Experimental Verification of Performance Improvement in Heat-Related Illness Prediction Using Clinical Significance-Based Binning
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset
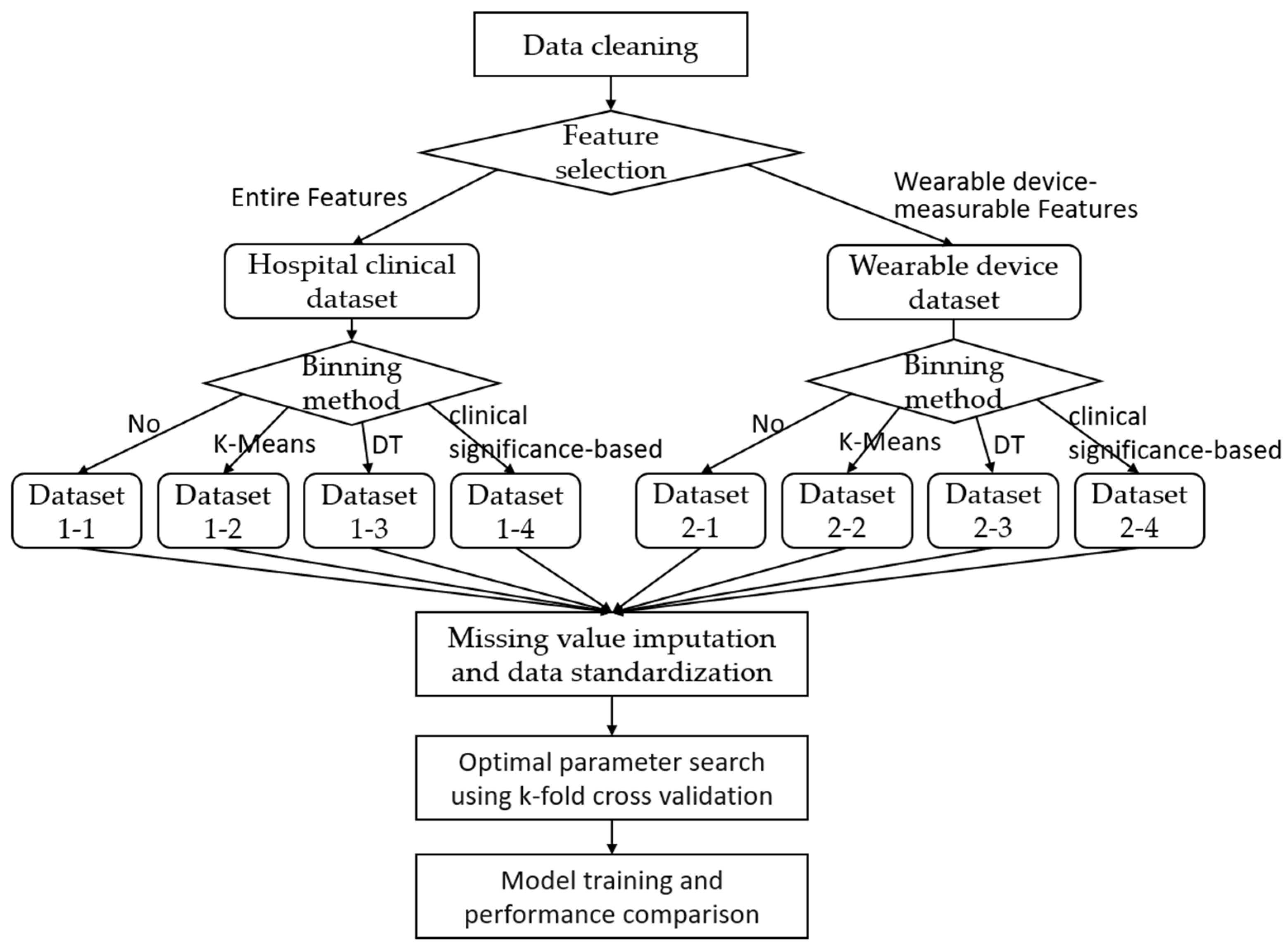
2.2. Experimental Strategy and Methods
- Utilizing all variables clinically collected in hospitals;
- Utilizing only variables measurable in real service environments through wearable devices.
