Interaction Between Human Microbiota, Immune System, and Hepatitis C Virus Infection: A Narrative Review
Abstract
1. Introduction
2. Methodology
2.1. Research Strategy
2.2. Eligibility Criteria
2.3. Data Extraction
3. HCV Infection
4. Microbiota and Viral Infections
5. HCV and Microbiota
6. Microbiota Alterations in HCV Patients
7. HCV and Immune System
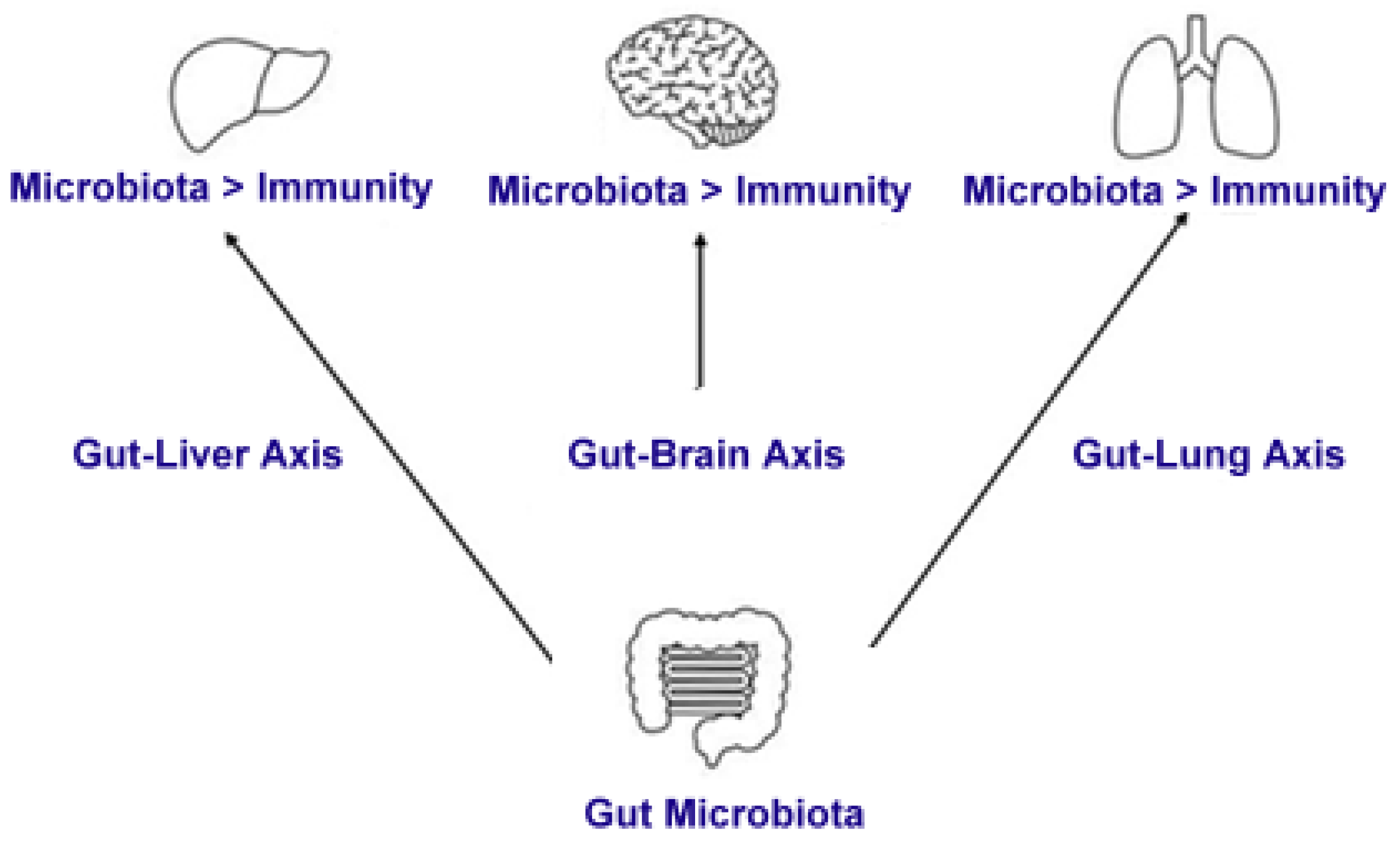
8. The Immune Response and Its Interaction with the Gut Microbiota
8.1. The Gut Microbiota and Immune System Development
8.2. Microbial Metabolites and Immune Modulation
8.3. Gut Microbiota and Systemic Immune Regulation
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| APCs | Antigen-Presenting Cells |
| CD | Cluster of Differentiation |
| CLRs | C-type Lectin Receptors |
| DCs | Dendritic Cells |
| HCC | Hepatocellular Carcinoma |
| HCV | Hepatitis C Virus |
| IECs | Intestinal Epithelial Cells |
| IL | Interleukin |
| KIR | Killer Immunoglobulin-like Receptors |
| NK | Natural Killer |
| NKT | Natural Killer T |
| NLRs | Nucleotide-binding Oligomerization Domain-like Receptors |
| PRRs | Pattern-Recognition Receptors |
| SCFAs | Short-Chain Fatty Acids |
| TLRs | Toll-like Receptors |
| Tregs | Regulatory T Cells |
| UNIFRAC | UniFrac distance |
References
- Bianconi, E.; Piovesan, A.; Facchin, F.; Beraudi, A.; Casadei, R.; Frabetti, F.; Vitale, L.; Pelleri, M.C.; Tassani, S.; Piva, F.; et al. An estimation of the number of cells in the human body. Ann. Hum. Biol. 2013, 40, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.L.; Lee, Y.K. Microflora of the gastrointestinal tract: A review. Methods Mol. Biol. 2004, 268, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Whitman, W.B.; Coleman, D.C.; Wiebe, W.J. Prokaryotes: The unseen majority. Proc. Natl. Acad. Sci. USA 1998, 95, 6578–6583. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; NISC Comparative Sequencing Program; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; et al. Topographical and temporal diversity of the human skin microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef]
- Keijser, B.J.; Zaura, E.; Huse, S.M.; van der Vossen, J.M.; Schuren, F.H.; Montijn, R.C.; ten Cate, J.M.; Crielaard, W. Pyrosequencing analysis of the oral microflora of healthy adults. J. Dent. Res. 2008, 87, 1016–1020. [Google Scholar] [CrossRef]
- Preveden, T.; Scarpellini, E.; Milić, N.; Luzza, F.; Abenavoli, L. Gut microbiota changes and chronic hepatitis C virus infection. Expert Rev. Gastroenterol. Hepatol. 2017, 11, 813–819. [Google Scholar] [CrossRef] [PubMed]
- Buffie, C.G.; Pamer, E.G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 2013, 13, 790–801. [Google Scholar] [CrossRef]
- Maynard, C.L.; Elson, C.O.; Hatton, R.D.; Weaver, C.T. Reciprocal interactions of the intestinal microbiota and immune system. Nature 2012, 489, 231–241. [Google Scholar] [CrossRef]
- Hill, D.A.; Hoffmann, C.; Abt, M.C.; Du, Y.; Kobuley, D.; Kirn, T.J.; Bushman, F.D.; Artis, D. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010, 3, 148–158. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef]
