Antimicrobial Susceptibility Testing in Chlamydia trachomatis: The Current State of Evidence and a Call for More National Surveillance Studies
Abstract
1. Introduction
2. Biology and Molecular Basis of Resistance
3. Methods for C. trachomatis Antimicrobial Susceptibility Testing
4. C. trachomatis Sensitivity Surveillance Efforts
5. Bridging Concept and Practice in the Implementation of National Surveillance Studies
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AMR | antimicrobial resistance |
| AST | antimicrobial susceptibility testing |
| ET | elementary body of Chlamydia trachomatis |
| kbp | kilobase pairs |
| Mbp | megabase pairs |
| MBC | minimal bactericidal concentration |
| MCC | minimal chlamydicidal concentration |
| MDA | mass drug administration |
| MFI | mean fluorescence intensity |
| MIC | minimal inhibitory concentration |
| NGS | next-generation sequencing |
| PCR | polymerase chain reaction |
| QRDR | quinolone-resistance determining region |
| RAM | resistance-associated mutations |
| RT | reticulate body of Chlamydia trachomatis |
| WHO | World Health Organization |
References
- World Health Organization. Chlamydia. Available online: https://www.who.int/news-room/fact-sheets/detail/chlamydia (accessed on 20 March 2025).
- Huai, P.; Li, F.; Chu, T.; Liu, D.; Liu, J.; Zhang, F. Prevalence of genital Chlamydia trachomatis infection in the general population: A meta-analysis. BMC Infect. Dis. 2020, 20, 589. [Google Scholar] [CrossRef] [PubMed]
- Jian, H.; Lu, W.J.; Chen, Z.W.; Liang, S.Q.; Yue, X.L.; Li, J.; Zhang, J.H.; Gong, X.D. Prevalence and trends of Chlamydia trachomatis infection in female sex workers and men who have sex with men in China: A systematic review and meta-analysis. BMC Public Health 2024, 24, 1579. [Google Scholar] [CrossRef] [PubMed]
- Risser, W.L.; Risser, J.M.; Risser, A.L. Current perspectives in the USA on the diagnosis and treatment of pelvic inflammatory disease in adolescents. Adolesc. Health Med. Ther. 2017, 8, 87–94. [Google Scholar] [CrossRef]
- Ljubin-Sternak, S.; Meštrović, T. Chlamydia trachomatis and Genital Mycoplasmas: Pathogens with an Impact on Human Reproductive Health. J. Pathog. 2014, 2014, 183167. [Google Scholar] [CrossRef]
- Tang, W.; Mao, J.; Li, K.T.; Walker, J.S.; Chou, R.; Fu, R.; Chen, W.; Darville, T.; Klausner, J.; Tucker, J.D. Pregnancy and fertility-related adverse outcomes associated with Chlamydia trachomatis infection: A global systematic review and meta-analysis. Sex. Transm. Infect. 2020, 96, 322–329. [Google Scholar] [CrossRef]
- Kristensen, T.S.; Foldager, A.; Laursen, A.S.D.; Mikkelsen, E.M. Sexually transmitted infections (Chlamydia trachomatis, genital HSV, and HPV) and female fertility: A scoping review. Sex. Reprod. Healthc. 2025, 43, 101067. [Google Scholar] [CrossRef]
- Jones, R.B.; Vander Der Pol, B.; Martin, D.H.; Shepard, M.K. Partial characterization of Chlamydia trachomatis isolates resistant to multiple antibiotics. J. Infect. Dis. 1990, 162, 1309–1315. [Google Scholar] [CrossRef]
- Lefèvre, J.C.; Lepargneur, J.P.; Guion, D.; Bei, S. Tetracycline-resistant Chlamydia trachomatis in Toulouse, France. Pathol. Biol. 1997, 45, 376–378. [Google Scholar]
- Somani, J.; Bhullar, V.B.; Workowski, K.A.; Farshy, C.E.; Black, C.M. Multiple drug-resistant Chlamydia trachomatis associated with clinical treatment failure. J. Infect. Dis. 2000, 181, 1421–1427. [Google Scholar] [CrossRef]
