Abstract
Hybrid CNN–Transformer networks seek to merge the local feature extraction capabilities of CNNs with the long-range dependency modeling abilities of Transformers, aiming to simultaneously address both local details and global contextual information. However, in many existing studies, CNNs and Transformers are often combined through the straightforward fusion of encoder features, which does not promote effective interaction between the two, thus limiting the potential benefits of each architecture. To overcome this shortfall, this study introduces a novel medical image segmentation (MIS) network, designated as DEFI-Net, which is based on dual-encoder interactive fusion. This network enhances segmentation performance by fostering interactive learning and feature fusion between the CNN and Transformer encoders. Specifically, during the encoding phase, DEFI-Net utilizes parallel encoding with both the CNN and Transformer to extract local and global features from the input images. The global–local interaction learning (GLIL) module then enables both the Transformer and CNN to assimilate global semantics and local details from each other, fully leveraging the strengths of the two encoders. In the feature fusion phase, the global–local feature fusion (GLFF) module integrates features from both encoders, using both global and local information to produce a more precise and comprehensive representation of features. Extensive experiments conducted on multiple public datasets, including multi-organ, cardiac, and colon polyp datasets, demonstrate that DEFI-Net surpasses several existing methods in terms of segmentation accuracy, thus highlighting its effectiveness and robustness in MIS tasks.
1. Introduction
Medical image segmentation (MIS) is crucial in aiding physicians to precisely ascertain the location, size, and shape of lesions, and it provides a robust foundation for accurate medical diagnostics and treatment planning. Medical images, unlike natural images, pose unique challenges such as intricate organ details, indistinct lesion boundaries, and low contrast. Therefore, achieving efficient and accurate segmentation is of critical importance.
With the rapid advancements in deep learning technology, convolutional neural networks (CNNs) have become the most prevalent approach for MIS tasks. Ronneberger et al. [1] introduced U-Net, a segmentation method based on a fully convolutional network that features a symmetric encoder–decoder structure with skip connections to preserve detailed information. The U-Net architecture has shown superior segmentation performance and its symmetric design has been widely adopted in various medical segmentation applications. Subsequent research has further refined U-Net to address diverse challenges in medical segmentation, yielding variants such as V-Net [2], attention U-Net [3], HADCNet [4], and I2U-Net [5]. However, the limited receptive field of convolutional kernels in CNNs allows for the excellent capture of local details but poses difficulties in modeling global contextual information, which limits the overall effectiveness of these segmentation methods [6,7,8,9,10].
Vision Transformers (ViTs) [11] have shown promise in enhancing the capability of networks to model long-range dependencies via self-attention mechanisms, effectively extracting global contextual information [12,13]. This has led to the development of improved methods based on ViTs, such as DeiT [14], PVT [15], and Swin-Transformer [16], which have been successfully applied in various vision-related tasks, demonstrating the efficacy of Transformers in visual applications. Inspired by these developments, several innovative approaches have integrated Transformers into networks specifically for MIS. Examples include Swin-UNet [17], TransDeepLab [18], and MISSFormer [19], which employ Transformers throughout both the encoding and decoding phases. While these methods excel at capturing global features, they often neglect local detail information. In response, some strategies have combined Transformers with CNN-based networks, such as TransUNet [20] and TransCeption [21], where Transformers are typically appended after the CNN encoding module. Although these hybrid approaches merge the strengths of CNNs and Transformers, they struggle to effectively integrate the features globally extracted by the Transformers with the local details captured by the CNNs, thereby compromising their segmentation capabilities. In contrast, other methods, like TransFuse [22], CTC-Net [23], and HiFormer [24], link CNNs and Transformers in parallel to concurrently extract global and local features. However, these methods frequently rely on simplistic feature fusion and lack interactive learning between the two encoder types, which prevents them from fully exploiting the complementary benefits of CNNs and Transformers.
Given the limitations inherent in existing hybrid architectures, which struggle to effectively combine the local detail extraction capabilities of CNNs with the global attention capabilities of Transformers, this study introduces a dual-encoder interactive learning network designed specifically for medical image segmentation. This network employs both Transformers and CNNs concurrently to extract image features, wherein the Transformers focus on global feature extraction and the CNNs concentrate on capturing local detail information. To better leverage the strengths of both architectures, we introduce the global–local interaction learning (GLIL) module, which facilitates interaction between the Transformer and CNN encoders. Additionally, a global–local feature fusion (GLFF) module is utilized to integrate features from both encoders, thereby enhancing the fusion process and achieving a more accurate feature representation. Extensive experiments were conducted on the Kvasir-SEG dataset [25], the ACDC cardiac dataset [26], and the Synapse multi-organ dataset [27] for validating the proposed method’s effectiveness.
The contributions of this study are presented below:
- The introduction of a novel network, DEFI-Net, notably enhancing the MIS performance through Transformer–CNN interactive learning. This interaction is facilitated by the GLIL module. The GLIL module consists of two parallel spatial attention modules: one module extracts global semantic information from the features of the Transformer encoder while the other captures local detail information from the features of the CNN encoder. Effective interaction between the two encoders is achieved by using the global semantic information to weight the features from the CNN encoder and refining the features from the Transformer encoder with local detail information.
- The development of a GLFF module that utilizes the global semantic information and local detail information produced by the GLIL module to integrate features from both the Transformer and CNN encoders. The GLFF module concatenates the features from the two encoders and employs a channel attention mechanism to enhance relevant channel features while suppressing less significant ones. The global semantic and local detail information from the GLIL module is then used to weight the concatenated features, resulting in a more precise feature representation.