- Kuss, S.K.; Best, G.T.; Etheredge, C.A.; Pruijssers, A.J.; Frierson, J.M.; Hooper, L.V.; Dermody, T.S.; Pfeiffer, J.K. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science 2011, 334, 249–252. [Google Scholar] [CrossRef]
- Robinson, C.M.; Jesudhasan, P.R.; Pfeiffer, J.K. Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell Host Microbe 2014, 15, 36–46. [Google Scholar] [CrossRef]
- Kane, M.; Case, L.K.; Kopaskie, K.; Kozlova, A.; MacDearmid, C.; Chervonsky, A.V.; Golovkina, T.V. Successful transmission of a retrovirus depends on the commensal microbiota. Science 2011, 334, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Miura, T.; Sano, D.; Suenaga, A.; Yoshimura, T.; Fuzawa, M.; Nakagomi, T.; Nakagomi, O.; Okabe, S. Histo-blood group antigen-like substances of human enteric bacteria as specific adsorbents for human noroviruses. J. Virol. 2013, 87, 9441–9451. [Google Scholar] [CrossRef] [PubMed]
- Vaarala, O. Is the origin of type 1 diabetes in the gut? Immunol. Cell Biol. 2012, 90, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Vaarala, O. Human intestinal microbiota and type 1 diabetes. Curr. Diab. Rep. 2013, 13, 601–607. [Google Scholar] [CrossRef]
- Zipris, D. The interplay between the gut microbiota and the immune system in the mechanism of type 1 diabetes. Curr. Opin. Endocrinol. Diabetes Obes. 2013, 20, 265–270. [Google Scholar] [CrossRef]
- Li, H.C.; Lo, S.Y. Hepatitis C virus: Virology, diagnosis and treatment. World J. Hepatol. 2015, 7, 1377–1389. [Google Scholar] [CrossRef]
- Alberti, A.; Chemello, L.; Benvegnù, L. Natural history of hepatitis C. J. Hepatol. 1999, 31 (Suppl. S1), 17–24. [Google Scholar] [CrossRef]
- Hoofnagle, J.H. Course and outcome of hepatitis C. Hepatology 2002, 36 (Suppl. S1), S21–S29. [Google Scholar] [CrossRef]
- Chen, S.L.; Morgan, T.R. The natural history of hepatitis C virus (HCV) infection. Int. J. Med. Sci. 2006, 3, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Carrozzo, M.; Scally, K. Oral manifestations of hepatitis C virus infection. World J. Gastroenterol. 2014, 20, 7534–7543. [Google Scholar] [CrossRef]
- Ozkok, A.; Yildiz, A. Hepatitis C virus associated glomerulopathies. World J. Gastroenterol. 2014, 20, 7544–7554. [Google Scholar] [CrossRef] [PubMed]
- Grimbert, S.; Valensi, P.; Lévy-Marchal, C.; Perret, G.; Richardet, J.P.; Raffoux, C.; Trinchet, J.C.; Beaugrand, M. High prevalence of diabetes mellitus in patients with chronic hepatitis C: A case-control study. Gastroenterol. Clin. Biol. 1996, 20, 544–548. [Google Scholar] [PubMed]
- Montenegro, L.; De Michina, A.; Misciagna, G.; Guerra, V.; Di Leo, A. Virus C hepatitis and type 2 diabetes: A cohort study in southern Italy. Am. J. Gastroenterol. 2013, 108, 1108–1111. [Google Scholar] [CrossRef]
- Preciado, M.V.; Valva, P.; Escobar-Gutierrez, A.; Rahal, P.; Ruiz-Tovar, K.; Yamasaki, L.; Vazquez-Chacon, C.; Martinez-Guarneros, A.; Carpio-Pedroza, J.C.; Fonseca-Coronado, S.; et al. Hepatitis C virus molecular evolution: Transmission, disease progression and antiviral therapy. World J. Gastroenterol. 2014, 20, 15992–16013. [Google Scholar] [CrossRef]