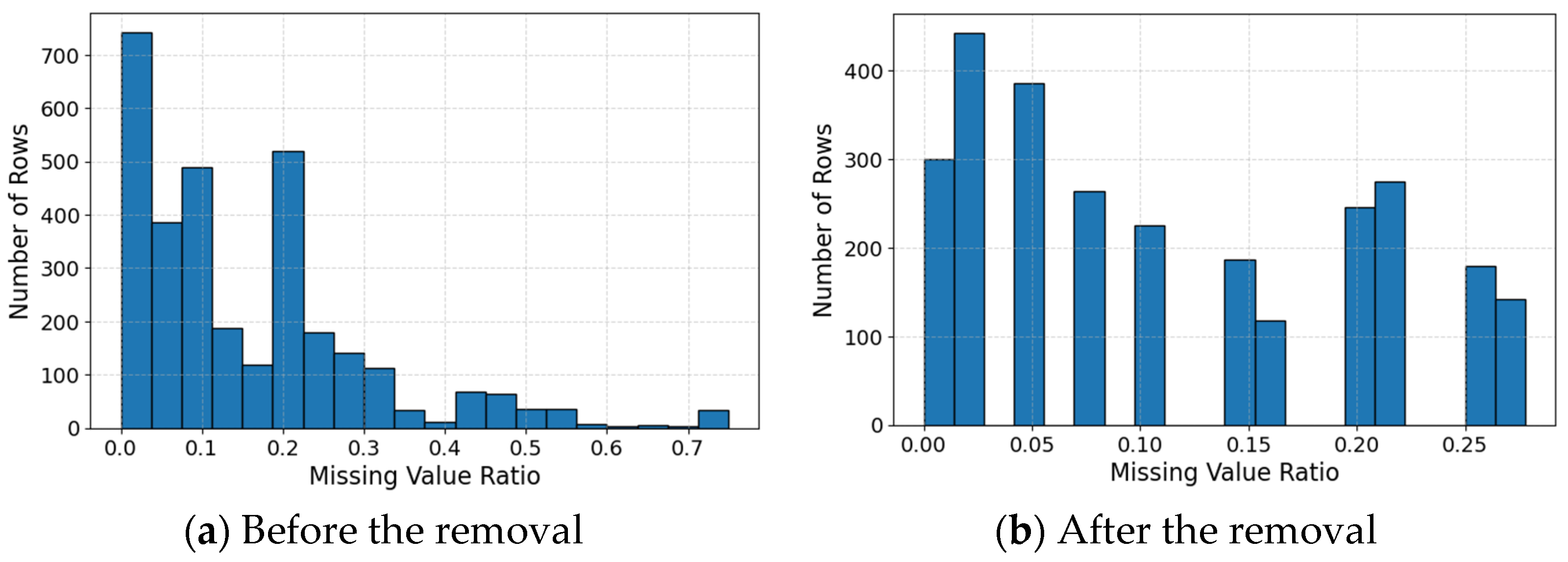
2.2.1. Data Cleaning
2.2.2. Feature Selection
2.2.3. Clinical Significance-Based Binning
- Age is categorized by 10-year intervals;
- Systolic blood pressure is categorized based on the Korea Disease Control and Prevention Agency’s National Health Information Portal [8];
- Heart rate is categorized based on the percentage of maximum heart rate (220 minus age), based on zone training criteria [23];
- Respiration rate is categorized based on basic nursing vital sign criteria [9];
- Body temperature is categorized based on basic nursing vital sign criteria [9];
- Glasgow coma scale is categorized based on the Glasgow coma scale [24].
