Graphene-Mediated Lubrication of Phospholipid Membranes: Insights from Molecular Dynamics Simulations
Abstract
1. Introduction
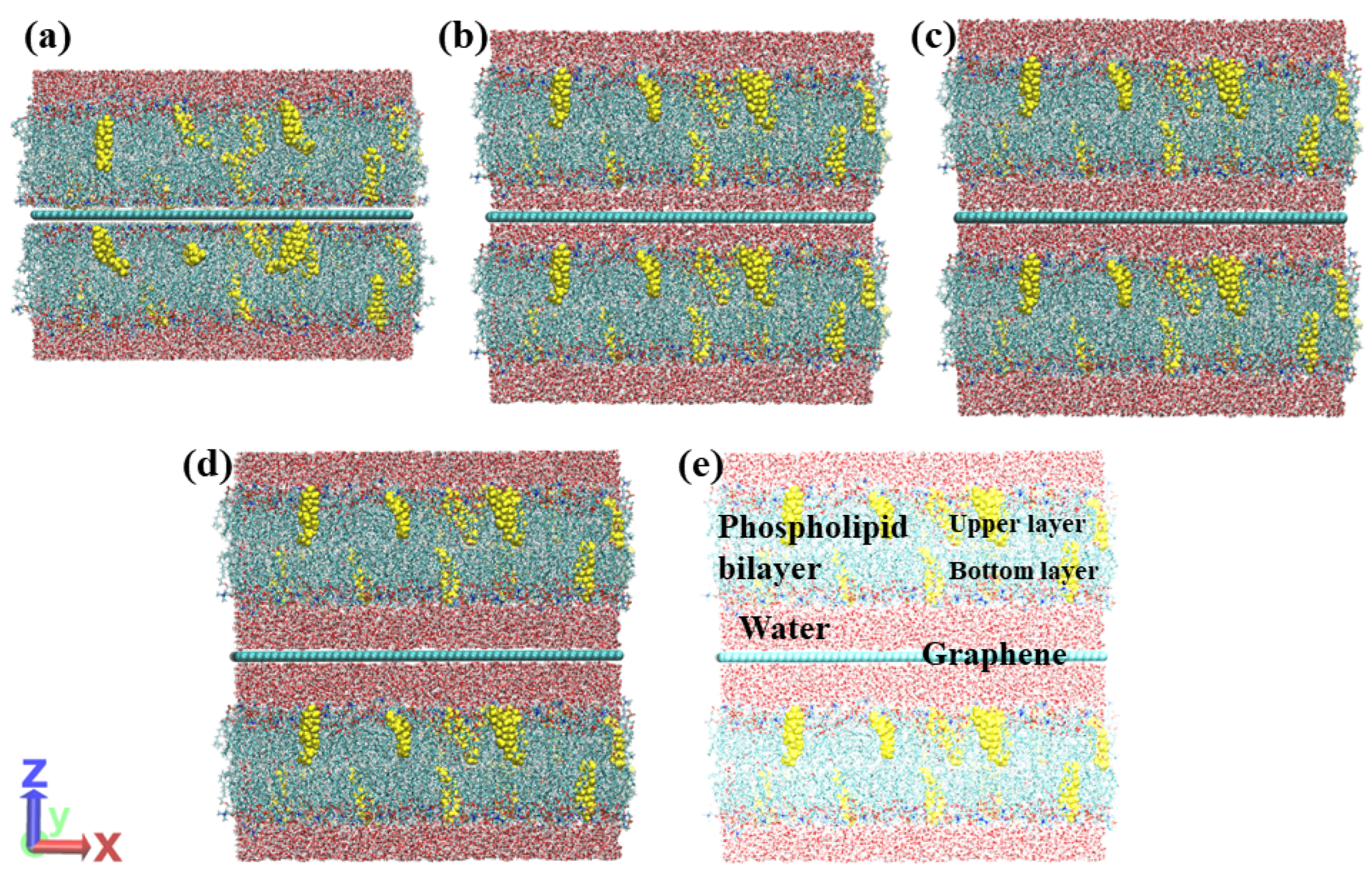
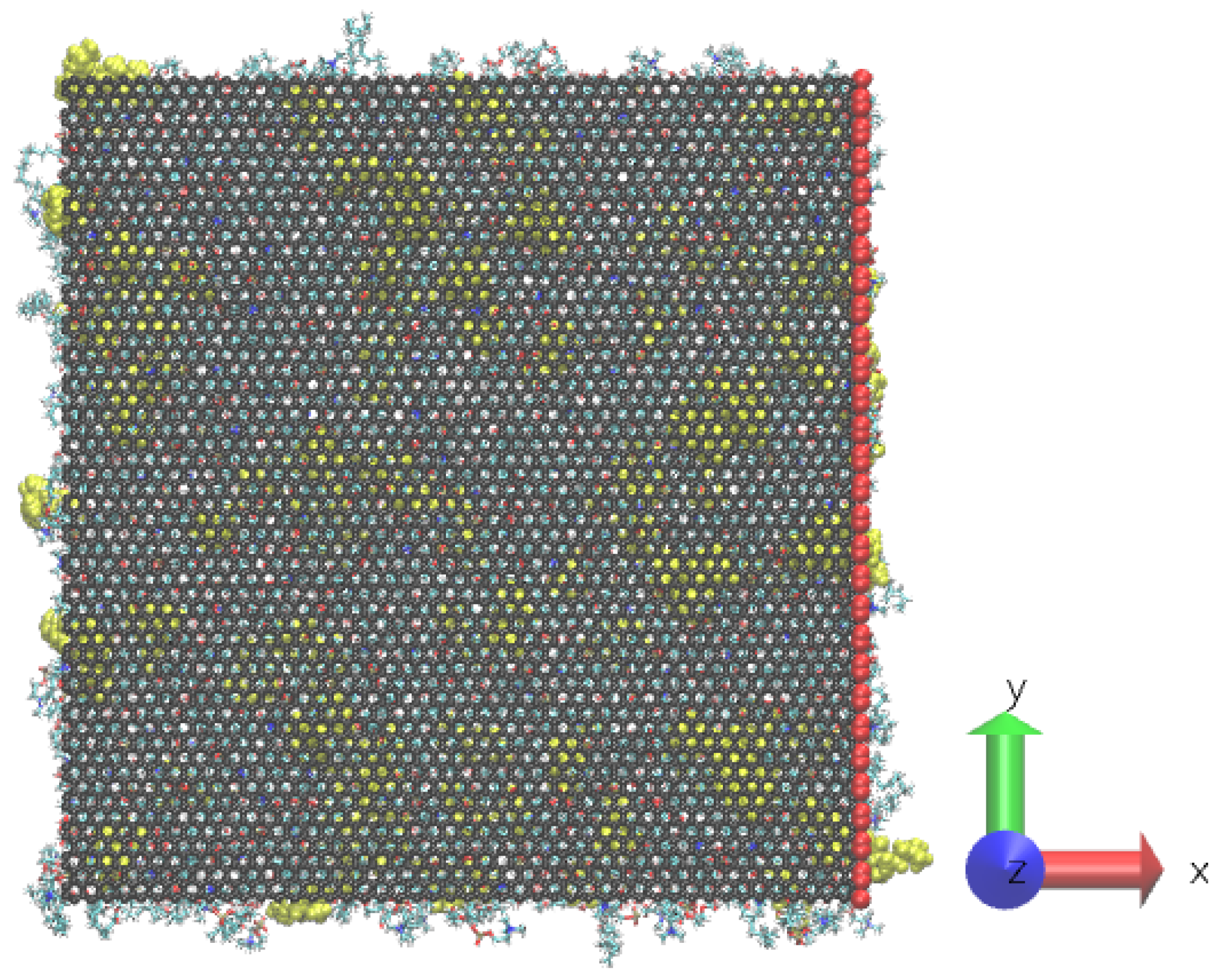
2. Materials and Methods
- DMPC with a 10 percent admixture of cholesterol;
- DMPC molecules only;
- DMPE (1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine) molecules only.