- Vercauteren, K.; de Jong, Y.P.; Meuleman, P. Animal models for the study of HCV. Curr. Opin. Virol. 2015, 13, 67–74. [Google Scholar] [CrossRef]
- Xi, Z.; Ramirez, J.L.; Dimopoulos, G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 2008, 4, e1000098. [Google Scholar] [CrossRef]
- Wilks, J.; Golovkina, T. Influence of microbiota on viral infections. PLoS Pathog. 2012, 8, e1002681. [Google Scholar] [CrossRef]
- Isaak, D.D.; Bartizal, K.F.; Caulfield, M.J. Decreased pathogenicity of murine leukemia virus-Moloney in gnotobiotic mice. Leukemia 1988, 2, 540–544. [Google Scholar] [PubMed]
- Kouttab, N.M.; Jutila, J.W. Friend leukemia virus infection in germfree mice following antigen stimulation. J. Immunol. 1972, 108, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Heidrich, B.; Vital, M.; Plumeier, I.; Döscher, N.; Kahl, S.; Kirschner, J.; Ziegert, S.; Solbach, P.; Lenzen, H.; Potthoff, A.; et al. Intestinal microbiota in patients with chronic hepatitis C with and without cirrhosis compared with healthy controls. Liver Int. 2018, 38, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, Y.; Bolgarina, Z.; Desai, H.N.; Senaratne, M.; Swami, S.S.; Aye, S.L.; Mohammed, L. The role of gut microbiome in hepatocellular carcinoma: A systematic review. Cureus 2023, 15, e43862. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Seo, G.S. Fecal Microbiota Transplantation: Is It Safe? Clin. Endosc. 2021, 54, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Zawistowska-Rojek, A.; Tyski, S. Are Probiotic Really Safe for Humans? Pol. J. Microbiol. 2018, 67, 251–258. [Google Scholar] [CrossRef]
- Adawi, D.; Ahrné, S.; Molin, G. Effects of different probiotic strains of Lactobacillus and Bifidobacterium on bacterial translocation and liver injury in an acute liver injury model. Int. J. Food Microbiol. 2001, 70, 213–220. [Google Scholar] [CrossRef]
- Bajaj, J.S.; Heuman, D.M.; Hylemon, P.B.; Sanyal, A.J.; Puri, P.; Sterling, R.K.; Luketic, V.; Stravitz, R.T.; Siddiqui, M.S.; Fuchs, M.; et al. Randomised clinical trial: Lactobacillus GG modulates gut microbiome, metabolome and endotoxemia in patients with cirrhosis. Aliment. Pharmacol. Ther. 2014, 39, 1113–1125. [Google Scholar] [CrossRef]
- Chiva, M.; Soriano, G.; Rochat, I.; Peralta, C.; Rochat, F.; Llovet, T.; Mirelis, B.; Schiffrin, E.J.; Guarner, C.; Balanzó, J. Effect of Lactobacillus johnsonii La1 and antioxidants on intestinal flora and bacterial translocation in rats with experimental cirrhosis. J. Hepatol. 2002, 37, 456–462. [Google Scholar] [CrossRef]
- Aly, A.M.; Adel, A.; El-Gendy, A.O.; Essam, T.M.; Aziz, R.K. Gut microbiome alterations in patients with stage 4 hepatitis C. Gut Pathog. 2016, 8, 42. [Google Scholar] [CrossRef]