- An evaluation of the robustness and generalizability of the proposed network through extensive experiments on three distinct medical image datasets: the Synapse multi-organ dataset, the ACDC cardiac dataset, and the Kvasir-SEG dataset. The experimental results demonstrate significant improvements over several existing MIS methods, thus further validating the effectiveness of the proposed approach.
2. Related Work
2.1. CNN for MIS
Convolution-based methods are widely used in medical image segmentation due to their outstanding performance. The U-Net architecture, introduced by Ronneberger et al. [1], features a symmetrical encoder–decoder structure. High-resolution features from the encoder are combined with low-resolution features from the decoder via skip connections, designed to retain detailed information. This has established U-Net as a leading framework for MIS due to its superior performance in this area. Building on this, Zhou et al. [28] developed UNet++, which enhances the U-Net architecture by adding additional skip connection pathways that link each decoder layer to multiple encoder layers. Additionally, a multilevel feature pyramid is created through the integration of cascading and skip connections. However, this architecture significantly increases the number of parameters. To overcome these drawbacks, Huang et al. [29] proposed Unet3+, which features full-scale skip connections and depth-supervised strategies to reduce the parameter count while effectively capturing both coarse and fine-grained features at multiple scales. With the growing use of attention mechanisms in various computer vision tasks, several researchers have explored their integration into medical segmentation networks. Oktay et al. [3] introduced the attention U-Net, which incorporates a gated attention mechanism within the skip connections to better segment small target areas. Similarly, Sinha et al. [30] developed the guided attention module, which includes spatial and channel self-attention submodules that help to adaptively integrate features across multiple scales. Drawing inspiration from ResNet [31], the Res-UNet [32] combines weighted attention mechanisms, skip connections, and residual connections within the U-Net architecture. This combination facilitates more effective feature modeling, enhancing the distinction between vascular and non-vascular structures and improving the accuracy of retinal vascular segmentation. Meanwhile, MultiResUNet [33] modifies the standard U-Net by replacing its convolutions with multi-scale convolution kernels and incorporating residual structures into the skip connections, achieving enhanced segmentation performance. Despite these advancements, convolutional neural networks, limited by the size of their convolutional kernels, still struggle to model long-range dependencies and effectively capture global contextual information.
2.2. Vision Transformer for MIS
In recent years, Transformer-based network architectures have proven to be highly effective in advancing the field of MIS. These architectures have attracted considerable attention for their capability to surmount the constraints traditionally associated with CNNs in capturing global contextual information. Among the purely Transformer-based approaches to MIS, TransDeepLab was introduced by Azad et al. [18]. This method, designed specifically for skin damage segmentation, incorporates various windowing strategies within the DeepLab framework. Similarly, Huang et al. [19] presented the MissFormer network, which employs a multi-scale global information fusion mechanism to substantially improve segmentation performance. Additionally, Swin-Unet, developed by Cao et al. [17], utilizes the Swin Transformer as a feature extractor. This model enhances its capability by incorporating a dynamic window mechanism, thus addressing the inherent limitations of the visual Transformer (ViT) in managing local inter-window interactions. Moreover, DS-TransUNet, proposed by Lin et al. [34], features a dual-branch Swin Transformer encoder to extract multi-scale features. It includes an innovative Transformer interactive fusion (TIF) module designed to effectively integrate multi-scale features from the encoder. This is achieved by fostering long-range dependencies between features of varying scales through a self-attention mechanism. In the realm of 3D medical image processing, the UNETR network introduced by Gao et al. [35] integrates a Transformer architecture with a U-shaped network design. This approach capitalizes on the Transformer encoder’s ability to serialize representations of 3D volumetric data, thereby significantly enhancing the capture of global multi-scale features. Despite these significant advancements, the application of Transformers in modeling long-range dependencies continues to face challenges, particularly in local feature extraction, where CNNs still hold a comparative advantage, especially in tasks involving MIS.
2.3. Combining CNN and Transformer for MIS
In recent years, efforts have been made to fully leverage the strengths of CNNs and Transformer networks by integrating these two architectures for MIS. One notable example is TransUnet [20], which innovatively combines Transformer networks and CNNs in a sequential manner. This model uses the Transformer network to process feature maps produced by the CNN, enhancing segmentation performance through capturing global information via a self-attention mechanism. Similarly, Gao et al. [36] introduced UTNet, which features an efficient self-attention module in its encoder to minimize the computational demands of capturing multi-scale long-range dependencies. Additionally, UTNet incorporates a unique self-attentive decoder designed to restore fine-grained details from skip connections. Contrasting with this serial integration, some recent studies have adopted a parallel approach to simultaneously extract both global and local features. For example, Zhang et al. [22] developed the TransFuse network, which uses separate encoders for CNNs and Transformers. These encoders independently extract image features, which are then fused and relayed to the decoder via a BiFusion module. In a similar vein, Yuan et al. [23] proposed a dual-encoder hybrid model combining CNN and Transformer technologies, where both encoders generate features that are subsequently fused and forwarded to the decoder for up-sampling through skip connections. Furthermore, Heidari et al. [24] introduced HiFormer, employing a combination of a Swin Transformer and a CNN-based encoder to create two distinct multi-scale feature representations. Additionally, Manzari et al. [37] developed BEFUnet, which enhances robustness and accuracy in edge segmentation through local cross-attention feature fusion and dual-encoder feature integration. Importantly, as depicted in Figure 1a, these parallel approaches typically fuse the features from both encoders in a straightforward manner and directly use them in the decoder, often lacking effective interaction between the different encoders.