- Bhengraj, A.R.; Srivastava, P.; Mittal, A. Lack of mutation in macrolide resistance genes in Chlamydia trachomatis clinical isolates with decreased susceptibility to azithromycin. Int. J. Antimicrob. Agents 2011, 38, 178–179. [Google Scholar] [CrossRef]
- Páez-Canro, C.; Alzate, J.P.; González, L.M.; Rubio-Romero, J.A.; Lethaby, A.; Gaitán, H.G. Antibiotics for treating urogenital Chlamydia trachomatis infection in men and non-pregnant women. Cochrane Database Syst. Rev. 2019, 1, CD010871. [Google Scholar] [CrossRef] [PubMed]
- Hufstetler, K.; Llata, E.; Miele, K.; Quilter, L.A.S. Clinical Updates in Sexually Transmitted Infections, 2024. J. Womens Health 2024, 33, 827–837. [Google Scholar] [CrossRef]
- Elwell, C.; Mirrashidi, K.; Engel, J. Chlamydia cell biology and pathogenesis. Nat. Rev. Microbiol. 2016, 14, 385–400. [Google Scholar] [CrossRef] [PubMed]
- Omsland, A.; Sixt, B.S.; Horn, M.; Hackstadt, T. Chlamydial metabolism revisited: Interspecies metabolic variability and developmental stage-specific physiologic activities. FEMS Microbiol. Rev. 2014, 38, 779–801. [Google Scholar] [CrossRef] [PubMed]
- Bavoil, P.; Ohlin, A.; Schachter, J. Role of disulfide bonding in outer membrane structure and permeability in Chlamydia trachomatis. Infect. Immun. 1984, 44, 479–485. [Google Scholar] [CrossRef]
- Abdelrahman, Y.M.; Belland, R.J. The chlamydial developmental cycle. FEMS Microbiol. Rev. 2005, 29, 949–959. [Google Scholar] [CrossRef]
- Christensen, S.; Halili, M.A.; Strange, N.; Petit, G.A.; Huston, W.M.; Martin, J.L.; McMahon, R.M. Oxidoreductase disulfide bond proteins DsbA and DsbB form an active redox pair in Chlamydia trachomatis, a bacterium with disulfide dependent infection and development. PLoS ONE 2019, 14, e0222595. [Google Scholar] [CrossRef]
- Nicholson, T.L.; Olinger, L.; Chong, K.; Schoolnik, G.; Stephens, R.S. Global Stage-Specific Gene Regulation during the Developmental Cycle of Chlamydia trachomatis. J. Bacteriol. 2003, 185, 3179–3189. [Google Scholar] [CrossRef]
- Jury, B.; Fleming, C.; Huston, W.M.; Luu, L.D.W. Molecular pathogenesis of Chlamydia trachomatis. Front. Cell. Infect. Microbiol. 2023, 13, 1281823. [Google Scholar] [CrossRef]
- Jacquier, N.; Viollier, P.H.; Greub, G. The role of peptidoglycan in chlamydial cell division: Towards resolving the chlamydial anomaly. FEMS Microbiol. Rev. 2015, 39, 262–275. [Google Scholar] [CrossRef]
- Liechti, G.W.; Kuru, E.; Hall, E.; Kalinda, A.; Brun, Y.V.; VanNieuwenhze, M.; Maurelli, A.T. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 2014, 506, 507–510. [Google Scholar] [CrossRef] [PubMed]
- Packiam, M.; Weinrick, B.; Jacobs, W.R.; Maurelli, A.T. Structural characterization of muropeptides from Chlamydia trachomatis peptidoglycan by mass spectrometry resolves “chlamydial anomaly”. Proc. Natl. Acad. Sci. USA 2015, 112, 11660–11665. [Google Scholar] [CrossRef] [PubMed]
- Wyrick, P.B. Chlamydia trachomatis Persistence In vitro: An Overview. J. Infect. Dis. 2010, 201 (Suppl. 2), S88–S95. [Google Scholar] [CrossRef] [PubMed]
- Kintner, J.; Lajoie, D.; Hall, J.; Whittimore, J.; Schoborg, R.V. Commonly prescribed β-lactam antibiotics induce C. trachomatis persistence/stress in culture at physiologically relevant concentrations. Front. Cell. Infect. Microbiol. 2014, 4, 44. [Google Scholar] [CrossRef]
