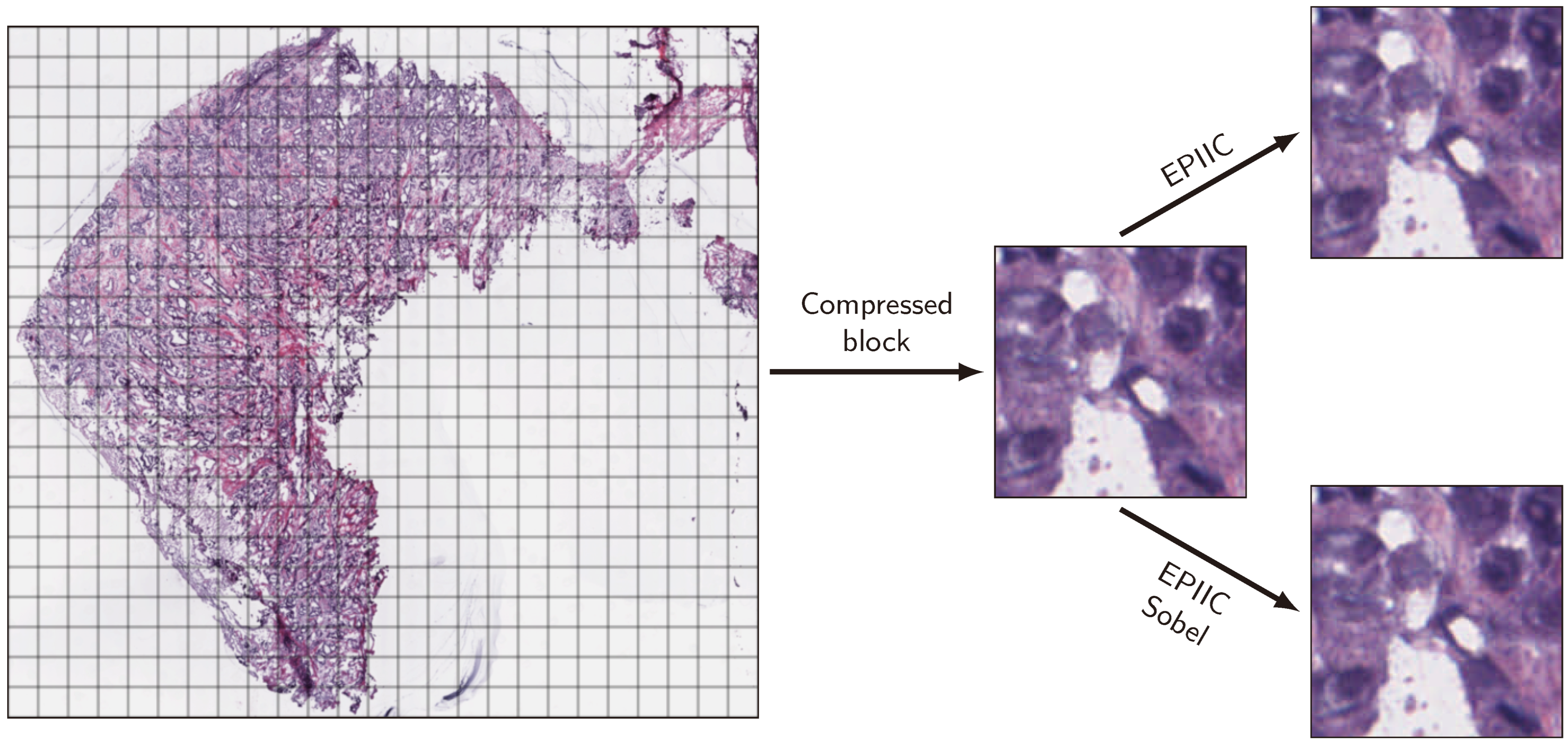
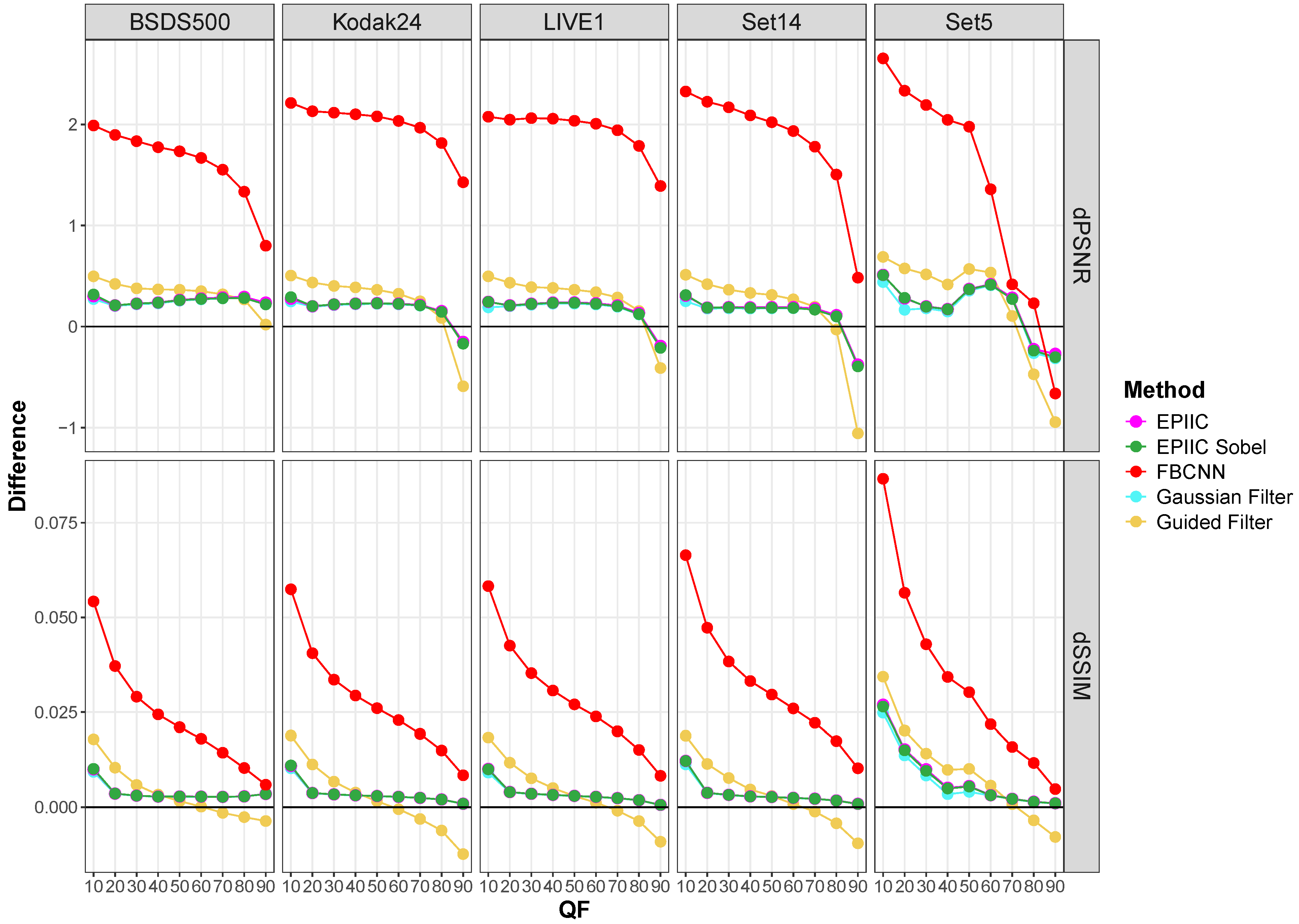
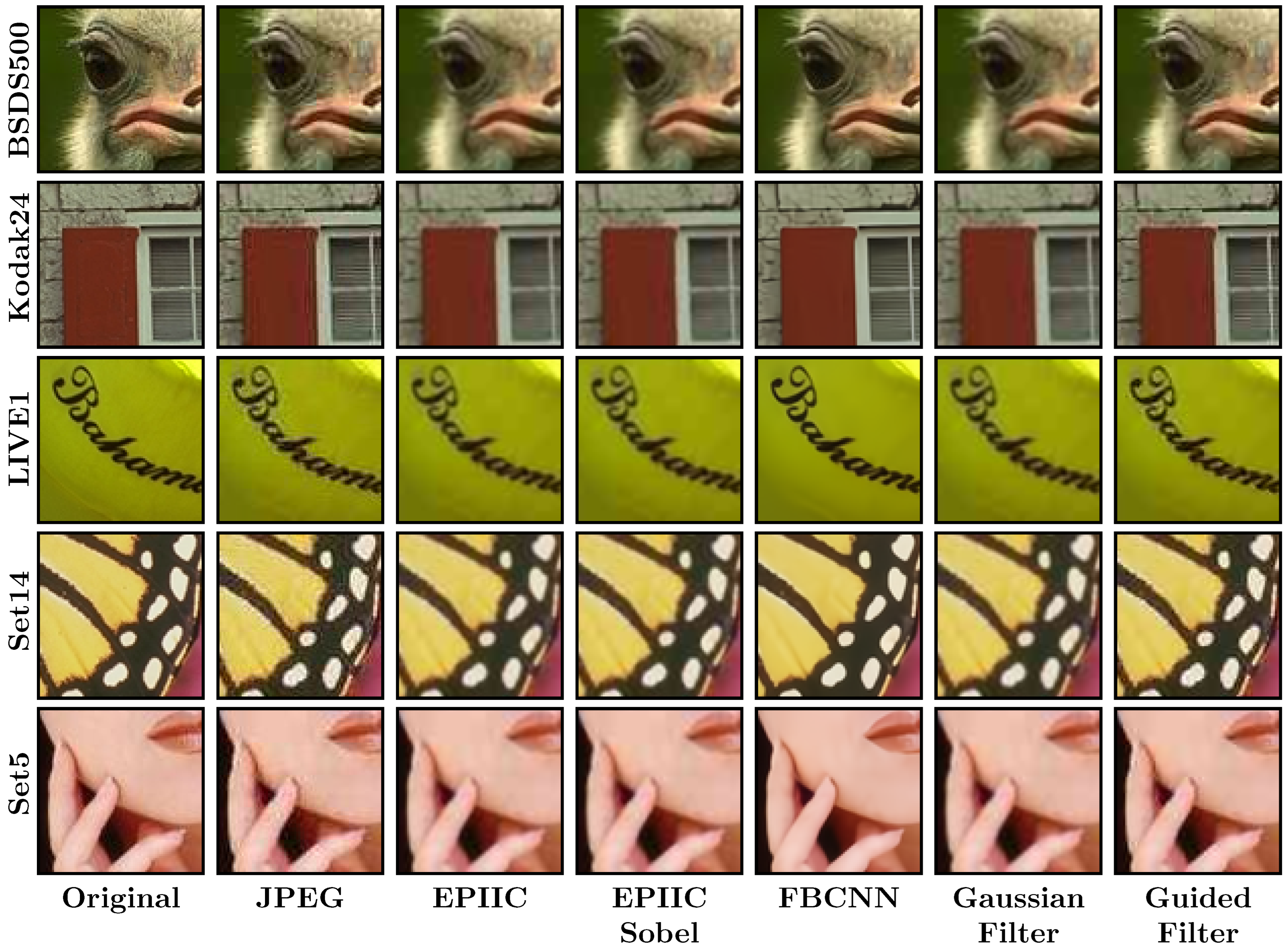
2.1. Proposed Methods
Removing compression artifacts by the EPIIC and EPIIC Sobel methods is divided into six main steps. A flowchart containing these successive steps and exemplary image fragments showing the results are shown in
Figure 1.