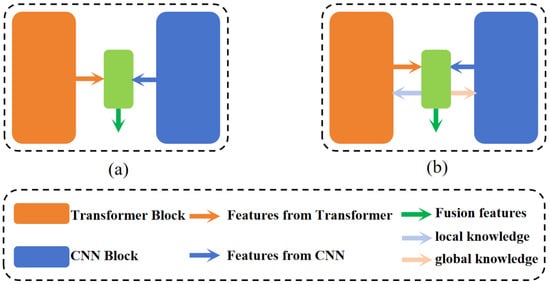
Figure 1.
Two network architectures based on parallel combination of CNN and Transformer. (a) Fusion of only features from different encoders. (b) The architecture of our proposed approach.
2.4. Channels and Spatial Attention Mechanisms
The channel attention mechanism is predominantly utilized to process feature maps produced by CNNs. It concentrates on discerning the significance of individual channels within the input feature maps, thereby selectively focusing on the channel dimensions. The initial implementation of this mechanism is exemplified by the SENet model, introduced by Hu et al. [38]. This model incorporates the squeeze-and-excitation (SE) module, which fundamentally computes the weights for each channel using global average pooling and fully connected layers. These weights are then leveraged to modulate the network’s focus across various channels. To mitigate the computational burden imposed by the linear layers in the SE module, Wang et al. [39] developed the efficient channel attention (ECA) module. This module utilizes 1D convolution with a specific kernel size, K, to efficiently derive channel attention from the feature map. In comparison to the SE block, the ECA module requires fewer parameters and its 1D convolution dynamically adjusts the channel weights. However, the channel attention mechanism predominantly centers on the relevance of individual channels, often overlooking spatial information. This oversight can lead to the omission of crucial structural details. Such limitations are effectively countered by the spatial attention mechanism. The pioneering application of spatial attention was the spatial Transformer network (STN), proposed by Jaderberg et al. [40] in 2015. This network allows the model to dynamically concentrate on varying regions of the input by learning spatial transformation parameters. To amalgamate both spatial and channel attention mechanisms, Woo et al. [41] introduced the convolutional block attention module (CBAM). This module is a sophisticated attention mechanism that sequentially integrates channel and spatial attention, enhancing the model’s ability to comprehend features more holistically and capture complex interdependencies among them.
3. Methodology
Figure 2 presents an MIS network, designated as DEFI-Net, which is founded on a dual-encoder interactive fusion approach. Distinct from conventional methodologies that employ Transformers and CNNs in parallel and directly harness the fused features, DEFI-Net not only amalgamates features from disparate encoders but also promotes interactive learning between the Transformer and CNN. During the encoding phase, we construct a parallel encoder featuring a three-layer pyramid structure that incorporates both CNN and Transformer technologies for extracting multi-scale features from the input image. Subsequently, the GLIL module is utilized to facilitate interaction between the Transformer encoder and the CNN encoder at each pyramid layer, with the learned features being transmitted to subsequent, deeper layers of the network. Concurrently, the global–local feature fusion (GLGF) module is applied to efficiently amalgamate features from both encoders. Additionally, skip connections are implemented to aggregate features across various scales, culminating in the decoding process to generate the final segmentation output.
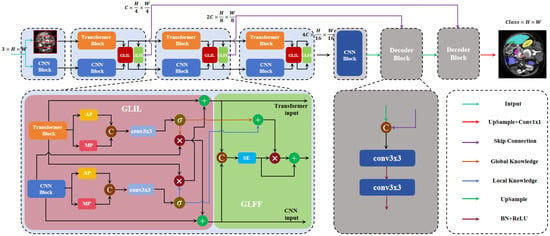
Figure 2.
The DEIF-Net architecture is composed of parallel encoders and GLIL and GLFF modules, as well as skip connections.
3.1. Transformer–CNN Dual-Encoder
Motivated by existing medical segmentation networks such as TransFuse [22] and CTC-Net [23], which utilize parallel encoders, we advance a dual-encoder architecture that capitalizes on interactive learning and feature fusion. This design is intended to enhance the network’s segmentation performance by exploiting the complementary strengths of different encoding systems. For the encoding phase, a dual-encoder network with a three-layer pyramid structure is established to effectively extract multi-scale features. In the Transformer encoder branch, for the input image , patch embedding layers with patch sizes of 4, 2, and 2 are sequentially applied between different Transformer blocks . This arrangement produces feature maps of sizes , , and , while the Transformer layers focus on extracting global features .
In the CNN encoder branch, ResNet-34 is directly implemented to extract multi-scale local features from images. The 2nd, 3rd, and 4th stages of ResNet align with the three blocks of the Transformer, maintaining consistent feature scales across both encoder branches.
3.2. Global–Local Interaction Learning Module
The exclusive use of either CNNs or Transformers as the encoding mechanism often fails to strike an optimal balance between extracting global features and capturing local detail features. Furthermore, existing medical segmentation networks that utilize hybrid CNN–Transformer architectures tend to merely fuse features from different encoders and directly employ them in the decoding phase, thereby overlooking the potential synergistic interactions among the encoders. To address this limitation and effectively harness the strengths of both CNNs and Transformers, the GLIL module has been developed. This module facilitates the exchange of information between the CNN and Transformer, promoting interactive learning. The GLIL module comprises two parallel spatial attention modules [41]: one extracts global information from the Transformer’s output features and the other captures detailed local information from the CNN’s output features. For an input feature F within the dimensions , the spatial attention module conducts average pooling and max pooling along the channel dimension to generate compressed feature maps and , respectively, both in .