- Bajaj, J.S.; Sterling, R.K.; Betrapally, N.S.; Nixon, D.E.; Fuchs, M.; Daita, K.; Heuman, D.M.; Sikaroodi, M.; Hylemon, P.B.; White, M.B.; et al. HCV eradication does not impact gut dysbiosis or systemic inflammation in cirrhotic patients. Aliment. Pharmacol. Ther. 2016, 44, 638–643. [Google Scholar] [CrossRef]
- Inoue, T.; Nakayama, J.; Moriya, K.; Kawaratani, H.; Momoda, R.; Ito, K.; Iio, E.; Nojiri, S.; Fujiwara, K.; Yoneda, M.; et al. Gut dysbiosis associated with hepatitis C virus infection. Clin. Infect. Dis. 2018, 67, 869–877. [Google Scholar] [CrossRef] [PubMed]
- El-Mowafy, M.; Elgaml, A.; El-Mesery, M.; Sultan, S.; Ahmed, T.A.E.; Gomaa, A.I.; Aly, M.; Mottawea, W. Changes of Gut-Microbiota-Liver Axis in Hepatitis C Virus Infection. Biology 2021, 10, 55. [Google Scholar] [CrossRef]
- Hsu, Y.C.; Chen, C.C.; Lee, W.H.; Chang, C.Y.; Lee, F.J.; Tseng, C.H.; Chen, T.H.; Ho, H.J.; Lin, J.T.; Wu, C.Y. Compositions of gut microbiota before and shortly after hepatitis C viral eradication by direct antiviral agents. Sci. Rep. 2022, 12, 5481. [Google Scholar] [CrossRef] [PubMed]
- Midori, Y.; Nosaka, T.; Hiramatsu, K.; Akazawa, Y.; Tanaka, T.; Takahashi, K.; Naito, T.; Matsuda, H.; Ohtani, M.; Nakamoto, Y. Isolation of mucosa-associated microbiota dysbiosis in the ascending colon in hepatitis C virus post-sustained virologic response cirrhotic patients. Front. Cell. Infect. Microbiol. 2024, 14, 1371429. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Yang, Y.; He, Q.; Zhu, P.; Liu, M.; Xu, J.; Zhao, M. Intestinal Microbiota—A Promising Target for Antiviral Therapy? Front. Immunol. 2021, 12, 676232. [Google Scholar] [CrossRef]
- Trebicka, J.; Macnaughtan, J.; Schnabl, B.; Shawcross, D.L.; Bajaj, J.S. The microbiota in cirrhosis and its role in hepatic decompensation. J. Hepatol. 2021, 75 (Suppl. S1), S67–S81. [Google Scholar] [CrossRef]
- Yang, X.; Mai, H.; Zhou, J.; Li, Z.; Wang, Q.; Lan, L.; Lu, F.; Yang, X.; Guo, B.; Ye, L.; et al. Alterations of the gut microbiota associated with the occurrence and progression of viral hepatitis. Front. Cell. Infect. Microbiol. 2023, 13, 1119875. [Google Scholar] [CrossRef]
- Soldán, M.; Argalášová, Ľ.; Hadvinová, L.; Galileo, B.; Babjaková, J. The Effect of Dietary Types on Gut Microbiota Composition and Development of Non-Communicable Diseases: A Narrative Review. Nutrients 2024, 16, 3134. [Google Scholar] [CrossRef]
- Taur, Y.; Pamer, E.G. The intestinal microbiota and susceptibility to infection in immunocompromised patients. Curr. Opin. Infect Dis. 2013, 26, 332–337. [Google Scholar] [CrossRef]
- Adhikary, S.; Esmeeta, A.; Dey, A.; Banerjee, A.; Saha, B.; Gopan, P.; Duttaroy, A.K.; Pathak, S. Impacts of gut microbiota alteration on age-related chronic liver diseases. Dig. Liver Dis. 2024, 56, 112–122. [Google Scholar] [CrossRef]
- Corado, J.; Toro, F.; Rivera, H.; Bianco, N.E.; Deibis, L.; De Sanctis, J.B. Impairment of natural killer (NK) cytotoxic activity in hepatitis C virus (HCV) infection. Clin. Exp. Immunol. 1997, 109, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Khakoo, S.I. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science 2004, 305, 872–874. [Google Scholar] [CrossRef]