The first step, common for both methods, involves the color deconvolution for hematoxylin-eosin staining. The algorithm explained in [
24] was applied to construct an
H channel image containing mostly nuclei stained by hematoxylin from eosin-stained tissue structures. A common Matlab implementation released for public use [
25] was used. For other images than H&E-stained slides, the
H channel image is replaced by converting the RGB image to grayscale.
The next step aims to preprocess the H image to increase the visibility of cell nuclei and reduce the noise present in the image caused by compression artifacts and the deconvolution process itself. In the EPIIC method, the threshold value is calculated first with the Otsu algorithm. Next, the contrast of the H image is adjusted by scaling the pixels with values from zero to the threshold value so that they occupy the entire allowed range of intensity values. As a result, only the most intensely colored elements are visible, and unnecessary background is removed. Then, the image is smoothed with the averaging filter to obtain more uniform shapes and colors of cell nuclei, which enables easier detection of the edges. In the EPIIC Sobel method, the image closing operation is performed on the H image with the structural element in the shape of a disk with a 3-pixel radius. After that, the contrast of an image is adjusted as before, but this time, the threshold value is fixed at 0.75. Next, the obtained image is smoothed with the averaging filter. The different threshold values make the EPIIC method better suited to images with more hematoxylin-stained structures, but in some cases, this results in the loss of poorly stained nuclei, which occurs less frequently in EPIIC Sobel.
In the next step, the gradient magnitude is calculated. In the EPIIC algorithm, the central difference method is used to calculate the gradient magnitude, which is blurred using the average smoothing filter. In the EPIIC Sobel, the gradient is calculated using the Sobel operator. The resulting image G is used to create an edge map.
In the following stage, the image
G is processed to obtain the binary edge map
. In the EPIIC method, the image is binarized with the Otsu method. The threshold value
T obtained by the Otsu algorithm is multiplied by the
parameter. This will ensure that weaker edges are not included in the map. This operation is followed by a morphological opening with a square-shaped structural element of size
to remove isolated pixels from the map. In the EPIIC Sobel algorithm, the threshold value
T is calculated based on the Root Mean Square (RMS) estimate of noise (the method described in [
26], pp. 466–469), and its original value is multiplied by the
parameter. The additional thinning stage is performed, and the opening operation is skipped. As a result of this method, the edges are much thinner than for EPIIC.
Since we are also interested in preserving information about the intensity of the edges, in the EPIIC method, the binary map is multiplied by the image G and the pixel values are then scaled. Pixels larger than zero are reduced by the threshold value and then divided by . Finally, the inverse image is used to obtain an edge map M. In the EPIIC Sobel variant, edge maps are created for each of the three RGB color channels. The gradient magnitudes of each channel, calculated with the central difference operator, are used instead of the image G in the multiplication. Pixel scaling is performed as in the EPIIC method. The threshold T is set to the original threshold value calculated in the previous step. To obtain the final edge maps, scaled images for each channel are inverted. Compared to the EPIIC method, which uses only the H channel to create the map, Sobel’s EPIIC method creates a separate edge map for each channel of the RGB color space.
The final step of both algorithms is weighted smoothing. First, weights for each pixel are calculated. In the EPIIC, only one weight matrix W is created by blurring the edge map M with a Gaussian filter. Then, each color channel of the compressed image is multiplied by the edge map, and the result of this operation is filtered with the same Gaussian filter used in the previous step. Pixel values are divided by corresponding weights from the W to obtain the final image without compression artifacts. In the EPIIC Sobel, the weight matrix is calculated for each color channel. Then, the same operations as above are performed using the edge maps and the weight matrices appropriate for the color channel.