| Feature | Binning Criteria |
|---|---|
| Age | 0: 0–9, 1: 10–19, 2: 20–29, 3: 30–39, etc. |
| Systolic Blood Pressure (SBP, PreSBP): | 0: Hypotension (<100 mmHg) 1: Normal (100–119 mmHg) 2: Prehypertension Warning (120–129 mmHg) 3: Prehypertension Stage (130–139 mmHg) 4: Mild Hypertension (140–159 mmHg) 5: Moderate or Severe Hypertension (≥160 mmHg) |
| Heart Rate (HR, PreHR) | 0: 50–59%, 1: 60–69%, 2: 70–79%, 3: 80–89%, 4: 90–100% |
| Respiration Rate (PreRR) | 0: Bradypnea (≤11 breaths/min) 1: Normal (12–20 breaths/min) 2: Tachypnea (≥21 breaths/min) |
| Body Temperature (BT, PreBT) | 0: Hypothermia (≤36.0 °C) 1: Normal (36.1–37.5 °C) 2: Fever (>37.5 °C) |
| Glasgow Coma Scale (PreGCS, GCS) | 0: Normal (15 points) 1: Mild Consciousness Impairment (13–14 points) 2: Moderate Coma (8–12 points) 3: Severe Coma (4–7 points) 4: Deep Coma (≤3 points) |
2.2.4. Missing Value Imputation and Data Standardization
2.2.5. Model Training
3. Results
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Korea Disease Control and Prevention Agency. Status of the Emergency Room Surveillance System for Heat-Related Illnesses. 2025. Available online: https://www.kdca.go.kr/board/board.es?mid=a20501010000&bid=0015&list_no=728181&act=view (accessed on 3 October 2025).
- Castineira, D.; Schlosser, K.R.; Geva, A.; Rahmani, A.R.; Fiore, G.; Walsh, B.K.; Smallood, C.D.; Arnold, J.H.; Santillana, M. Adding continuous vital sign information to static clinical data improves the prediction of length of stay after intubation: A data-driven machine learning approach. Respir. Care 2020, 65, 1367–1377. [Google Scholar] [CrossRef] [PubMed]
- Mayampurath, A.; Jani, P.; Dai, Y.; Gibbons, R.; Edelson, D.; Churpek, M.M. A vital sign-based model to predict clinical deterioration in hospitalized children. Pediatr. Crit. Care Med. 2020, 21, 820–826. [Google Scholar] [CrossRef] [PubMed]
- Sundrani, S.; Chen, J.; Jin, B.T.; Abad, Z.S.H.; Rajpurkar, P.; Kim, D. Predicting patient decompensation from continuous physiologic monitoring in the emergency department. NPJ Digit. Med. 2023, 6, 60. [Google Scholar] [CrossRef] [PubMed]
- Candel, B.G.; Duijzer, R.; Gaakeer, M.I.; Ter Avest, E.; Sir, Ö. The association between vital signs and clinical outcomes in emergency department patients of different age categories. Emerg. Med. J. 2022, 39, 903–911. [Google Scholar] [CrossRef]
- van Rossum, M.C.; Vlaskamp, L.B.; Posthuma, L.M.; Visscher, M.J.; Breteler, M.J.; Hermens, H.J.; Kalkman, C.J.; Preckel, B. Adaptive threshold-based alarm strategies for continuous vital signs monitoring. J. Clin. Monit. Comput. 2022, 36, 407–417. [Google Scholar] [CrossRef]
- Haahr-Raunkjaer, C.; Mølgaard, J.; Elvekjaer, M.; Rasmussen, S.M.; Achiam, M.P.; Jorgensen, L.N.; Søgaard, M.I.; Grønbaek, K.K.; Oxbøll, A.B.; Sørensen, H.B.; et al. Continuous monitoring of vital sign abnormalities; association to clinical complications in 500 postoperative patients. Acta Anaesthesiol. Scand. 2022, 66, 552–562. [Google Scholar] [CrossRef]
- Korea Disease Control and Prevention Agency. Korea National Health Information Portal: Classification of Blood Pressure According to Hypertension Treatment Guidelines. Available online: https://health.kdca.go.kr/healthinfo/biz/health/gnrlzHealthInfo/gnrlzHealthInfo/gnrlzHealthInfoView.do?cntnts_sn=5300 (accessed on 3 October 2025).