- Brown, M.G. Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science 2001, 292, 934–937. [Google Scholar] [CrossRef]
- Golden-Mason, L.; Rosen, H.R.; Golden-Mason, L. Natural killer cells: Primary target for hepatitis C virus immune evasion strategies? Liver Transplant. 2006, 12, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Guidotti, L.G.; Chisari, F.V. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu. Rev. Immunol. 2001, 19, 65–91. [Google Scholar] [CrossRef]
- Lunemann, S.; Schlaphoff, V.; Cornberg, M.; Wedemeyer, H. NK cells in hepatitis C: Role in disease susceptibility and therapy. Dig. Dis. 2012, 30, 48–54. [Google Scholar] [CrossRef]
- Doherty, D.G.; O’Farrelly, C. Innate and adaptive lymphoid cells in the human liver. Immunol. Rev. 2000, 174, 5–20. [Google Scholar] [CrossRef]
- Ahmad, A.; Alvarez, F. Role of NK and NKT cells in the immunopathogenesis of HCV-induced hepatitis. J. Leukoc. Biol. 2004, 76, 743–759. [Google Scholar] [CrossRef] [PubMed]
- Swain, M.G. Hepatic NKT cells: Friend or foe? Clin. Sci. 2008, 114, 457–466. [Google Scholar] [CrossRef]
- Lucas, M.; Gadola, S.; Meier, U.; Young, N.T.; Harcourt, G.; Karadimitris, A.; Coumi, N.; Brown, D.; Dusheiko, G.; Cerundolo, V.; et al. Frequency and phenotype of circulating Valpha24/Vbeta11 double-positive natural killer T cells during hepatitis C virus infection. J. Virol. 2003, 77, 2251–2257. [Google Scholar] [CrossRef]
- Deignan, T.; Curry, M.P.; Doherty, D.G.; Golden-Mason, L.; Volkov, Y.; Norris, S.; Nolan, N.; Traynor, O.; McEntee, G.; Hegarty, J.E.; et al. Decrease in hepatic CD56(+) T cells and V alpha 24(+) natural killer T cells in chronic hepatitis C viral infection. J. Hepatol. 2002, 37, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Inoue, M.; Kanto, T.; Miyatake, H.; Itose, I.; Miyazaki, M.; Yakushijin, T.; Sakakibara, M.; Kuzushita, N.; Hiramatsu, N.; Takehara, T.; et al. Enhanced ability of peripheral invariant natural killer T cells to produce IL-13 in chronic hepatitis C virus infection. J. Hepatol. 2006, 45, 190–196. [Google Scholar] [CrossRef]
- Wynn, T.A. IL-13 effector functions. Annu. Rev. Immunol. 2003, 21, 425–456. [Google Scholar] [CrossRef]
- Parkin, J.; Cohen, B. An overview of the immune system. Lancet 2001, 357, 1777–1789. [Google Scholar] [CrossRef] [PubMed]
- Hillion, S.; Arleevskaya, M.I.; Blanco, P.; Bordron, A.; Brooks, W.H.; Cesbron, J.Y.; Kaveri, S.; Vivier, E.; Renaudineau, Y. The Innate Part of the Adaptive Immune System. Clin. Rev. Allergy Immunol. 2020, 58, 151–154. [Google Scholar] [CrossRef]
- Bonilla, F.A.; Oettgen, H.C. Adaptive immunity. J. Allergy Clin. Immunol. 2010, 125, S33–S40. [Google Scholar] [CrossRef]
- Fiebiger, U.; Bereswill, S.; Heimesaat, M.M. Dissecting the interplay between intestinal microbiota and host immunity in health and disease: Lessons learned from germfree and gnotobiotic animal models. Eur. J. Microbiol. Immunol. 2016, 6, 253–271. [Google Scholar] [CrossRef] [PubMed]