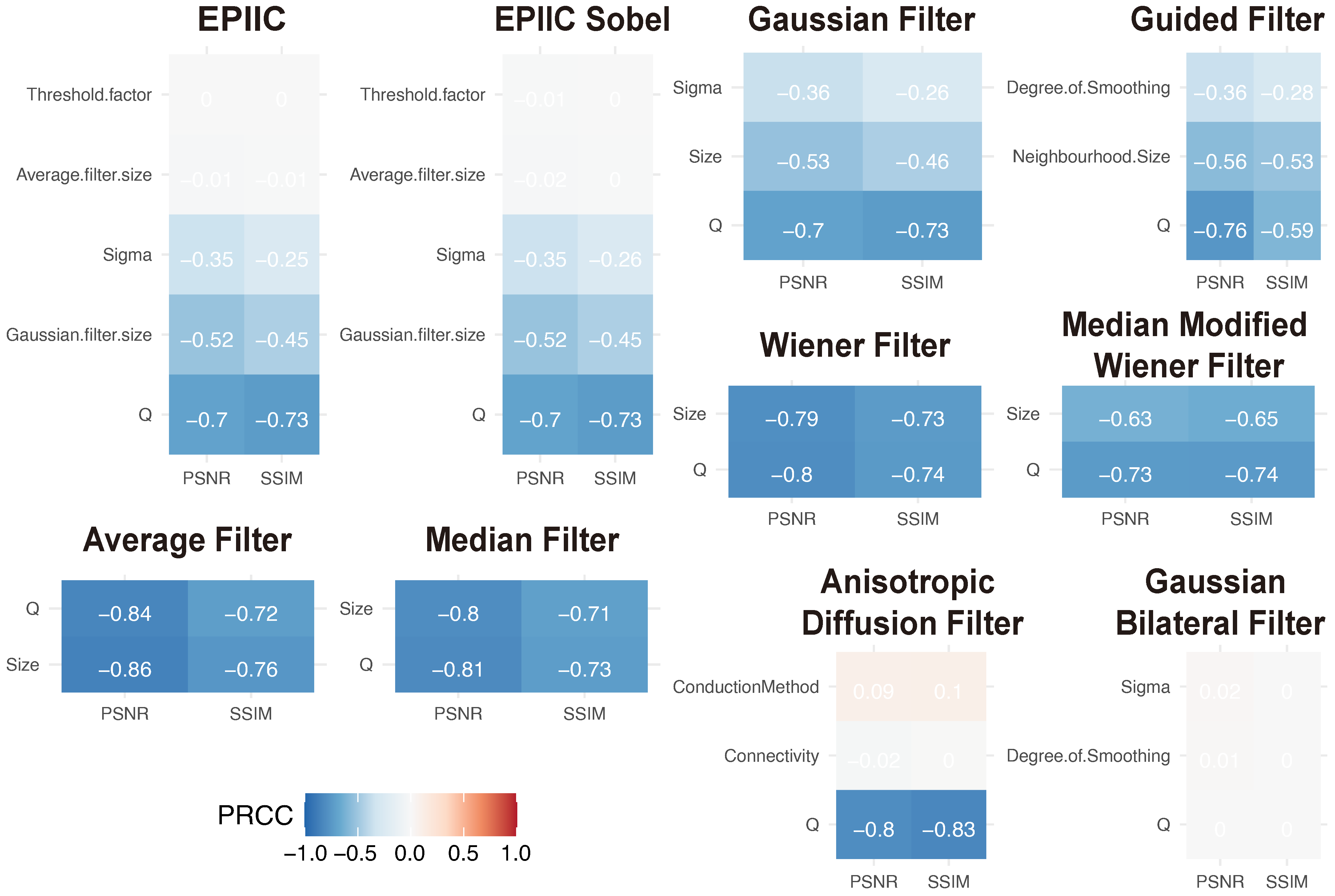
We tested both the averaging filter and the Gaussian filter with sizes ranging from to . The standard deviation of the Gaussian filter varied between and . Additionally, we examined the performance of different parameter values, which ranged from to .
2.2. Other Edge-Preserving Methods
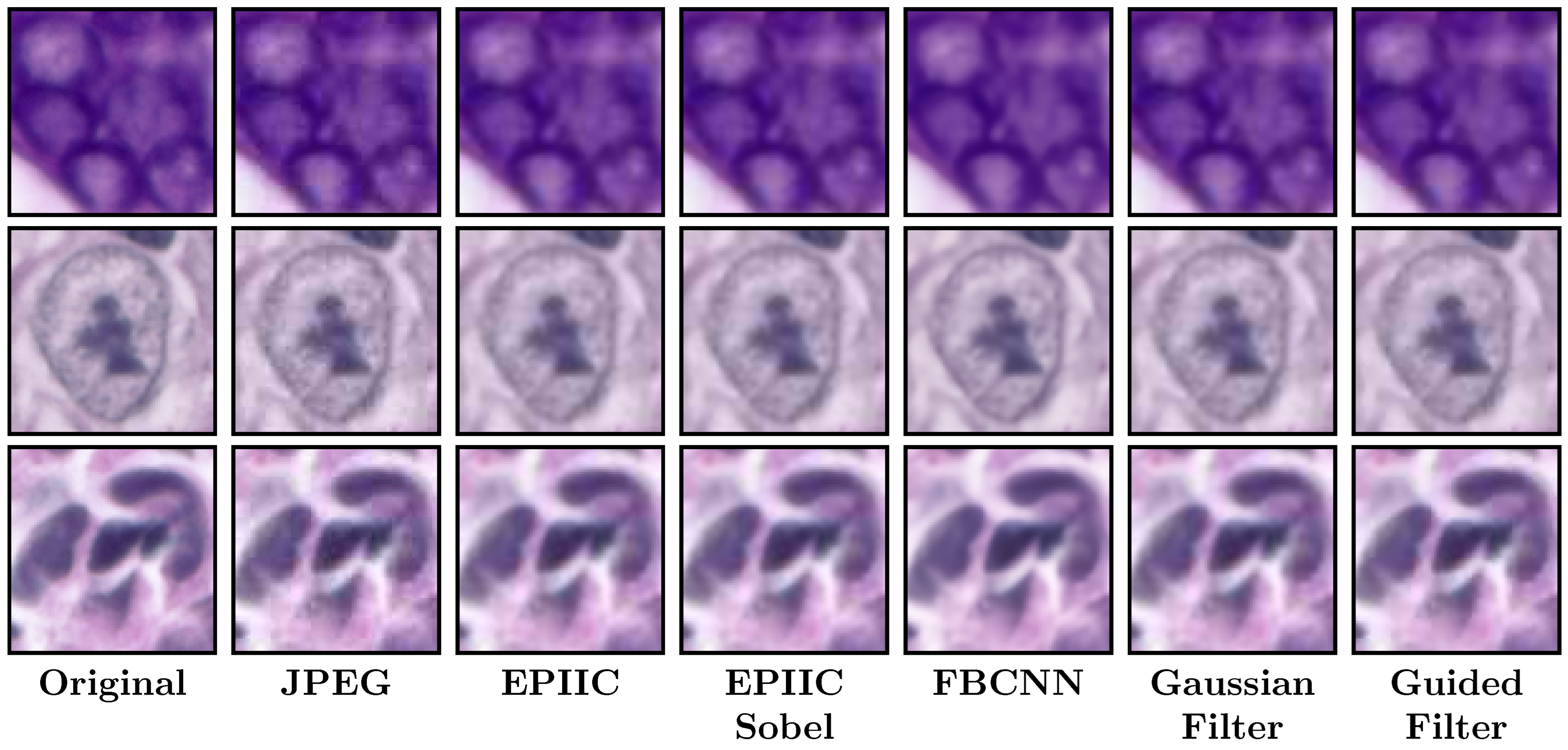
Having a strong interest in the best possible nuclei edge retention, the results of the developed methods were compared with the performance of dedicated filters for edge preservation and noise reduction.
The following filters were considered: the average (mean) filter, the median filter, the Wiener filter, which derives from the mean operator, and the Median Modified Wiener filter (MMWF), which is a nonlinear adaptive spatial filter that uses the quality and abilities of both median and Wiener filters [
27]. MMWF provides global denoising, eliminating spikes and Gaussian noise while preserving the structures visible in the image. For each of the filters listed, the only parameter is the window size, which was tested in the range from 3 to 7, depending on the type of filter.
In addition, the performance of a Gaussian filter of size 3, 5, and 7 with standard deviation
varying from 0.4 to 2.8, and the Guided filter [
28] was tested. The input image was used as a guidance image with the size of the rectangular neighborhood around each pixel from 3 to 7 and the amount of smoothing ranged from 0.001 to 0.05 (when the value is small, only the neighborhood with a small variance will be smoothed and the areas with stronger edges will be preserved).
In [
29], the authors proposed a class of algorithms that realizes image denoising using a diffusion process. An anisotropic diffusion filter was used as a speckle removal technique to improve the quality of ultrasonography [