These compressed maps, and , are then concatenated along the channel dimension to form . Next, a convolutional operation followed by a sigmoid activation function is used for deriving the attention weights for the input feature map F.
This architecture allows for the parallel extraction of global information from the Transformer and local detail knowledge from the CNN. To facilitate effective interaction between the Transformer and CNN, the global information is utilized for modulating the features extracted by the CNN while the local detail knowledge is used for modulating the features extracted by the Transformer. Additionally, residual connections are employed to enhance the stability of the interactive learning process.
Through the implementation of the GLIL module, the output features of the Transformer and CNN are enabled to mutually enhance their learning and propagate effectively to the subsequent layer of the encoder. This method significantly enriches the interaction between the Transformer and CNN encoders, facilitating a more integrated and effective use of their respective advantages within the network.
3.3. Global–Local Feature Fusion Module
To enhance the integration of features from disparate encoders, we have developed a GLFF module. This module capitalizes on both global and local information to facilitate a more effective amalgamation of features. Specifically, the feature outputs from the Transformer, denoted as , and the CNN, denoted as , are combined through concatenation to yield the fused feature :
The GLFF module incorporates a channel attention mechanism [38] that prioritizes salient channel features while attenuating the less relevant ones. Initially, average pooling is applied along the spatial dimension to derive the channel-wise representation of the fused features, denoted as . Following this, a nonlinear transformation is applied to using the sigmoid function and a multi-layer perceptron (MLP), resulting in the generation of channel attention weights . These weights are then applied element-wise to the concatenated features to highlight significant channel characteristics:
Here, and are the weight matrices corresponding to the fully connected layers, with denoting the ReLU activation function.
Subsequently, the integrated features are modulated using the global semantic information and local detail knowledge derived from the GLFF module, leading to a richer feature representation:
3.4. Loss Function
The model employs a composite loss function, which is a weighted sum of the CE loss and Dice loss, designated as the total loss:
Here, represents the total loss, denotes the cross-entropy loss, and refers to the Dice loss. For the purposes of our experiment, we set to 0.5.
4. Experiments and Results
4.1. Implementation Details
The experiments conducted utilized the DEIF-Net on a single NVIDIA RTX 3090 GPU. Optimization of the network parameters was achieved using the SGD optimizer. The experiments were run with a batch size of four across 150 epochs. The initial learning rate was set at 0.01, which was reduced by multiplying it by 0.9 at regular intervals during the training process. All inputs were uniformly resized to 224 × 224 pixels. We applied simple data augmentations such as random rotation and flip. The hyperparameters within the network were set as . In the Transformer block, the channel dimension was set to 320.
4.2. Datasets
4.2.1. Synapse Multi-Organ Segmentation
The Synapse multi-organ dataset [27], originating from the MICCAI 2015 Multi-Atlas Abdomen Labeling Challenge, consists of 30 abdominal CT scans. Each scan is annotated to identify eight distinct organs: the stomach, spleen, pancreas, liver, right kidney, left kidney, gallbladder, and aorta. Importantly, each CT scan comprises between 85 and 198 axial slices, totaling 3779 slices across the dataset. In alignment with the data configuration used by TransUnet [20], the Synapse dataset was divided into 18 training cases (encompassing 2211 axial slices) and 12 test cases.
4.2.2. Automated Cardiac Diagnosis Challenge (ACDC) Segmentation
The ACDC [26], initiated by MICCAI in 2017, focuses on the segmentation of the myocardium (Myo), right ventricle (RV), and left ventricle (LV) in cardiac MRI data. The dataset features short-axis slices, extending from the base to the apex of the heart, with slice thicknesses ranging from 5 to 8 mm and spatial resolutions between 0.83 and 1.75 mm2 per pixel. For experimental purposes, the dataset was segmented as 20 test cases, 10 validation cases, and 70 training cases (comprising 1930 axial slices), following the data setup of TranUnet [20].
4.2.3. Kvasir-SEG Dataset
The Kvasir-SEG dataset [25] consists of 1000 annotated images of colon polyps, featuring a range of resolutions from 332 × 487 to 1920 × 1072 pixels. The dataset was partitioned such that eighty percent was randomly allocated as the training set while the remaining twenty percent was evenly divided between the validation and test sets. These subsets facilitated the evaluation of the model’s performance.
4.3. Evaluation Metrics
In this research, several evaluation metrics were utilized to ascertain the effectiveness of the model. These metrics included the Dice similarity coefficient (DSC), the 95th percentile Hausdorff distance (HD95), and the intersection over union (IoU) coefficient. Each metric is elaborated upon in the subsequent sections.
4.3.1. Dice Similarity Coefficient (DSC) and Intersection over Union (IoU)
The DSC and the IoU are pivotal metrics for assessing the performance of segmentation models. These metrics measure the extent of overlap between the predicted outcomes and the ground truth labels, with values ranging from 0 to 1. A value of 0 indicates no overlap, whereas a value of 1 denotes perfect alignment, with higher values indicative of superior model performance.