- Riffaud-Widner, C.M.; Widner, R.E.; Ouellette, S.P.; Rucks, E.A. Effect of tryptophan starvation on inclusion membrane composition and chlamydial-host interactions. Infect. Immun. 2025, 93, e0053224. [Google Scholar] [CrossRef]
- Rodrigues, R.; Marques, L.; Vieira-Baptista, P.; Sousa, C.; Vale, N. Therapeutic Options for Chlamydia trachomatis Infection: Present and Future. Antibiotics 2022, 11, 1634. [Google Scholar] [CrossRef]
- Xue, Y.; Zheng, H.; Mai, Z.; Qin, X.; Chen, W.; Huang, T.; Chen, D.; Zheng, L. An in vitro model of azithromycin-induced persistent Chlamydia trachomatis infection. FEMS Microbiol. Lett. 2017, 364, fnx145. [Google Scholar] [CrossRef]
- Nunes, A.; Gomes, J.P. Evolution, phylogeny, and molecular epidemiology of Chlamydia. Infect. Genet. Evol. 2014, 23, 49–64. [Google Scholar] [CrossRef]
- Sigalova, O.M.; Chaplin, A.V.; Bochkareva, O.O.; Shelyakin, P.V.; Filaretov, V.A.; Akkuratov, E.E.; Burskaia, V.; Gelfand, M.S. Chlamydia pan-genomic analysis reveals balance between host adaptation and selective pressure to genome reduction. BMC Genom. 2019, 20, 710. [Google Scholar] [CrossRef]
- Luu, L.D.W.; Kasimov, V.; Phillips, S.; Myers, G.S.A.; Jelocnik, M. Genome organization and genomics in Chlamydia: Whole genome sequencing increases understanding of chlamydial virulence, evolution, and phylogeny. Front. Cell. Infect. Microbiol. 2023, 13, 1178736. [Google Scholar] [CrossRef]
- Szabo, K.V.; O’Neill, C.E.; Clarke, I.N. Diversity in chlamydial plasmids. PLoS ONE 2020, 15, e0233298. [Google Scholar] [CrossRef] [PubMed]
- Sigar, I.M.; Schripsema, J.H.; Wang, Y.; Clarke, I.N.; Cutcliffe, L.T.; Seth-Smith, H.M.; Thomson, N.R.; Bjartling, C.; Unemo, M.; Persson, K.; et al. Plasmid deficiency in urogenital isolates of Chlamydia trachomatis reduces infectivity and virulence in a mouse model. Pathog. Dis. 2014, 70, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.A.; Hadfield, J.; Thomson, N.R.; Cleary, D.W.; Marsh, P.; Clarke, I.N.; O’Neill, C.E. The nature and extent of plasmid variation in Chlamydia trachomatis. Microorganisms 2020, 8, 373. [Google Scholar] [CrossRef]
- Benamri, I.; Azzouzi, M.; Sanak, K.; Moussa, A.; Radouani, F. An overview of genes and mutations associated with Chlamydiae species’ resistance to antibiotics. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 59. [Google Scholar] [CrossRef] [PubMed]
- Mestrovic, T.; Ljubin-Sternak, S. Molecular mechanisms of Chlamydia trachomatis resistance to antimicrobial drugs. Front. Biosci. (Landmark Ed.) 2018, 23, 656–670. [Google Scholar] [CrossRef]
- Misyurina, O.Y.; Chipitsyna, E.V.; Finashutina, Y.P.; Lazarev, V.N.; Akopian, T.A.; Savicheva, A.M.; Govorun, V.M. Mutations in a 23S rRNA gene of Chlamydia trachomatis associated with resistance to macrolides. Antimicrob. Agents. Chemother. 2004, 48, 1347–1349. [Google Scholar] [CrossRef]
- Dugan, J.; Rockey, D.D.; Jones, L.; Andersen, A.A. Tetracycline resistance in Chlamydia suis mediated by genomic islands inserted into the chlamydial inv-like gene. Antimicrob. Agents Chemother. 2004, 48, 3989–3995. [Google Scholar] [CrossRef]
- Marti, H.; Kim, H.; Joseph, S.J.; Dojiri, S.; Read, T.D.; Dean, D. Tet(C) Gene Transfer between Chlamydia suis Strains Occurs by Homologous Recombination after Co-infection: Implications for Spread of Tetracycline-Resistance among Chlamydiaceae. Front. Microbiol. 2017, 8, 156. [Google Scholar] [CrossRef]