- Song, K.A.; Choi, D.W.; Kang, M.S.; Kang, H.J.; Gong, K.R.; Kwon, M.J.; Kwon, Y.S.; Kwon, J.O.; Kim, K.M.; Kim, D.Y.; et al. Fundamentals of Nursing I, 9th ed.; Soomoonsa: Paju, Republic of Korea, 2021; p. 338. [Google Scholar]
- Hayashida, K.; Kondo, Y.; Hifumi, T.; Shimazaki, J.; Oda, Y.; Shiraishi, S.; Fukuda, T.; Sasaki, J.; Shimizu, K. A novel early risk assessment tool for detecting clinical outcomes in patients with heat-related illness (J-ERATO score): Development and validation in independent cohorts in Japan. PLoS ONE 2018, 13, e0197032. [Google Scholar] [CrossRef]
- Hirano, Y.; Kondo, Y.; Hifumi, T.; Yokobori, S.; Kanda, J.; Shimazaki, J.; Hayashida, K.; Moriya, T.; Yagi, M.; Takauji, S.; et al. Machine learning-based mortality prediction model for heat-related illness. Sci. Rep. 2021, 11, 9501. [Google Scholar] [CrossRef]
- Kuo, W.Y.; Huang, C.C.; Liu, C.F.; Sung, M.I.; Hsu, C.C.; Lin, H.J.; Su, S.B.; Guo, H.R. Utilizing machine learning for predicting mortality in patients with heat-related illness who visited the emergency department. Int. J. Med. Inform. 2025, 201, 105951. [Google Scholar] [CrossRef]
- Shakerian, S.; Habibnezhad, M.; Ojha, A.; Lee, G.; Liu, Y.; Jebelli, H.; Lee, S. Assessing occupational risk of heat stress at construction: A worker-centric wearable sensor-based approach. Saf. Sci. 2021, 142, 105395. [Google Scholar] [CrossRef]
- Jang, J.; Lee, K.H.; Joo, S.; Kwon, O.; Yi, H.; Lee, D. Smart Helmet for Vital Sign-Based Heatstroke Detection Using Support Vector Machine. J. Sens. Sci. Technol. 2022, 31, 433–440. [Google Scholar] [CrossRef]
- Shimazaki, T.; Anzai, D.; Watanabe, K.; Nakajima, A.; Fukuda, M.; Ata, S. Heat stroke prevention in hot specific occupational environment enhanced by supervised machine learning with personalized vital signs. Sensors 2022, 22, 395. [Google Scholar] [CrossRef]
- Yaldiz, C.O.; Buller, M.J.; Richardson, K.L.; An, S.; Lin, D.J.; Satish, A. Early prediction of impending Exertional heat stroke with Wearable Multimodal sensing and anomaly detection. IEEE J. Biomed. Health Inform. 2023, 27, 5803–5814. [Google Scholar] [CrossRef]
- Khanmohammadi, S.; Adibeig, N.; Shanehbandy, S. An improved overlapping k-means clustering method for medical applications. Expert Syst. Appl. 2017, 67, 12–18. [Google Scholar] [CrossRef]
- Yang, Z.; Zou, W.; Liu, H.; Sharma, R.P.; Zhang, M.; Hu, Z. The Effect of Soil and Topography Factors on Larix gmelinii var. Principis-rupprechtii Forest Mortality and Capability of Decision Tree Binning Method and Generalized Linear Models in Predicting Tree Mortality. Forests 2024, 15, 2060. [Google Scholar] [CrossRef]