The DSC is computed according to the following formula:
Here, P and T represent the sets of predicted and ground truth pixels, respectively, is the intersection of these sets, and is the sum of pixels in both sets.
The IoU is defined by the subsequent equation:
In this formula, represents the union of the predicted and ground truth pixel sets.
4.3.2. Hausdorff Distance at the 95th Percentile (HD95)
The Hausdorff distance (HD) serves as a metric for evaluating the congruence of boundaries between segmentation results and their corresponding ground truths. It measures the distance between the peripheries of these entities, thereby focusing on the precision of boundary delineation. In contrast to the DSC, the HD places a greater emphasis on the accuracy of boundary localization.
For two non-empty sets, A and B, the HD is defined by the following equation:
Here, represents the Euclidean distance between points a and b. Essentially, HD computes the greatest of all the shortest distances from points in one set to the nearest points in the other set. In the context of segmentation tasks, it is common to use HD95, which quantifies the maximum distance for 95% of the points in set A to their nearest points in set B. A superior alignment of the segmentation result with the ground truth can be indicated by a lower HD95 value.
4.4. Results on Synapse Multi-Organ Segmentation
For evaluating the segmentation efficacy of DEFI-Net on multi-organ medical images, we carried out a series of experiments using the Synapse dataset. The performance of DEFI-Net was benchmarked against numerous leading-edge segmentation methods. The comparative analysis included various architectures, such as CNN-based networks (e.g., U-Net [1], Att-UNet [3], Res-UNet [32]) and purely Transformer-based networks (e.g., Swin U-Net [17], TransDeepLab [18], MISSFormer [19])), along with hybrid models employing serial (e.g., TransUNet [20], TransCeption [21]) and parallel CNN–Transformer configurations (e.g., CTC-Net [23], TransFuse [22], HiFormer [24]). As depicted in Table 1, methods such as U-Net and Att-U-Net, primarily based on CNNs, effectively segment the aorta but struggle with organs that exhibit large variations in shape and size. DEFI-Net demonstrated superior segmentation accuracy across multiple organs, achieving the highest average Dice similarity coefficient (DSC) of 83.29% and a competitive 95% Hausdorff distance (HD) of 18.15 mm. This performance surpasses that of both purely CNN-based and Transformer-based approaches, underscoring the synergistic benefits derived from integrating CNN and Transformer encoders, which significantly enhance segmentation accuracy across most organs. Moreover, DEFI-Net displayed superior segmentation outcomes when compared to hybrid models using both serial and parallel combinations. This highlights the critical role of effective encoder interactions in achieving enhanced model performance. Notably, DEFI-Net exhibited remarkable improvements in the segmentation of the pancreas, spleen, and stomach. The most significant enhancement was observed in the pancreas segmentation, where the DSC increased by 3.02% relative to other evaluated methods. In contrast to models like Swin-UNet [17] and HiFormer [24], DEFI-Net does not rely on pre-trained weights for initializing the Transformer module, further emphasizing its robust feature extraction capabilities and the efficacy of its feature interaction and fusion strategies at each stage.

Table 1.
Comparison results of the proposed method on the Synapse dataset, with the best results indicated in red and the second best in blue.
Figure 3 presents various methods’ segmentation results. It is apparent that conventional convolution-based networks like U-Net [1] and Att-UNet [3] are less effective in accurately identifying and segmenting organs of varying sizes, such as the pancreas and stomach, often leading to under-segmentation or erroneous delineations. Similarly, Swin-UNet [17], TransUNet [20], and HiFormer [24] falter in stomach and pancreatic segmentation, failing to completely delineate the organs, showing issues such as under-segmentation and irregular boundaries. In stark contrast, DEFI-Net exhibits better performance in multi-organ segmentation tasks, accurately identifying and segmenting the pancreas and stomach with more precise, smooth-edged, and complete segmentation areas. These visualization results are consistent with the empirical data, further validating DEFI-Net’s substantial improvements in segmenting organs of varying sizes, particularly the pancreas and stomach.
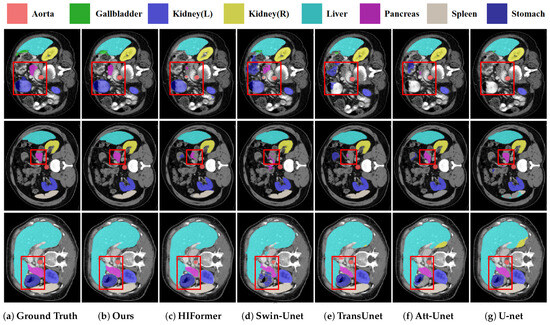
Figure 3.
Qualitative results of multiple models on the Synapse dataset, with ground truth, our DEIF-Net, HiFormer, Swin U-Net, TransUnet, Att-Unet, and U-Net visualization indicated by (a–g), separately.
4.5. Results on ACDC Segmentation
The DEIF-Net method was assessed using the ACDC dataset and compared with various other segmentation approaches. The results of these comparisons are detailed in Table 2. DEIF-Net outperformed others in DSC and HD95, achieving values of 92.02% and 1.08 mm, respectively. It also recorded the highest DSC accuracy for ventricular segmentation. In particular, DEIF-Net demonstrated improvements of 2.66% and 1.74% in DSC accuracy over the Unet [1] and Att-Unet [3] models, both of which employ convolutional network architectures. Furthermore, in contrast with Transformer-based models such as Swin-Unet [17] and MISSFormer [19], DEIF-Net showed enhancements of 2.02% and 4.12%, respectively. Additionally, it outperformed the hybrid Transformer and CNN-based models TransUnet [20] and HiFormer [24] by 2.31% and 1.59%, respectively. It is important to note that several methods utilizing hybrid Transformer and CNN architectures did not demonstrate significant advancements over traditional convolutional networks like Unet and Att-Unet. By contrast, DEIF-Net capitalizes on the synergistic interaction and feature fusion between different encoder types, effectively harnessing the strengths of both, which culminates in marked improvements in segmentation efficacy.