30] and the magnetic resonance images [
31]. In this work, different connectivity values were examined: four or eight neighborhoods and conduction methods: exponential and quadratic. In the diffusion process, five iterations were used.
In [
32], the authors introduced a non-iterative bilateral filter to preserve edges through a nonlinear combination of nearby pixels. Noise removal is achieved by combining two Gaussian filters: the first works in the spatial domain, and the second filter works in the intensity domain. This filter applies spatially weighted averaging to smooth edges and preserve their shape. This method was also used to eliminate distortions from medical images [
30]. Here, degrees of smoothing from 0.005 to 0.02 and standard deviations of spatial Gaussian smoothing kernel from 0.4 to 1.3 were tested.
Another tested algorithm, the non-local means (NL-means), based on a non-local averaging of all pixels in the image, was presented in [
33]. It can smooth blocking artifacts and preserve the edges simultaneously. Due to the nature of the algorithm, the most favorable case is the textured or periodic image, but some natural images also have sufficient redundancy for the filtering action to have a positive impact on image quality.
Finally, the deep convolutional neural network FBCNN [
20], which achieves state-of-the-art performance in the blind JPEG artifacts removal on single and multiple compressed color and grayscale images, was tested. Trained weights were used for a model designed to remove single compression artifacts from color images provided by the authors (
https://github.com/jiaxi-jiang/FBCNN/releases/tag/v1.0, accessed on 4 February 2025).