- Takegawa, R.; Kanda, J.; Yaguchi, A.; Yokobori, S.; Hayashida, K. A prehospital risk assessment tool predicts clinical outcomes in hospitalized patients with heat-related illness: A Japanese nationwide prospective observational study. Sci. Rep. 2023, 13, 1189. [Google Scholar] [CrossRef] [PubMed]
- Japanese Association for Acute Medicine, Heatstroke Surveillance Committee. Heat related illness in Japan: The final report of Heatstroke STUDY 2012. Jpn. J. Acute Med. 2012, 23, 211–229. [Google Scholar]
- Japanese Association for Acute Medicine, Heatstroke Surveillance Committee. Characteristics of elderly heat illness patients in Japan—Analysis from Heatstroke STUDY 2010. Jpn. J. Acute Med. 2011, 22, 331–340. [Google Scholar]
- Lin, W.C.; Tsai, C.F. Missing value imputation: A review and analysis of the literature (2006–2017). Artif. Intell. Rev. 2020, 53, 1487–1509. [Google Scholar] [CrossRef]
- McArdle, W.D.; Katch, F.I.; Katch, V.L. Essentials of Exercise Physiology, 5th ed.; Wolters Kluwer Health: Philadelphia, PA, USA, 2015. [Google Scholar]
- Korean Neurological Association. Neurology, 5th ed.; Gunja Publishing: Seoul, Republic of Korea, 2017. [Google Scholar]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the dimensionality of data with neural networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef]
- Shrivastava, A.; Rameshan, R.; Agnihotri, S. Latent space characterization of autoencoder variants. arXiv 2024, arXiv:2412.04755. [Google Scholar] [CrossRef]
- Beaulieu-Jones, B.K.; Moore, J.H. Missing data imputation in the electronic health record using deeply learned autoencoders. In Proceedings of the Pacific Symposium on Biocomputing, Kohala Coast, HI, USA, 3–7 January 2017; pp. 207–218. [Google Scholar]
- Gondara, L.; Wang, K. Mida: Multiple imputation using denoising autoencoders. In Proceedings of the Pacific-Asia Conference on Knowledge Discovery and Data Mining, Melbourne, VIC, Australia, 3–6 June 2018; pp. 260–272. [Google Scholar]
- Darmawan, H.; Yuliana, M.; Hadi, M. GRU and XGBoost Performance with Hyperparameter Tuning Using GridSearchCV and Bayesian Optimization on an IoT-Based Weather Prediction System. Int. J. Adv. Sci. Eng. Inf. Technol. 2023, 13, 851–862. [Google Scholar] [CrossRef]
- Tompra, K.V.; Papageorgiou, G.; Tjortjis, C. Strategic machine learning optimization for cardiovascular disease prediction and high-risk patient identification. Algorithms 2024, 17, 178. [Google Scholar] [CrossRef]
| Category | Features | Description |
|---|---|---|
| Dependent Features | Admission | Hospital admission status (0: not admitted, 1: admitted) |
| Demographic Information | Age, Sex | Age and gender of patients |