- Suchland, R.J.; Sandoz, K.M.; Jeffrey, B.M.; Stamm, W.E.; Rockey, D.D. Horizontal transfer of tetracycline resistance among Chlamydia spp. in vitro. Antimicrob. Agents. Chemother. 2009, 53, 4604–4611. [Google Scholar] [CrossRef]
- Binet, R.; Maurelli, A.T. Frequency of development and associated physiological cost of azithromycin resistance in Chlamydia psittaci 6BC and C. trachomatis L2. Antimicrob. Agents Chemother. 2007, 51, 4267–4275. [Google Scholar] [CrossRef]
- Zhu, H.; Wang, H.P.; Jiang, Y.; Hou, S.P.; Liu, Y.J.; Liu, Q.Z. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in Chlamydia trachomatis strains selected in vitro by macrolide passage. Andrologia 2010, 42, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhu, H.; Yang, L.N.; Liu, Y.J.; Hou, S.P.; Qi, M.L.; Liu, Q.Z. Differences in 23S ribosomal RNA mutations between wild-type and mutant macrolide-resistant Chlamydia trachomatis isolates. Exp. Ther. Med. 2015, 10, 1189–1193. [Google Scholar] [CrossRef] [PubMed]
- Deguchi, T.; Hatazaki, K.; Ito, S.; Kondo, H.; Horie, K.; Nakane, K.; Mizutani, K.; Tsuchiya, T.; Yasuda, M.; Yokoi, S.; et al. Macrolide and fluoroquinolone resistance is uncommon in clinical strains of Chlamydia trachomatis. J. Infect. Chemother. 2018, 24, 610–614. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, C.E.; Seth-Smith, H.M.B.; Van Der Pol, B.; Harris, S.R.; Thomson, N.R.; Cutcliffe, L.T.; Clarke, I.N. Chlamydia trachomatis clinical isolates identified as tetracycline resistant do not exhibit resistance in vitro: Whole-genome sequencing reveals a mutation in porB but no evidence for tetracycline resistance genes. Microbiology 2013, 159 Pt 4, 748–756. [Google Scholar] [CrossRef]
- Li, M.; Zhang, X.; Huang, K.; Qiu, H.; Zhang, J.; Kang, Y.; Wang, C. Presence of Chlamydia trachomatis and Mycoplasma spp., but not Neisseria gonorrhoeae and Treponema pallidum, in women undergoing an infertility evaluation: High prevalence of tetracycline resistance gene tet(M). AMB Express 2017, 7, 206. [Google Scholar] [CrossRef]
- Shao, L.; You, C.; Cao, J.; Jiang, Y.; Liu, Y.; Liu, Q. High treatment failure rate is better explained by resistance gene detection than by minimum inhibitory concentration in patients with urogenital Chlamydia trachomatis infection. Int. J. Infect. Dis. 2020, 96, 121–127. [Google Scholar] [CrossRef]
- Dessus-Babus, S.; Bébéar, C.M.; Charron, A.; Bébéar, C.; De Barbeyrac, B. Sequencing of gyrase and topoisomerase IV quinolone-resistance-determining regions of Chlamydia trachomatis and characterization of quinolone-resistant mutants obtained in vitro. Antimicrob. Agents Chemother. 1998, 42, 2474–2481. [Google Scholar] [CrossRef]
- Morrissey, I.; Salman, H.; Bakker, S.; Farrell, D.; Bébéar, C.M.; Ridgway, G. Serial passage of Chlamydia spp. in sub-inhibitory fluoroquinolone concentrations. J. Antimicrob. Chemother. 2002, 49, 757–761. [Google Scholar] [CrossRef]
- Yokoi, S.; Yasuda, M.; Ito, S.; Takahashi, Y.; Ishihara, S.; Deguchi, T.; Maeda, S.; Kubota, Y.; Tamaki, M.; Fukushi, K. Uncommon occurrence of fluoroquinolone resistance-associated alterations in GyrA and ParC in clinical strains of Chlamydia trachomatis. J. Infect. Chemother. 2004, 10, 262–267. [Google Scholar] [CrossRef]
- Misiurina, O.I.; Shipitsina, E.V.; Finashutina, I.P.; Lazarev, V.N.; Akopian, T.A.; Savicheva, A.M.; Govorun, V.M. Analysis of point mutations in the ygeD, gyrA and parC genes in fluoroquinolones resistant clinical isolates of Chlamydia trachomatis. Mol. Gen. Mikrobiol. Virusol. 2004, 3, 3–7. [Google Scholar]