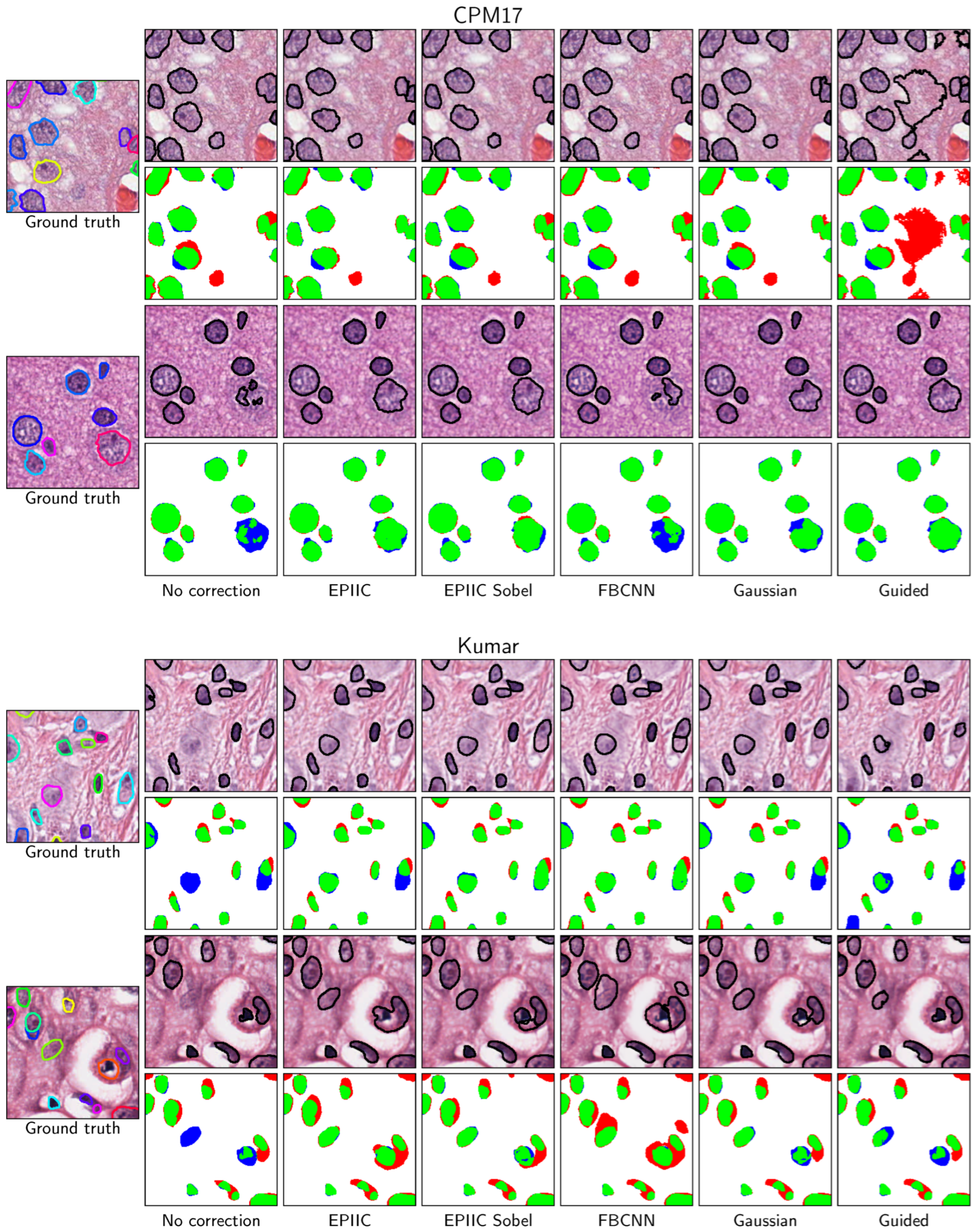
2.5. Nuclei Segmentation Methods
Two state-of-the-art nuclei instance segmentation methods, Hover-Net [
36] and STARDIST [
37], were employed to assess the impact of developed methods on the segmentation quality.
The Hover-Net network received an input RGB patch of
, with values normalized between 0 and 1. Since we wanted to verify whether removing compression artifacts would cause segmentation improvement, reference models were trained for each dataset separately using original images without any modification for training and validation. These models were marked as no correction method. For reference models, in online augmentation, identical techniques were used as in the original paper [
36], that is, flip, rotation, Gaussian blur, median blur, additive Gaussian noise, modification of contrast, hue, and saturation.
Next, we trained models for each selected artifact removal method for each dataset. First, compression artifacts were removed from images from the training and test sets. The EPIIC and EPIIC Sobel methods used a Gaussian filter with a window size of and a standard deviation , a parameter , and an averaging filter of size . For the Gaussian filtering, the same and kernel size were used as in the previous methods. For the Guided filter, we set the degree of smoothing to and the neighborhood size to 3. During model training, the augmentation was limited to flip, rotation, contrast modification, hue, and saturation.
All models described above were initialized with pre-trained weights on the ImageNet dataset [
38]. Only decoders were trained for the first 50 epochs, and then all layers were fine-tuned for 75 epochs. The Adam optimizer was used with the following learning schedule: for each stage, the initial value of the learning rate was
, and after every 25 epochs, it was reduced ten times. Batch size was set to 10 and 2 for the first and second stages, respectively. An early stopping method with patience equal to 50 was used by selecting the model that achieved the best
score on the validation dataset.
The STARDIST model with 64 rays and a U-Net [
39] backbone of depth 4 was used. The dropout equal to 0.7 was added to the network to avoid overfitting, and loss functions specified by the authors were used. The weights of the network were initialized randomly. Each model was trained for 400 epochs with a batch size equal to 2 using the Adam optimizer, starting with a learning rate of 0.0003. If no progress was made for 80 epochs, the learning rate was reduced by half. The weights for the final model were selected based on the smallest validation loss during training. The network received an RGB input patch with a size of
normalized to the range 0–1. The same online augmentation procedure as for the Hover-Net model was applied.