| Environmental Information | Location, Weather | Location and weather information of heat illness occurrence |
| Underlying Diseases | HT, DM, Heart Disease, Dementia, etc. | Underlying diseases of patients, such as hypertension (HT), diabetes (DM), heart disease, and dementia |
| Pre-admission Vital Signs | PreSBP, PreHR, PreBT, PreGCS, etc. | Vital signs such as systolic blood pressure (PreSBP), heart rate (PreHR), body temperature (PreBT), and consciousness level (PreGCS) before hospital admission |
| Hospital-measured Vital Data | SBP, HR, BT, WBC, Cre, AST, CK, etc. | Systolic blood pressure (SBP), heart rate (HR), body temperature (BT); function indicators of blood, liver and kidney, such as WBC, Cre, AST, CK, etc. |
| Dataset | Description |
|---|---|
| Dataset 1-1 | Raw dataset based on all variables |
| Dataset 1-2 | Binning dataset based on all variables using K-Means clustering |
| Dataset 1-3 | Binning dataset based on all variables using decision tree |
| Dataset 1-4 | Binning dataset based on all variables using the clinical significance-based binning method |
| Dataset 2-1 | Raw dataset based on wearable device variables |
| Dataset 2-2 | Binning dataset based on wearable device variables using K-Means clustering |
| Dataset 2-3 | Binning dataset based on wearable device variables using decision tree |
| Dataset 2-4 | Binning dataset based on wearable device variables using the clinical significance-based binning method |
| Model | Parameter | Search Range | Optimal Values | |||
|---|---|---|---|---|---|---|
| Dataset 1 | Dataset 2 | Dataset 3 | Dataset 4 | |||
| KNN | Number of neighbors (k) | 3, 5, 7, 9, 11 | 11 | 11 | 11 | 11 |
| Distance metrics | Euclidean, Manhattan | Manhattan | Manhattan | Manhattan | Manhattan | |
| SVM | Kernel | Linear, rbf | rbf | rbf | rbf | rbf |
| Regularization Parameter | 0.1, 1, 10, 100 | 100 | 1 | 1 | 10 | |
| Gamma | 0.001, 0.01, 0.1, 1 | 0.001 | 0.1 | 0.1 | 0.1 | |
| Random Forest | Number of trees | 100, 200, 500 | 500 | 200 | 500 | 500 |
| Maximum depth | None, 10, 20, 30 | 20 | None | 20 | 10 | |
| Minimum samples split | 2, 5, 10 | 2 | 2 | 2 | 2 | |
| Minimum samples of leaf nodes | 1, 2, 4 | 1 | 1 | 1 | 1 | |
| MLP | Number of hidden layers | 2, 3, 4, 5, 6 | 2 | 3 | 3 | 5 |
| Learning rate | 1 × 10−4, 5 × 10−4, 1 × 10−3, 2 × 10−3, 5 × 10−3 | 1 × 10−3 | 1 × 10−4 | 5 × 10−3 | 2 × 10−3 | |
| Batch size | 16, 32, 64, 128 | 128 | 128 | 16 | 16 | |
| CNN | Number of filters | 32, 64, 128 | 128 | 128 | 64 | 64 |
| Kernel size | 3 × 3, 5 × 5 | 5 × 5 | 5 × 5 | 3 × 3 | 3 × 3 | |
| Learning rate | 1 × 10−4, 1 × 10−3, 1 × 10−2 | 1 × 10−3 | 1 × 10−3 | 1 × 10−3 | 1 × 10−2 | |