- Fukata, M.; Arditi, M. The role of pattern recognition receptors in intestinal inflammation. Mucosal Immunol. 2013, 6, 451–463. [Google Scholar] [CrossRef]
- Frantz, A.L.; Rogier, E.W.; Weber, C.R.; Shen, L.; Cohen, D.A.; Fenton, L.A.; Bruno, M.E.C.; Kaetzel, C.S. Targeted deletion of MyD88 in intestinal epithelial cells results in compromised antibacterial immunity associated with downregulation of polymeric immunoglobulin receptor, mucin-2, and antibacterial peptides. Mucosal Immunol. 2012, 5, 501–512. [Google Scholar] [CrossRef]
- Araki, A.; Kanai, T.; Ishikura, T.; Makita, S.; Uraushihara, K.; Iiyama, R.; Totsuka, T.; Takeda, K.; Akira, S.; Watanabe, M. MyD88-deficient mice develop severe intestinal inflammation in dextran sodium sulfate colitis. J. Gastroenterol. 2005, 40, 16–23. [Google Scholar] [CrossRef]
- Kubinak, J.L.; Petersen, C.; Stephens, W.Z.; Soto, R.; Bake, E.; O’Connell, R.M.; Round, J.L. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe 2015, 17, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Cao, X. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat. Rev. Immunol. 2016, 16, 35–50. [Google Scholar] [CrossRef]
- Vandamme, D.; Landuyt, B.; Luyten, W.; Schoofs, L. A comprehensive summary of LL-37, the factotum human cathelicidin peptide. Cell. Immunol. 2012, 280, 22–35. [Google Scholar] [CrossRef]
- Chen, F.; Stappenbeck, T.S. Microbiome control of innate reactivity. Curr. Opin. Immunol. 2019, 56, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Schnupf, P.; Gaboriau-Routhiau, V.; Cerf-Bensussan, N. Modulation of the gut microbiota to improve innate resistance. Curr. Opin. Immunol. 2018, 54, 137–144. [Google Scholar] [CrossRef]
- Chun, E.; Lavoie, S.; Fonseca-Pereira, D.; Bae, S.; Michaud, M.; Hoveyda, H.R.; Fraser, G.L.; Comeau, C.A.G.; Glickman, J.N.; Fuller, M.H.; et al. Metabolite-sensing receptor Ffar2 regulates colonic group 3 innate lymphoid cells and gut immunity. Immunity 2019, 51, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Rannug, A. How the AHR became important in intestinal homeostasis-a diurnal FICZ/AHR/CYP1A1 feedback controls both immunity and immunopathology. Int. J. Mol. Sci. 2020, 21, 5681. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; IBDMDB Investigators; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef]
- Baars, A.; Oosting, A.; Knol, J.; Garssen, J.; Van Bergenhenegouwen, J. The gut microbiota as a therapeutic target in IBD and metabolic disease: A role for the bile acid receptors FXR and TGR5. Microorganisms 2015, 3, 641–666. [Google Scholar] [CrossRef]
- Labbé, A.; Ganopolsky, J.G.; Martoni, C.J.; Prakash, S.; Jones, M.L. Bacterial bile metabolising gene abundance in Crohn’s, ulcerative colitis and type 2 diabetes metagenomes. PLoS ONE 2014, 9, e115175. [Google Scholar] [CrossRef]
- Biagi, E.; Candela, M.; Turroni, S.; Garagnani, P.; Franceschi, C.; Brigidi, P. Ageing and gut microbes: Perspectives for health maintenance and longevity. Pharmacol. Res. 2013, 69, 11–20. [Google Scholar] [CrossRef]