- Dreses-Werringloer, U.; Padubrin, I.; Köhler, L.; Hudson, A.P. Detection of nucleotide variability in rpoB in both rifampin-sensitive and rifampin-resistant strains of Chlamydia trachomatis. Antimicrob. Agents Chemother. 2003, 47, 2316–2318. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kutlin, A.; Kohlhoff, S.; Roblin, P.; Hammerschlag, M.R.; Riska, P. Emergence of resistance to rifampin and rifalazil in Chlamydophila pneumoniae and Chlamydia trachomatis. Antimicrob. Agents Chemother. 2005, 49, 903–907. [Google Scholar] [CrossRef] [PubMed]
- Suchland, R.J.; Bourillon, A.; Denamur, E.; Stamm, W.E.; Rothstein, D.M. Rifampin resistant RNA polymerase mutants of Chlamydia trachomatis remain susceptible to the ansamycin rifalazil. Antimicrob. Agents Chemother. 2005, 49, 1120–1126. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rupp, J.; Solbach, W.; Gieffers, J. Variation in the mutation frequency determining quinolone resistance in Chlamydia trachomatis serovars L2 and D. J. Antimicrob. Chemother. 2008, 61, 91–94. [Google Scholar] [CrossRef][Green Version]
- Suchland, R.J.; Geisler, W.M.; Stamm, W.E. Methodologies and cell lines used for antimicrobial susceptibility testing of Chlamydia spp. Antimicrob. Agents Chemother. 2003, 47, 636–642. [Google Scholar] [CrossRef]
- Meštrović, T.; Ljubin-Sternak, S.; Bedenić, B. Technical aspects of Chlamydia trachomatis antimicrobial susceptibility testing in cell culture system. Tech. J. 2015, 9, 136–141. [Google Scholar]
- Meštrović, T.; Virok, D.P.; Ljubin-Sternak, S.; Raffai, T.; Burián, K.; Vraneš, J. Antimicrobial Resistance Screening in Chlamydia trachomatis by Optimized McCoy Cell Culture System and Direct qPCR-Based Monitoring of Chlamydial Growth. Methods Mol. Biol. 2019, 2042, 33–43. [Google Scholar]
- Pitt, R.; Alexander, S.; Ison, C.; Horner, P.; Hathorn, E.; Goold, P.; Woodford, N.; Cole, M.J. Phenotypic antimicrobial susceptibility testing of Chlamydia trachomatis isolates from patients with persistent or successfully treated infections. J. Antimicrob. Chemother. 2018, 73, 680–686. [Google Scholar] [CrossRef]
- You, C.; Liao, M.; Wang, M.; Zhao, L.; Li, L.; Ye, X.; Yang, T. The Effect of Amoxicillin Pre-Exposure on Treatment Outcomes and Antimicrobial Susceptibility in Patients with Urogenital Chlamydia trachomatis Infection. Infect. Drug Resist. 2023, 16, 3575–3587. [Google Scholar] [CrossRef]
- Cross, N.A.; Kellock, D.J.; Kinghorn, G.R.; Taraktchoglou, M.; Bataki, E.; Oxley, K.M.; Hawkey, P.M.; Eley, A. Antimicrobial susceptibility testing of Chlamydia trachomatis using a reverse transcriptase PCR-based method. Antimicrob. Agents Chemother. 1999, 43, 2311–2313. [Google Scholar] [CrossRef]
- Eszik, I.; Lantos, I.; Önder, K.; Somogyvári, F.; Burián, K.; Endrész, V.; Virok, D.P. High dynamic range detection of Chlamydia trachomatis growth by direct quantitative PCR of the infected cells. J. Microbiol. Methods 2016, 120, 15–22. [Google Scholar] [CrossRef] [PubMed]
- van Niekerk, J.M.; van Loo, I.H.M.; Lucchesi, M.; Morré, S.A.; Hoebe, C.J.P.A.; Dukers-Muijrers, N.H.T.M.; Wolffs, P.F.G. Direct assessment of possible mutations in the 23S rRNA gene encoding macrolide resistance in Chlamydia trachomatis. PLoS ONE 2022, 17, e0265229. [Google Scholar] [CrossRef] [PubMed]