2.6. Nuclear Segmentation Evaluation Metrics
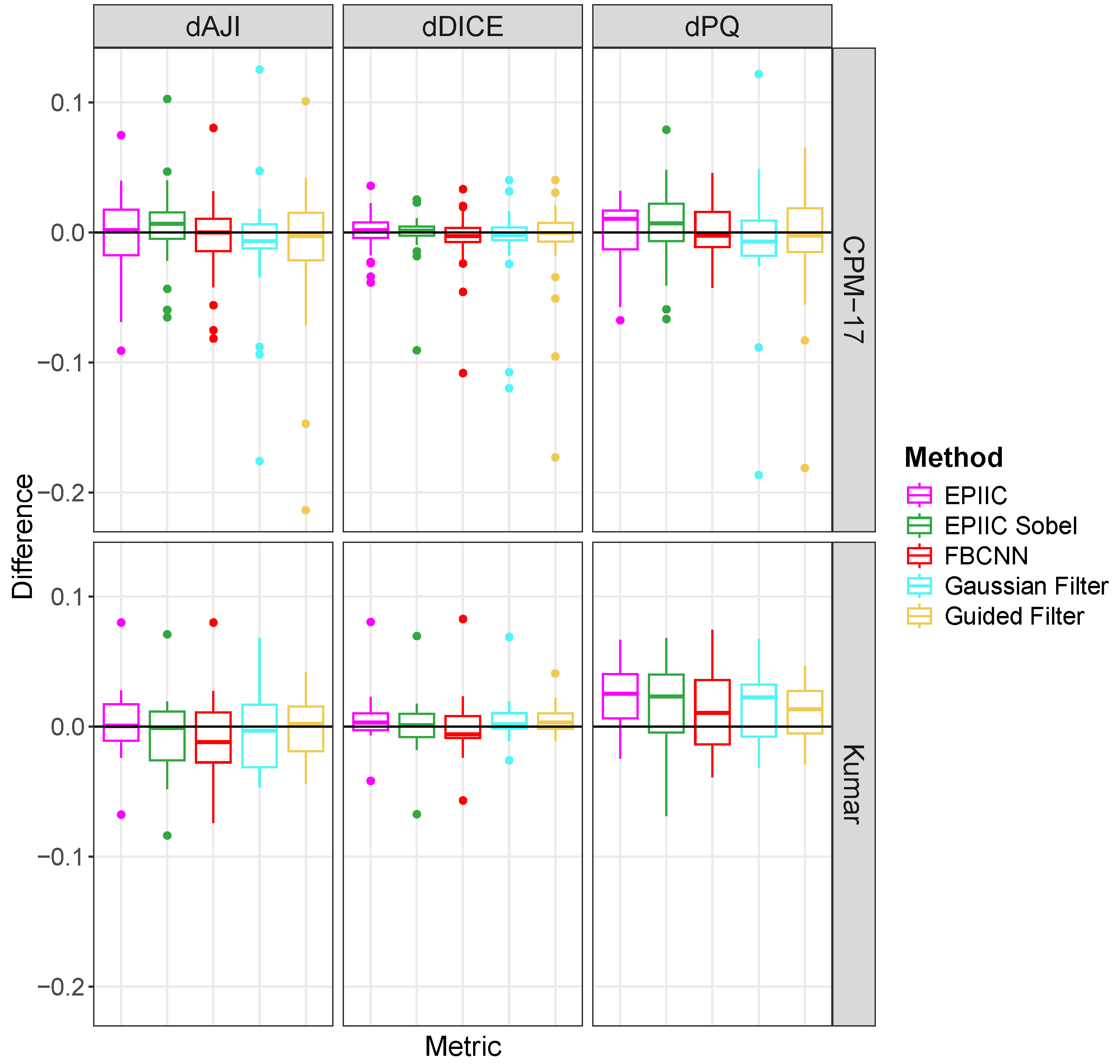
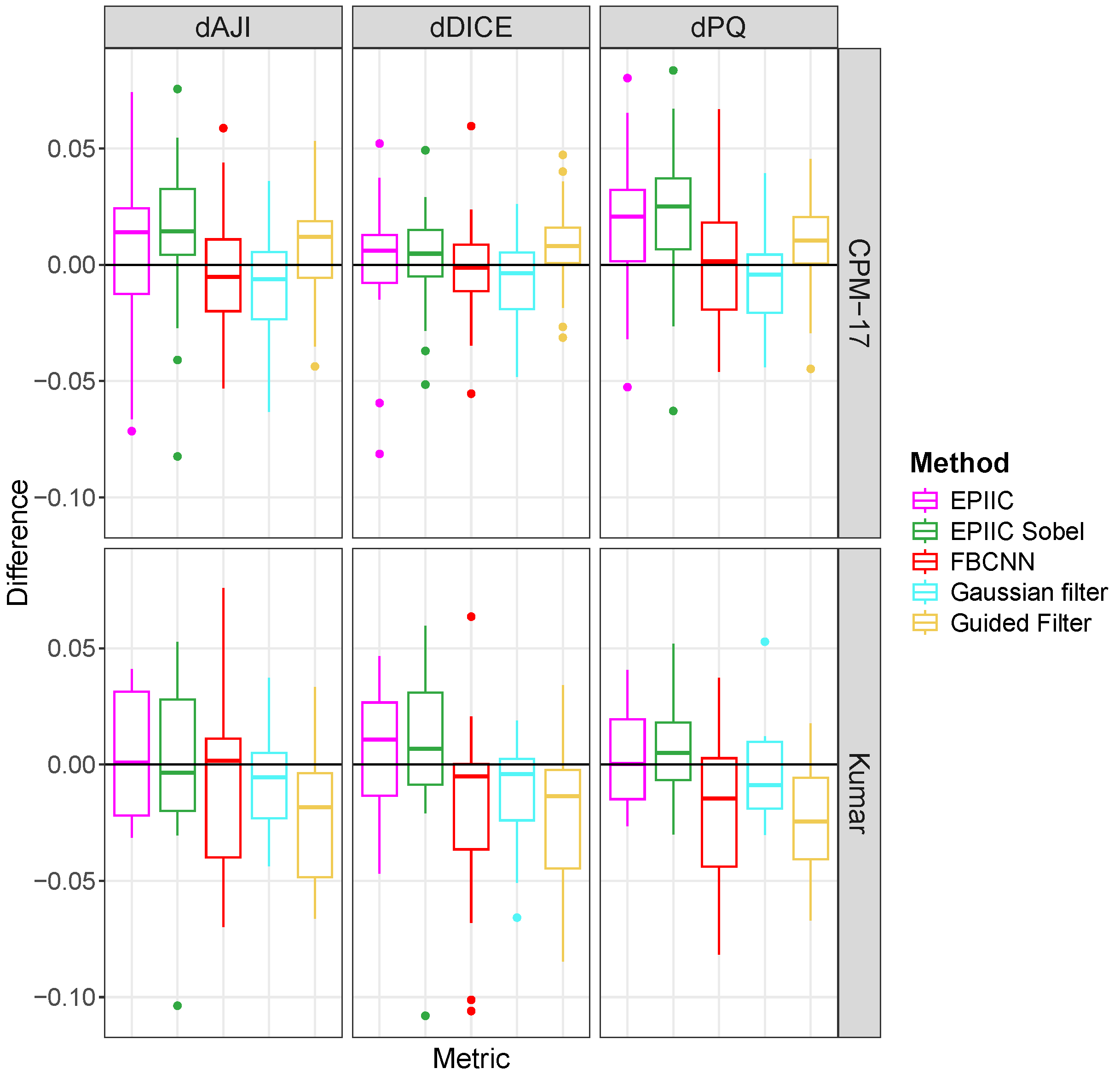
To evaluate the overall score for nuclear instance segmentation, three metrics widely used in the literature were used: Ensemble Dice [
40], Aggregated Jaccard Index (
) [
41], and Panoptic Quality (
) [
42]. Ensemble Dice computes and aggregates Dice coefficient (
) per nucleus, which is defined as:
where
X denotes the ground truth and
Y is a prediction. It is usually used to calculate the similarity of two samples.
calculates the ratio of an aggregated intersection cardinality and an aggregated union cardinality between the ground truth and the prediction.
for nuclear segmentation is defined as the product of the Detection Quality (
) and the Segmentation Quality (
):
The Detection Quality (
) can be interpreted as the F1-score, which is widely used to evaluate instance detection. At the same time, segmentation quality (
) informs how close each correctly detected instance is to its matched ground truth. The following formulas describe these measures:
where
x stands for a ground truth segment,
y is a prediction segment and
denotes intersection over union.
provides a direct insight into the detection quality of individual nuclear instances and the segmentation of each detected nucleus. If the
each
pair is unique [
42]. This property allows to divide the predicted and ground truth segments into the following sets: true positives (TP), false positives (FP), and false negatives (FN), which represent correctly matched pairs of segments, mismatched predicted segments, and mismatched ground truth segments, respectively.
2.7. Datasets