- Negi, S.; Das, D.K.; Pahari, S.; Nadeem, S.; Agrewala, J.N. Potential role of gut microbiota in induction and regulation of innate immune memory. Front. Immunol. 2019, 10, 2441. [Google Scholar] [CrossRef] [PubMed]
- Blander, J.M.; Longman, R.S.; Iliev, I.D.; Sonnenberg, G.F.; Artis, D. Regulation of inflammation by microbiota interactions with the host. Nat. Immunol. 2017, 18, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Littman, D.R. The microbiota in adaptive immune homeostasis and disease. Nature 2016, 535, 75–84. [Google Scholar] [CrossRef]
- Francino, M.P. Early development of the gut microbiota and immune health. Pathogens 2014, 3, 769–790. [Google Scholar] [CrossRef] [PubMed]
- Owaga, E.; Hsieh, R.-H.; Mugendi, B.; Masuku, S.; Shih, C.-K.; Chang, J.-S. Th17 cells as potential probiotic therapeutic targets in inflammatory bowel diseases. Int. J. Mol. Sci. 2015, 16, 20841–20858. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; Van Der Veeken, J.; DeRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Luu, M.; Pautz, S.; Kohl, V.; Singh, R.; Romero, R.; Lucas, S.; Hofmann, J.; Raifer, H.; Vachharajani, N.; Carrascosa, L.C.; et al. The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nat. Commun. 2019, 10, 760. [Google Scholar] [CrossRef]
- Ahern, P.P.; Maloy, K.J. Understanding immune-microbiota interactions in the intestine. Immunology 2020, 159, 4–14. [Google Scholar] [CrossRef]
- Grainger, J.; Daw, R.; Wemyss, K. Systemic instruction of cell-mediated immunity by the intestinal microbiome. F1000Research 2018, 7, 1910. [Google Scholar] [CrossRef]
- Doskali, M.; Tanaka, Y.; Ohira, M.; Ishiyama, K.; Tashiro, H.; Chayama, K.; Ohdan, H. Possibility of adoptive immunotherapy with peripheral blood-derived CD3−CD56+ and CD3+CD56+ cells for inducing antihepatocellular carcinoma and antihepatitis C virus activity. J. Immunother. 2011, 34, 129–138. [Google Scholar] [CrossRef] [PubMed]

| Study/Reference | Population | Key Findings |
|---|---|---|
| Aly et al., 2016 [39] | 6 HCV-infected cirrhotic patients vs. 8 healthy controls | Significant reduction in gut microbiome diversity in HCV-infected patients. The potential alteration of the microbiome in chronic hepatitis C may be influenced by bacterial translocation, alongside the liver’s compromised functions in digestion and protein synthesis. |
| Inoue et al., 2018 [41] | 166 HCV-infected patients vs. 23 healthy controls | Gut dysbiosis observed even in asymptomatic patients; progression of clinical stage associated with further dysbiosis. In individuals infected with HCV, there was a notable reduction in bacterial diversity when contrasted with healthy subjects. This decline was particularly evident in the Clostridiales order, while an increase was observed in the populations of Streptococcus and Lactobacillus. |