- Lodhia, Z.; Costa Da Silva, J.; Correia, C.; Cordeiro, D.; João, I.; Carreira, T.; Schäfer, S.; Aliyeva, E.; Portugal, C.; Monge, I.; et al. Surveying genetic markers of antibiotic resistance and genomic background in Chlamydia trachomatis: Insights from a multiplex NGS-based approach in clinical strains from Portugal. J. Antimicrob. Chemother. 2025, 17, dkaf036. [Google Scholar] [CrossRef] [PubMed]
- Büttner, K.A.; Bregy, V.; Wegner, F.; Purushothaman, S.; Imkamp, F.; Roloff Handschin, T.; Puolakkainen, M.H.; Hiltunen-Back, E.; Braun, D.; Kisakesen, I.; et al. Evaluating methods for genome sequencing of Chlamydia trachomatis and other sexually transmitted bacteria directly from clinical swabs. Microb. Genom. 2025, 11, 001353. [Google Scholar] [CrossRef]
- Dessus-Babus, S.; Belloc, F.; Bébéar, C.M.; Poutiers, F.; Lacombe, F.; Bébéar, C.; de Barbeyrac, B. Antibiotic susceptibility testing for Chlamydia trachomatis using flow cytometry. Cytometry 1998, 31, 37–44. [Google Scholar] [CrossRef]
- Sandoz, K.M.; Rockey, D.D. Antibiotic resistance in Chlamydiae. Future Microbiol. 2010, 5, 1427–1442. [Google Scholar] [CrossRef]
- Samra, Z.; Rosenberg, S.; Soffer, Y.; Dan, M. In vitro susceptibility of recent clinical isolates of Chlamydia trachomatis to macrolides and tetracyclines. Diagn. Microbiol. Infect. Dis. 2001, 39, 177–179. [Google Scholar] [CrossRef]
- Takahashi, S.; Hamasuna, R.; Yasuda, M.; Ishikawa, K.; Hayami, H.; Uehara, S.; Yamamoto, S.; Minamitani, S.; Kadota, J.; Iwata, S.; et al. Nationwide surveillance of the antimicrobial susceptibility of Chlamydia trachomatis from male urethritis in Japan. J. Infect. Chemother. 2016, 22, 581–586. [Google Scholar] [CrossRef]
- Takahashi, S.; Yasuda, M.; Wada, K.; Matsumoto, M.; Hayami, H.; Kobayashi, K.; Miyazaki, J.; Kiyota, H.; Matsumoto, T.; Yotsuyanagi, H.; et al. Nationwide surveillance of the antimicrobial susceptibility of Chlamydia trachomatis from male urethritis in Japan: Comparison with the first surveillance report. J. Infect. Chemother. 2022, 28, 1–5. [Google Scholar] [CrossRef]
- Ljubin-Sternak, S.; Mestrovic, T.; Vilibic-Cavlek, T.; Mlinaric-Galinovic, G.; Sviben, M.; Markotic, A.; Skerk, V. In vitro susceptibility of urogenital Chlamydia trachomatis strains in a country with high azithromycin consumption rate. Folia Microbiol. 2013, 58, 361–365. [Google Scholar] [CrossRef]
- Mestrović, T.; Ljubin-Sternak, S.; Sviben, M.; Bedenić, B.; Vranes, J.; Markotić, A.; Skerk, V. Antimicrobial Sensitivity Profile of Chlamydia trachomatis isolates from Croatia in McCoy Cell Culture System and Comparison with the Literature. Clin. Lab. 2016, 62, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Solomon, A.W.; Mohammed, Z.; Massae, P.A.; Shao, J.F.; Foster, A.; Mabey, D.C.; Peeling, R.W. Impact of mass distribution of azithromycin on the antibiotic susceptibilities of ocular Chlamydia trachomatis. Antimicrob. Agents Chemother. 2005, 49, 4804–4806. [Google Scholar] [CrossRef] [PubMed]
- West, S.K.; Moncada, J.; Munoz, B.; Mkocha, H.; Storey, P.; Hardick, J.; Gaydos, C.A.; Quinn, T.C.; Schachter, J. Is there evidence for resistance of ocular Chlamydia trachomatis to azithromycin after mass treatment for trachoma control? J. Infect. Dis. 2014, 210, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Foschi, C.; Salvo, M.; Cevenini, R.; Marangoni, A. Chlamydia trachomatis antimicrobial susceptibility in colorectal and endocervical cells. J. Antimicrob. Chemother. 2018, 73, 409–413. [Google Scholar] [CrossRef]