Table 2.
Comparative results of the developed method on the ACDC dataset, with the second highest performance indicated by blue and the highest performance indicated by red.
Further evaluations of DEIF-Net’s performance in cardiac organ segmentation were conducted through visual assessments, as depicted in Figure 4. DEIF-Net and other methods were generally successful in accurately segmenting the left ventricle and myocardium. However, the segmentation of the right ventricle posed challenges for several models, which exhibited issues such as incomplete segmentation and non-smooth segmentation boundaries. For example, Att-Unet [3] consistently failed to fully segment the right ventricle in various cases, while HiFormer [24], Swin-Unet [17], and TransUnet [20] struggled with effectively delineating the segmentation edges in multiple instances. In contrast, DEIF-Net not only accomplished complete segmentation of the right ventricle but also excelled at delineating the edges of various tissues, demonstrating reduced misdetection rates compared to the models mentioned.
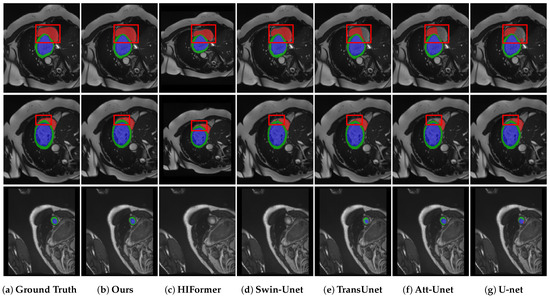
Figure 4.
Qualitative results of multiple models on the ACDC dataset, with the right ventricle highlighted in red, the left ventricle in blue, and the myocardium in green.
4.6. Results on Kvasir-SEG Segmentation
For the evaluation of the efficacy of DEFI-Net, we carried out a series of experiments using the Kvasir-SEG dataset, with performance benchmarks compared against multiple existing approaches. DEFI-Net achieved the highest scores in both DSC and IoU, with results of 87.45% and 80.51%, respectively (Table 3). In comparison to convolutional-network-based methods, including Att-UNet [3] and U-Net [1], DEFI-Net exhibited significant enhancements, improving DSC by 4.46% over U-Net and 4.60% over Att-UNet. Additionally, DEFI-Net surpassed hybrid CNN–Transformer methods, including TransUNet [20] and HiFormer [24]. Specifically, it improved DSC and IoU by 1.84% and 2.36% over TransUNet and by 1.49% and 2.25% over HiFormer, respectively. These findings underscore DEFI-Net’s superior segmentation capabilities on the Kvasir-SEG dataset.

Table 3.
Comparison results of the proposed method on the Kvasir-SEG dataset, with the best outcomes highlighted in red and the second best in blue.
Furthermore, the segmentation results of DEFI-Net and other comparative methods on the Kvasir-SEG dataset were visualized. As depicted in Figure 5, DEFI-Net demonstrated better performance in segmenting colon polyp images, outperforming other methods. Notably, while most competing methods encountered issues with incomplete segmentation, DEFI-Net effectively mitigated this challenge, thereby reinforcing its effectiveness in colon polyp image segmentation.
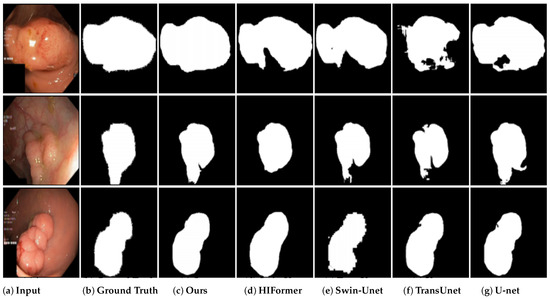
Figure 5.
Qualitative results of different models on the Kvasir-SEG dataset.
4.7. Ablation Experiments
For ascertaining the effectiveness of specific components within the DEFI-Net architecture, a series of ablation experiments were performed across various datasets. These experiments focused on evaluating the impact of disabling the GLIL and GLFF modules. A baseline network was established by directly concatenating the outputs of the encoder to assess the contributions of these modules. As demonstrated in Table 4, integrating the GLIL module significantly enhanced segmentation performance, as evidenced by notable improvements in DSC and HD on the Synapse and ACDC datasets, and in DSC and IoU on the Kvasir-SEG dataset. These results highlight the critical role of encoder interactions and confirm that the GLIL module effectively integrates complementary features from both CNN and Transformer architectures. Conversely, when the GLIL module was disabled and replaced with the GLFF module, there was an improvement in segmentation accuracy, indicating that the GLFF module effectively amalgamates multi-scale encoder features to augment model performance. Ultimately, the highest segmentation performance was realized when all proposed modules were integrated, further validating the synergistic effect of combining the GLIL and GLFF components within the DEFI-Net framework.

Table 4.
Ablation experiments on three datasets.