| Hsu et al., 2022 [43] | 42 HCV-infected patients vs. 84 healthy controls | Altered gut microbiome composition observed; specific bacterial taxa changes noted. The gut microbiota in patients infected with HCV exhibits alterations when compared to uninfected controls; however, the overall composition of the microbiome does not show significant changes in the short term following the eradication of HCV. |
| Aspect | Details |
|---|---|
| Innate Immune Response in HCV | Cellular innate immune response is crucial in host defense against HCV infection |
| Role of NK Cells | NK cells are essential for early antiviral immunity, comprising 30–50% of liver lymphocytes |
| NK Cell Classification | CD16 + CD56dim NK cells: cytotoxic, strong effector functions (target cell lysis) CD16 − CD56bright NK cells: immunoregulatory, cytokine-producing |
| Impact of Other Infections on NK Cells | Infections like cytomegalovirus, Epstein-Barr, herpes virus reduce NK cell cytotoxic activity |
| NK Cell Functions in HCV Infection | Secrete pro-inflammatory cytokines (TNF-α, IFN-γ) to suppress HCV replication. |
| NK-Cell-Induced Liver Injury | Cytotoxic response can lead to unintended liver injury and inflammation |
| Killer Immunoglobulin-like Receptors (KIRs) | KIRs interact with MHC class I molecules, regulating NK cell activity |
| NK Cells in Early HCV Infection | Contribute to early viral load reduction through cytolysis, cytokine secretion, and activation of other immune cells |
| Adaptive Immune Response Activation | Dendritic cells (APCs) process and present viral antigens to activate CD4+ helper T cells, CD8+ cytotoxic T cells, and B cells |
| Interaction between Dendritic Cells and NK Cells | NKp30 receptor triggers IL-12 and IL-15 release from dendritic cells, enhancing NK cell function and cytokine production (IFN-γ, TNF-α) |
| Natural Killer T (NKT) Cells | NKT cells comprise 26% of intrahepatic lymphocytes, secreting IFN-γ, TNF-α, IL-2 |
| NKT Cells in Chronic HCV | Conflicting findings on NKT cell frequency: some studies show increased, others show decreased NKT cells in the liver of chronic HCV patients |
| NKT Cells Cytokine Production in HCV | Altered cytokine profiles, particularly IL-13, which regulates cell-mediated immunity and may influence chronic HCV progression |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frumento, D.; Țălu, Ș. Interaction Between Human Microbiota, Immune System, and Hepatitis C Virus Infection: A Narrative Review. Appl. Sci. 2025, 15, 3157. https://doi.org/10.3390/app15063157
Frumento D, Țălu Ș. Interaction Between Human Microbiota, Immune System, and Hepatitis C Virus Infection: A Narrative Review. Applied Sciences. 2025; 15(6):3157. https://doi.org/10.3390/app15063157
Chicago/Turabian StyleFrumento, Davide, and Ștefan Țălu. 2025. "Interaction Between Human Microbiota, Immune System, and Hepatitis C Virus Infection: A Narrative Review" Applied Sciences 15, no. 6: 3157. https://doi.org/10.3390/app15063157
APA StyleFrumento, D., & Țălu, Ș. (2025). Interaction Between Human Microbiota, Immune System, and Hepatitis C Virus Infection: A Narrative Review. Applied Sciences, 15(6), 3157. https://doi.org/10.3390/app15063157