- Villa, L.; Boga, J.A.; Otero, L.; Vazquez, F.; Milagro, A.; Salmerón, P.; Vall-Mayans, M.; Maciá, M.D.; Bernal, S.; Piñeiro, L. Phenotypic and Genotypic Antimicrobial Susceptibility Testing of Chlamydia trachomatis Isolates from Patients with Persistent or Clinical Treatment Failure in Spain. Antibiotics 2023, 12, 975. [Google Scholar] [CrossRef]
- Vojvodić, Ž.; Daus Šebeđak, D. Outpatient Antibiotic Consumption for Urinary Infections in Croatia 2005–2014: What can be Learned from Utilization Trends. Zdr Varst. 2018, 57, 183–191. [Google Scholar] [CrossRef]
- Willmington, C.; Vainieri, M.; Seghieri, C. Estimating variations in the use of antibiotics in primary care: Insights from the Tuscany region, Italy. Int. J. Health Plann. Manag. 2022, 37, 1049–1060. [Google Scholar] [CrossRef]
- Albiger, B.; Revez, J.; Leitmeyer, K.C.; Struelens, M.J. Networking of Public Health Microbiology Laboratories Bolsters Europe’s Defenses against Infectious Diseases. Front. Public Health 2018, 6, 46. [Google Scholar] [CrossRef]
- Shaw, D.; Torreblanca, R.A.; Amin-Chowdhury, Z.; Bautista, A.; Bennett, D.; Broughton, K.; Casanova, C.; Choi, E.H.; Claus, H.; Corcoran, M.; et al. The importance of microbiology reference laboratories and adequate funding for infectious disease surveillance. Lancet Digit. Health 2024, 7, e275–e281. [Google Scholar] [CrossRef]
- Dumm, R.E.; Marlowe, E.M.; Patterson, L.; Larkin, P.M.K.; She, R.C.; Filkins, L.M. The foundation for the microbiology laboratory’s essential role in diagnostic stewardship: An ASM Laboratory Practices Subcommittee report. J. Clin. Microbiol. 2024, 62, e0096024. [Google Scholar] [CrossRef]
- Rodríguez-Domínguez, M.; Casabona, J.; Galán, J.C. A challenging future in the sexually transmitted infection diagnostics landscape: Chlamydia trachomatis as model. Enferm. Infecc. Microbiol. Clin. (Engl. Ed.) 2022, 40, 470–472. [Google Scholar] [CrossRef] [PubMed]
- Allison, J.J.; Kiefe, C.I.; Wall, T.; Casebeer, L.; Ray, M.N.; Spettell, C.M.; Hook, E.W., 3rd; Oh, M.K.; Person, S.D.; Weissman, N.W. Multicomponent Internet continuing medical education to promote chlamydia screening. Am. J. Prev. Med. 2005, 28, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Kahlmeter, G.; Turnidge, J. Wild-type distributions of minimum inhibitory concentrations and epidemiological cut-off values-laboratory and clinical utility. Clin. Microbiol. Rev. 2023, 36, e0010022. [Google Scholar] [CrossRef] [PubMed]
- Rodino, K.G.; Simner, P.J. Status check: Next-generation sequencing for infectious-disease diagnostics. J. Clin. Investig. 2024, 134, e178003. [Google Scholar] [CrossRef]
- Postiglione, U.; Batisti Biffignandi, G.; Corbella, M.; Merla, C.; Olivieri, E.; Petazzoni, G.; Feil, E.J.; Bandi, C.; Cambieri, P.; Gaiarsa, S.; et al. Combining Genome Surveillance and Metadata To Characterize the Diversity of Staphylococcus aureus Circulating in an Italian Hospital over a 9-Year Period. Microbiol. Spectr. 2023, 11, e0101023. [Google Scholar] [CrossRef]
| Antimicrobial Drug | Genes and Regions with Detected Mutations | References |
|---|---|---|
| Macrolides | 23S rRNA, rpID L4 protein gene, rpIV L22 protein gene | Misyurina 2004 [37], Binet 2007 [41], Zhu 2010 [42], Yiang 2015 [43], Deguchi 2018 [44] |