4.8. Conclusions
A hybrid CNN–Transformer architecture named DEFI-Net that exhibits superior segmentation performance in MIS tasks is introduced. This advantage arises from effective interaction and feature fusion between the CNN and Transformer encoders. The interaction within DEFI-Net is facilitated by the GLIL, which allows the CNN to absorb global information from the Transformer and vice versa, enabling the Transformer to acquire local detail knowledge from the CNN. This promotes a robust interaction between the two encoders. Feature fusion is accomplished through the GLFF, which harnesses both global and local information to enhance the integration of features. Evaluations conducted on three distinct datasets underscore the superior performance of DEFI-Net compared to other methods. Additionally, ablation experiments validate the critical role of encoder interactions in achieving high segmentation accuracy. However, the dual-encoder structure of DEFI-Net introduces significant computational overhead. Future work will therefore focus on developing a more efficient network structure inspired by DEFI-Net, aiming to maintain high accuracy while reducing computational costs.
Author Contributions
Conceptualization, H.Y., Y.F. and P.Y.; data curation, H.Y., Y.F. and P.Y.; formal analysis, H.Y. and P.Y.; funding acquisition, H.Y.; investigation, H.Y. and P.Y.; methodology, H.Y.; project administration, H.Y.; resources, H.Y.; software, H.Y.; supervision, Y.F.; validation, H.Y.; visualization, H.Y. and P.Y.; writing—original draft, H.Y.; writing—review and editing, H.Y., Y.F. and P.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The dataset is made available online at https://www.synapse.org/Synapse:syn3193805 (accessed on 10 February 2025), https://www.creatis.insa-lyon.fr/Challenge/acdc/databases.html (accessed on 10 February 2025), https://datasets.simula.no/kvasir-seg/ (accessed on 10 February 2025).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; proceedings, part III 18; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D vision (3DV), Stanford, CA, USA, 25–28 October 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 565–571. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Xu, Q.; Ma, Z.; Na, H.; Duan, W. DCSAU-Net: A deeper and more compact split-attention U-Net for medical image segmentation. Comput. Biol. Med. 2023, 154, 106626. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.; Dong, C.; Yan, Q.; Sun, Y.; Zhang, C.; Li, Z.; Xu, S. I2U-Net: A dual-path U-Net with rich information interaction for medical image segmentation. Med. Image Anal. 2024, 97, 103241. [Google Scholar] [PubMed]
- Shamshad, F.; Khan, S.; Zamir, S.W.; Khan, M.H.; Hayat, M.; Khan, F.S.; Fu, H. Transformers in medical imaging: A survey. Med. Image Anal. 2023, 88, 102802. [Google Scholar] [CrossRef] [PubMed]
- Pu, Q.; Xi, Z.; Yin, S.; Zhao, Z.; Zhao, L. Advantages of transformer and its application for medical image segmentation: A survey. Biomed. Eng. Online 2024, 23, 14. [Google Scholar] [CrossRef] [PubMed]
- Thirunavukarasu, R.; Kotei, E. A comprehensive review on transformer network for natural and medical image analysis. Comput. Sci. Rev. 2024, 53, 100648. [Google Scholar] [CrossRef]
- He, K.; Gan, C.; Li, Z.; Rekik, I.; Yin, Z.; Ji, W.; Gao, Y.; Wang, Q.; Zhang, J.; Shen, D. Transformers in medical image analysis. Intell. Med. 2023, 3, 59–78. [Google Scholar]
- Karimi, D.; Vasylechko, S.D.; Gholipour, A. Convolution-free medical image segmentation using transformers. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; proceedings, part I 24; Springer: Berlin/Heidelberg, Germany, 2021; pp. 78–88. [Google Scholar]
- Alexey, D. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Khan, S.; Naseer, M.; Hayat, M.; Zamir, S.W.; Khan, F.S.; Shah, M. Transformers in vision: A survey. ACM Comput. Surv. (CSUR) 2022, 54, 1–41. [Google Scholar] [CrossRef]
- Parvaiz, A.; Khalid, M.A.; Zafar, R.; Ameer, H.; Ali, M.; Fraz, M.M. Vision Transformers in medical computer vision—A contemplative retrospection. Eng. Appl. Artif. Intell. 2023, 122, 106126. [Google Scholar] [CrossRef]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jégou, H. Training data-efficient image transformers & distillation through attention. In Proceedings of the International Conference on Machine Learning, Virtual, 18–24 July 2021; PMLR: New York, NY, USA, 2021; pp. 10347–10357. [Google Scholar]
- Wang, W.; Xie, E.; Li, X.; Fan, D.P.; Song, K.; Liang, D.; Lu, T.; Luo, P.; Shao, L. Pyramid vision transformer: A versatile backbone for dense prediction without convolutions. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 11–17 October 2021; pp. 568–578. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the European Conference on Computer Vision, Tel Aviv, Israel, 23–27 October 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 205–218. [Google Scholar]
- Azad, R.; Heidari, M.; Shariatnia, M.; Aghdam, E.K.; Karimijafarbigloo, S.; Adeli, E.; Merhof, D. Transdeeplab: Convolution-free transformer-based deeplab v3+ for medical image segmentation. In Proceedings of the InternationalWorkshop on PRedictive Intelligence in MEdicine, Singapore, 22 September 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 91–102. [Google Scholar]