| Batch size | 32, 64 | 64 | 64 | 64 | 32 | |
| Hidden dimension | 64, 128, 256 | 256 | 64 | 128 | 64 | |
| Transformer | Number of attention heads | 2, 4, 8 | 4 | 2 | 2 | 4 |
| Number of encoder layers | 2, 4, 6 | 2 | 6 | 4 | 4 | |
| Learning rate | 1 × 10−4, 5 × 10−4, 1 × 10−3, 2 × 10−3, 5 × 10−3 | 1 × 10−3 | 1 × 10−3 | 1 × 10−3 | 1 × 10−3 | |
| Dropout | 0.1, 0.3 | 0.1 | 0.1 | 0.3 | 0.1 | |
| Model | Dataset | Accuracy | Precision | Recall | F1-Score | AUC |
|---|---|---|---|---|---|---|
| KNN | ||||||
| Dataset 1-1 | 81.4 | 85.2 | 60.8 | 70.9 | 89.6 | |
| Dataset 1-2 | 80.6 | 84.9 | 58.8 | 69.5 | 87.9 | |
| Dataset 1-3 | 81.7 | 84.9 | 61.9 | 71.6 | 87.8 | |
| Dataset 1-4 | 80.2 | 87.6 | 54.0 | 66.8 | 90.2 | |
| SVM | ||||||
| Dataset 1-1 | 84.1 | 85.1 | 69.6 | 76.6 | 89.5 | |
| Dataset 1-2 | 83.2 | 83.3 | 68.9 | 75.4 | 89 | |
| Dataset 1-3 | 82.5 | 81.1 | 69.1 | 74.6 | 89.6 | |
| Dataset 1-4 | 87.9 | 88.0 | 77.9 | 82.6 | 94.4 | |
| Random Forest | ||||||
| Dataset 1-1 | 85.5 | 82.5 | 77.6 | 80.0 | 92.8 | |
| Dataset 1-2 | 86.3 | 84.8 | 77.3 | 80.9 | 92.5 | |
| Dataset 1-3 | 85.2 | 82.9 | 75.9 | 79.2 | 92.8 | |
| Dataset 1-4 | 91.7 | 91.4 | 85.5 | 88.4 | 96.3 | |
| MLP | ||||||
| Dataset 1-1 | 84.4 | 81.9 | 74.7 | 78.2 | 88.9 | |
| Dataset 1-2 | 80.6 | 74.9 | 72.7 | 73.8 | 88.3 | |
| Dataset 1-3 | 84.4 | 83.1 | 72.9 | 77.7 | 89.2 | |
| Dataset 1-4 | 83.3 | 81.2 | 71.5 | 76.0 | 90.6 | |
| CNN | ||||||
| Dataset 1-1 | 81.6 | 82.2 | 66.4 | 73.5 | 88.3 | |
| Dataset 1-2 | 82.2 | 82.4 | 66.8 | 73.8 | 88.1 | |
| Dataset 1-3 | 83.5 | 82.6 | 70.3 | 76 | 88.7 | |
| Dataset 1-4 | 81.7 | 87.0 | 59.6 | 70.7 | 90.3 | |
| Transformer | ||||||
| Dataset 1-1 | 84.7 | 86.1 | 70.5 | 77.5 | 92.1 | |
| Dataset 1-2 | 84.1 | 74.6 | 87.4 | 80.5 | 91.7 | |
| Dataset 1-3 | 82.1 | 81.1 | 67.4 | 73.6 | 91 | |
| Dataset 1-4 | 86.1 | 79.3 | 84.7 | 81.9 | 93.4 |
| Model/ | Dataset 1-2 | Dataset 1-3 | Dataset 1-4 | |||
|---|---|---|---|---|---|---|
| Metrics | F1-Score | AUC | F1-Score | AUC | F1-Score | AUC |
| KNN | −2.0% | −1.9% | 1.0% | −2.0% | −5.8% | 0.7% |
| SVM | −1.6% | −0.6% | −2.6% | 0.1% | 7.8% | 5.5% |
| Random Forest | 1.1% | −0.3% | −1.0% | 0.0% | 10.5% | 3.8% |
| MLP | −5.6% | −0.7% | −0.6% | 0.3% | −2.8% | 1.9% |
| CNN | 0.4% | −0.2% | 3.4% | 0.5% | −3.8% | 2.3% |
| Transformer | 3.9% | −0.4% | −5.0% | −1.2% | 5.7% | 1.4% |
| Model | Dataset | Accuracy | Precision | Recall | F1-Score | AUC |
|---|---|---|---|---|---|---|
| KNN | ||||||
| Dataset 2-1 | 84.3 | 84.9 | 70.6 | 77.1 | 90.2 | |
| Dataset 2-2 | 83.6 | 82.3 | 72 | 76.8 | 90 | |
| Dataset 2-3 | 84.3 | 84.2 | 71.4 | 77.3 | 91.2 | |