Several datasets were used to evaluate the impact of selected artifacts removal methods on the image quality. The BreCaHAD dataset [
43] includes 162 uncompressed histopathological RGB images showing tissue sections with breast cancer cells. The images are divided into 17 different cases. Each image was taken with 40× magnification and then divided into smaller tiles saved as
TIFF files. The proposed methods were also tested on the datasets commonly used in a task of compression artifacts removal: the test set of BSDS500 [
44], LIVE1 [
45], Kodak24 (
https://r0k.us/graphics/kodak/, accessed on 4 February 2025) Set5 [
46], and Set14 [
47]. All of these datasets consist of uncompressed RGB images.
Since the BreCaHAD dataset is not labeled for the nuclear segmentation task, two additional datasets were used: Kumar [
41] and CPM-17 [
40]. Both datasets contain compressed images of varying quality, with visible JPEG compression artifacts, but the information about the compression quality factor is not included. The Kumar dataset consists of 30 image tiles of size
. Slides were acquired at
magnification and come from seven organs (6 breasts, 6 livers, 6 kidneys, 6 prostates, 2 bladders, 2 colons, and 2 stomachs) of The Cancer Genome Atlas (TCGA) database. We split this dataset into the test set with 14 images (2 prostates, 2 kidneys, 2 livers, 2 breasts, 2 bladders, 2 colons, and 2 stomachs) and the training set with 16 image tiles (4 prostates, 4 kidneys, 4 livers, and 4 breasts). The CPM-17 dataset includes 32 images from TCGA varying in size from
to
in both the training and test datasets. Images were taken at
and
magnification and included four different types of cancer. Note, we utilize the same split used in the [
36] for both datasets.