- Huang, X.; Deng, Z.; Li, D.; Yuan, X.; Fu, Y. Missformer: An effective transformer for 2d medical image segmentation. IEEE Trans. Med. Imaging 2022, 42, 1484–1494. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Azad, R.; Jia, Y.; Aghdam, E.K.; Cohen-Adad, J.; Merhof, D. Enhancing medical image segmentation with TransCeption: A multi-scale feature fusion approach. arXiv 2023, arXiv:2301.10847. [Google Scholar]
- Zhang, Y.; Liu, H.; Hu, Q. Transfuse: Fusing transformers and cnns for medical image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; proceedings, Part I 24; Springer: Berlin/Heidelberg, Germany, 2021; pp. 14–24. [Google Scholar]
- Yuan, F.; Zhang, Z.; Fang, Z. An effective CNN and Transformer complementary network for medical image segmentation. Pattern Recognit. 2023, 136, 109228. [Google Scholar]
- Heidari, M.; Kazerouni, A.; Soltany, M.; Azad, R.; Aghdam, E.K.; Cohen-Adad, J.; Merhof, D. Hiformer: Hierarchical multi-scale representations using transformers for medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 2–7 January 2023; pp. 6202–6212. [Google Scholar]
- Jha, D.; Smedsrud, P.; Riegler, M.; Halvorsen, P.; De Lange, T.; Johansen, D.; Johansen, H. Kvasir-SEG: A Segmented Polyp Dataset. In MultiMedia Modeling; MMM 2020. Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2019; p. 11962. [Google Scholar]
- Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.A.; Cetin, I.; Lekadir, K.; Camara, O.; Ballester, M.A.G.; et al. Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: Is the problem solved? IEEE Trans. Med. Imaging 2018, 37, 2514–2525. [Google Scholar] [CrossRef] [PubMed]
- Landman, B.; Xu, Z.; Igelsias, J.; Styner, M.; Langerak, T.; Klein, A. Miccai multi-atlas labeling beyond the cranial vault–workshop and challenge. In Proceedings of the MICCAI Multi-Atlas Labeling Beyond Cranial Vault—Workshop Challenge, Munich, Germany, 9 October 2015; Volume 5, p. 12. [Google Scholar]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 20 September 2018; Proceedings 4; Springer: Berlin/Heidelberg, Germany, 2018; pp. 3–11. [Google Scholar]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.; Chen, Y.W.; Wu, J. Unet 3+: A full-scale connected unet for medical image segmentation. In Proceedings of the ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1055–1059. [Google Scholar]
- Sinha, A.; Dolz, J. Multi-scale self-guided attention for medical image segmentation. IEEE J. Biomed. Health Inform. 2020, 25, 121–130. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Xiao, X.; Lian, S.; Luo, Z.; Li, S. Weighted res-unet for high-quality retina vessel segmentation. In Proceedings of the 2018 9th International Conference on Information Technology in Medicine and Education (ITME), Hangzhou, China, 19–21 October 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 327–331. [Google Scholar]
- Ibtehaz, N.; Rahman, M.S. MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw. 2020, 121, 74–87. [Google Scholar] [PubMed]
- Lin, A.; Chen, B.; Xu, J.; Zhang, Z.; Lu, G.; Zhang, D. Ds-transunet: Dual swin transformer u-net for medical image segmentation. IEEE Trans. Instrum. Meas. 2022, 71, 1–15. [Google Scholar]
- Hatamizadeh, A.; Tang, Y.; Nath, V.; Yang, D.; Myronenko, A.; Landman, B.; Roth, H.R.; Xu, D. Unetr: Transformers for 3d medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Virtual, 3–8 January 2022; pp. 574–584. [Google Scholar]
- Gao, Y.; Zhou, M.; Metaxas, D.N. UTNet: A hybrid transformer architecture for medical image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part III 24; Springer: Berlin/Heidelberg, Germany, 2021; pp. 61–71. [Google Scholar]
- Manzari, O.N.; Kaleybar, J.M.; Saadat, H.; Maleki, S. BEFUnet: A Hybrid CNN-Transformer Architecture for Precise Medical Image Segmentation. arXiv 2024, arXiv:2402.08793. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Wang, Q.; Wu, B.; Zhu, P.; Li, P.; Zuo, W.; Hu, Q. ECA-Net: Efficient channel attention for deep convolutional neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Virtual, 14–19 June 2020; pp. 11534–11542. [Google Scholar]
- Jaderberg, M.; Simonyan, K.; Zisserman, A. Spatial transformer networks. In Proceedings of the 29th Annual Conference on Neural Information Processing Systems 2015, Montreal, QC, Canada, 7–12 December 2015. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Azad, R.; Al-Antary, M.T.; Heidari, M.; Merhof, D. Transnorm: Transformer provides a strong spatial normalization mechanism for a deep segmentation model. IEEE Access 2022, 10, 108205–108215. [Google Scholar] [CrossRef]
- Azad, R.; Arimond, R.; Aghdam, E.K.; Kazerouni, A.; Merhof, D. Dae-former: Dual attention-guided efficient transformer for medical image segmentation. In Proceedings of the InternationalWorkshop on PRedictive Intelligence in MEdicine, Vancouver, BC, Canada, 8 October 2023; Springer: Berlin/Heidelberg, Germany, 2023; pp. 83–95. [Google Scholar]
- Liu, Q.; Kaul, C.; Wang, J.; Anagnostopoulos, C.; Murray-Smith, R.; Deligianni, F. Optimizing vision transformers for medical image segmentation. In Proceedings of the ICASSP 2023-2023 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Rhodes Island, Greece, 4–10 June 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 1–5. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).