| Dataset 2-4 | 86.6 | 85.3 | 77.7 | 81.3 | 93.9 | |
| SVM | ||||||
| Dataset 2-1 | 82.4 | 81.8 | 68.1 | 74.3 | 83.1 | |
| Dataset 2-2 | 85 | 84.3 | 74.1 | 78.8 | 88.6 | |
| Dataset 2-3 | 83 | 81.9 | 70.2 | 75.6 | 86.7 | |
| Dataset 2-4 | 86.6 | 83.4 | 80.3 | 81.8 | 91.7 | |
| Random Forest | ||||||
| Dataset 2-1 | 86.8 | 86.1 | 77.5 | 81.6 | 94.1 | |
| Dataset 2-2 | 87.2 | 86.9 | 77.8 | 82.1 | 93.6 | |
| Dataset 2-3 | 89.1 | 87.2 | 83.2 | 85.2 | 95.4 | |
| Dataset 2-4 | 93.4 | 90.8 | 91.6 | 91.2 | 98.1 | |
| MLP | ||||||
| Dataset 2-1 | 85.7 | 85.3 | 75.0 | 79.8 | 92.1 | |
| Dataset 2-2 | 85.2 | 85.7 | 72.8 | 78.7 | 88.7 | |
| Dataset 2-3 | 83.9 | 85.4 | 68.9 | 76.3 | 93 | |
| Dataset 2-4 | 89.8 | 83.9 | 89.9 | 86.8 | 95.2 | |
| CNN | ||||||
| Dataset 2-1 | 84.7 | 84.1 | 73.1 | 78.2 | 91.4 | |
| Dataset 2-2 | 86 | 87.9 | 72.8 | 79.6 | 92.9 | |
| Dataset 2-3 | 84.7 | 84.1 | 73.1 | 78.2 | 92.4 | |
| Dataset 2-4 | 88.5 | 83.1 | 86.9 | 85.0 | 95.7 | |
| Transformer | ||||||
| Dataset 2-1 | 84.7 | 82.2 | 75.6 | 78.8 | 92.2 | |
| Dataset 2-2 | 85.8 | 82.5 | 79.1 | 80.8 | 93.3 | |
| Dataset 2-3 | 84.7 | 89 | 67.7 | 76.9 | 92.4 | |
| Dataset 2-4 | 92.0 | 89.7 | 88.6 | 89.2 | 97.6 |
| Model/ | Dataset 2-2 | Dataset 2-3 | Dataset 2-4 | |||
|---|---|---|---|---|---|---|
| Metrics | F1-Score | AUC | F1-Score | AUC | F1-Score | AUC |
| KNN | −0.4% | −0.2% | 0.3% | 1.1% | 5.4% | 4.1% |
| SVM | 6.1% | 6.6% | 1.7% | 4.3% | 10.1% | 10.3% |
| Random Forest | 0.6% | −0.5% | 4.4% | 1.4% | 11.8% | 4.3% |
| MLP | −1.4% | −3.7% | −4.4% | 1.0% | 8.8% | 3.4% |
| CNN | 1.8% | 1.6% | 0.0% | 1.1% | 8.7% | 4.7% |
| Transformer | 2.5% | 1.2% | −2.4% | 0.2% | 13.2% | 5.9% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, J.; Jeong, H. Experimental Verification of Performance Improvement in Heat-Related Illness Prediction Using Clinical Significance-Based Binning. Appl. Sci. 2025, 15, 12500. https://doi.org/10.3390/app152312500
Shin J, Jeong H. Experimental Verification of Performance Improvement in Heat-Related Illness Prediction Using Clinical Significance-Based Binning. Applied Sciences. 2025; 15(23):12500. https://doi.org/10.3390/app152312500
Chicago/Turabian StyleShin, Jeongwoo, and Hanjo Jeong. 2025. "Experimental Verification of Performance Improvement in Heat-Related Illness Prediction Using Clinical Significance-Based Binning" Applied Sciences 15, no. 23: 12500. https://doi.org/10.3390/app152312500
APA StyleShin, J., & Jeong, H. (2025). Experimental Verification of Performance Improvement in Heat-Related Illness Prediction Using Clinical Significance-Based Binning. Applied Sciences, 15(23), 12500. https://doi.org/10.3390/app152312500