| Tetracyclines | ompB/porB gene, tet(M) gene for ribosomal protection protein | O’Neill 2013 [45], Li 2017 [46], Shao 2020 [47] |
| Fluoroquionolones | gyrA quinolone-resistance-determining region (QRDR), ParC gene for topoisomerase IV subunit C, ygeD gene for efflux protein | Dessus-Babus 1998 [48], Morrissey 2002 [49], Yokoi 2004 [50], Misiurina 2004 [51], Deguchi 2018 [44] |
| Rifampin | rpoB gene coding beta-subunit of DNA-dependent RNA polymerase | Dreses-Werringloer 2003 [52], Kutlin 2005 [53], Suchland 2005 [54], Rupp 2008 [55] |
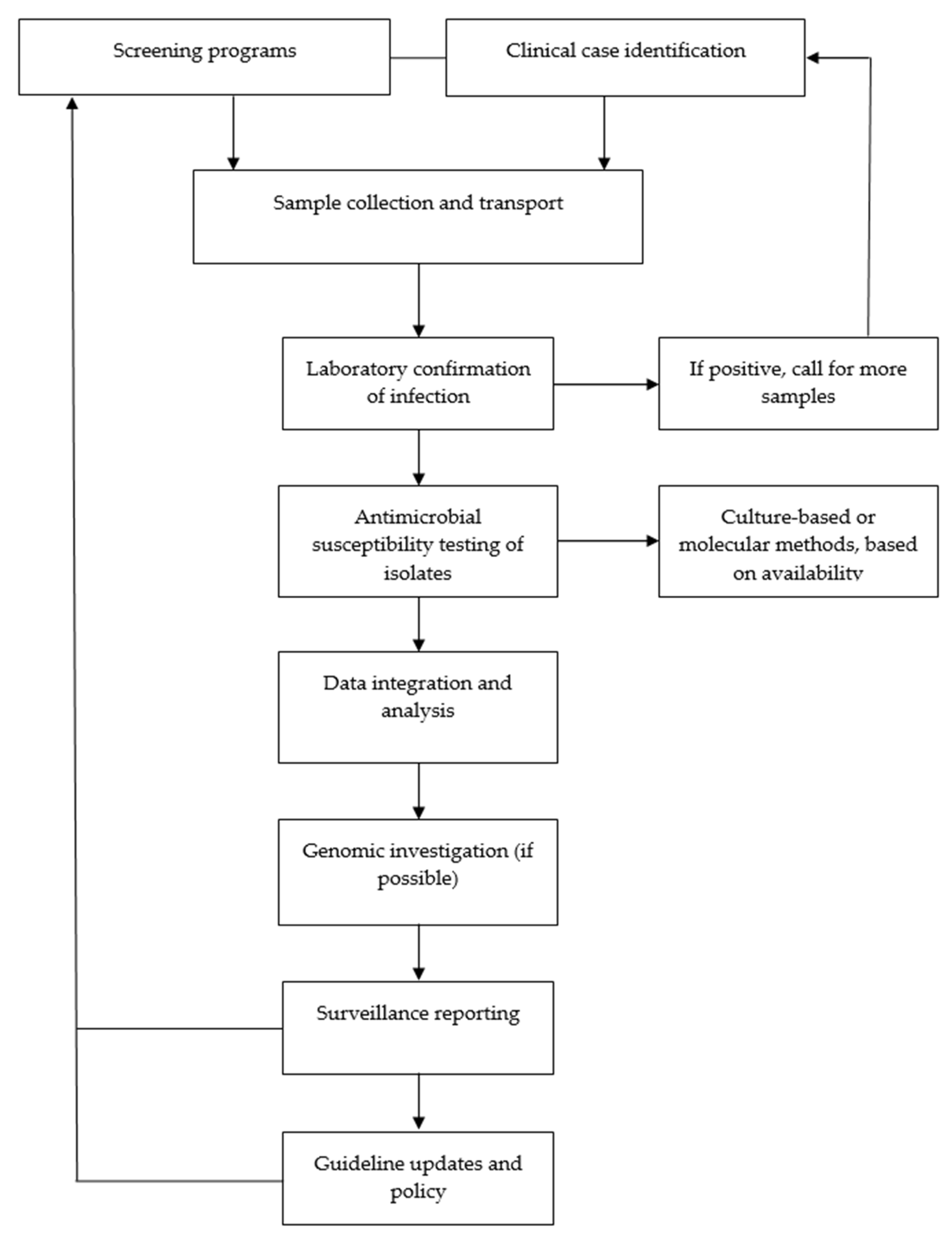
| Step | Process | Description |
|---|---|---|
| 1 | Clinical case identification or screening programs | Recognizing suspected C. trachomatis infections in primary, secondary, and tertiary care as well as in STI clinics in symptomatic individuals; conducting targeted screening programs to detect C. trachomatis infections in asymptomatic individuals |
| 2 | Sample collection and transport | Collecting urogenital, anorectal, and pharyngeal specimen for testing while ensuring proper storage and transport to clinical microbiology laboratories |
| 3 | Laboratory confirmation of infection | Performing nucleic acid amplification tests for diagnosis; if positive, alerting clinicians in step 1 for more samples (if necessary) to conduct AST |
| 4 | Conducting AST of isolates | Conducting culture-based AST and/or molecular assays to detect resistance markers (contingent upon availability in the reference laboratory) |
| 5 | Data integration and analysis | Aggregating results to identify shifts in MICs and other resistance patterns/trends across different settings; developing regional and national antibiograms |
| 6 | Genomic investigation | Employing whole-genome sequencing and next-generation sequencing to monitor resistance-associated mutations and strain evolution (contingent upon availability) |
| 7 | Surveillance reporting | Sharing findings with healthcare providers on all levels, public health agencies, and relevant stakeholders to inform further decisions |
| 8 | Guideline updates and policy | Adjusting treatment protocols (if necessary) and national healthcare policies based on the analysis of surveillance data to guide clinical action and policy development |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ljubin-Sternak, S.; Meštrović, T. Antimicrobial Susceptibility Testing in Chlamydia trachomatis: The Current State of Evidence and a Call for More National Surveillance Studies. Appl. Sci. 2025, 15, 4322. https://doi.org/10.3390/app15084322
Ljubin-Sternak S, Meštrović T. Antimicrobial Susceptibility Testing in Chlamydia trachomatis: The Current State of Evidence and a Call for More National Surveillance Studies. Applied Sciences. 2025; 15(8):4322. https://doi.org/10.3390/app15084322
Chicago/Turabian StyleLjubin-Sternak, Sunčanica, and Tomislav Meštrović. 2025. "Antimicrobial Susceptibility Testing in Chlamydia trachomatis: The Current State of Evidence and a Call for More National Surveillance Studies" Applied Sciences 15, no. 8: 4322. https://doi.org/10.3390/app15084322
APA StyleLjubin-Sternak, S., & Meštrović, T. (2025). Antimicrobial Susceptibility Testing in Chlamydia trachomatis: The Current State of Evidence and a Call for More National Surveillance Studies. Applied Sciences, 15(8), 4322. https://doi.org/10.3390/app15084322