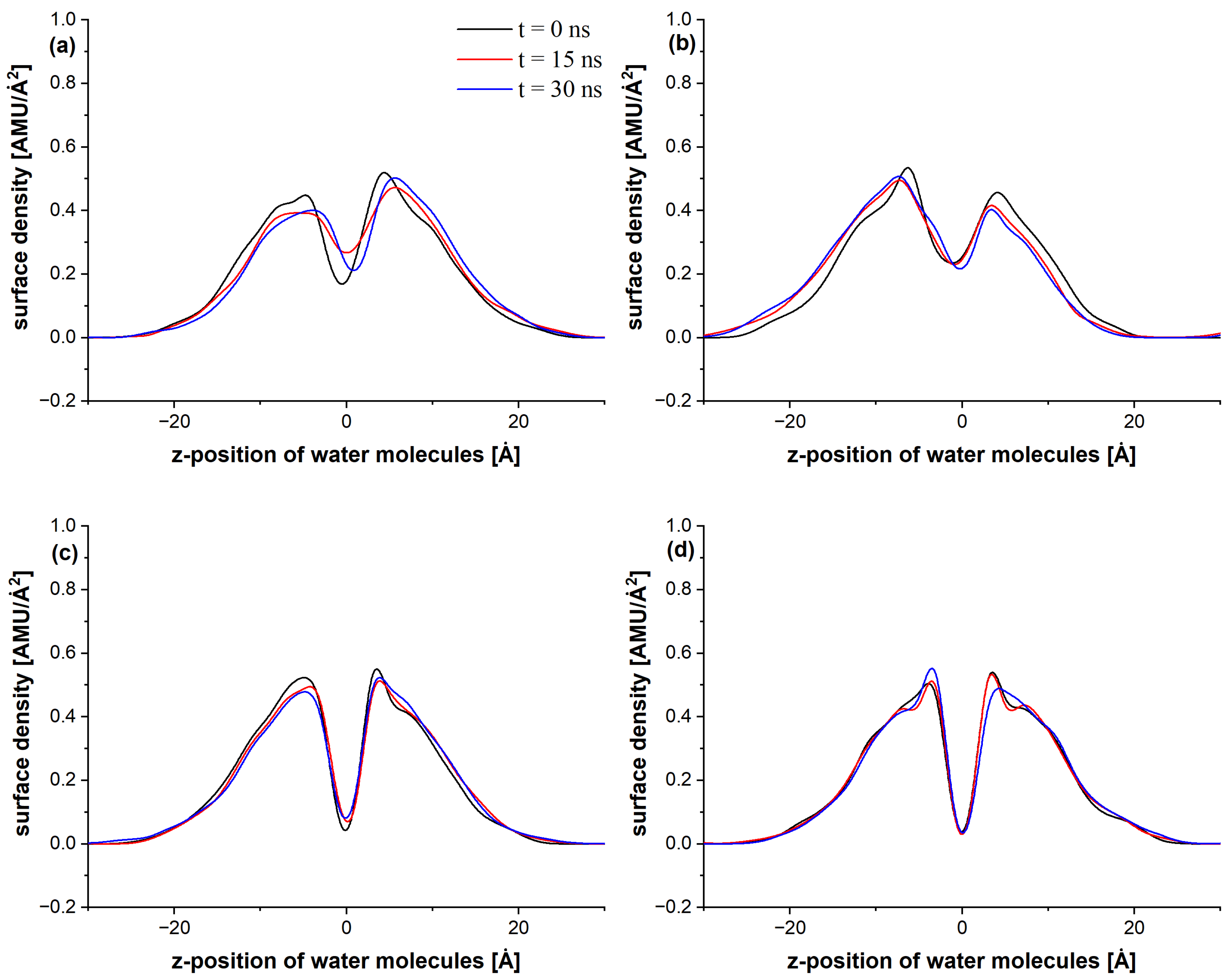
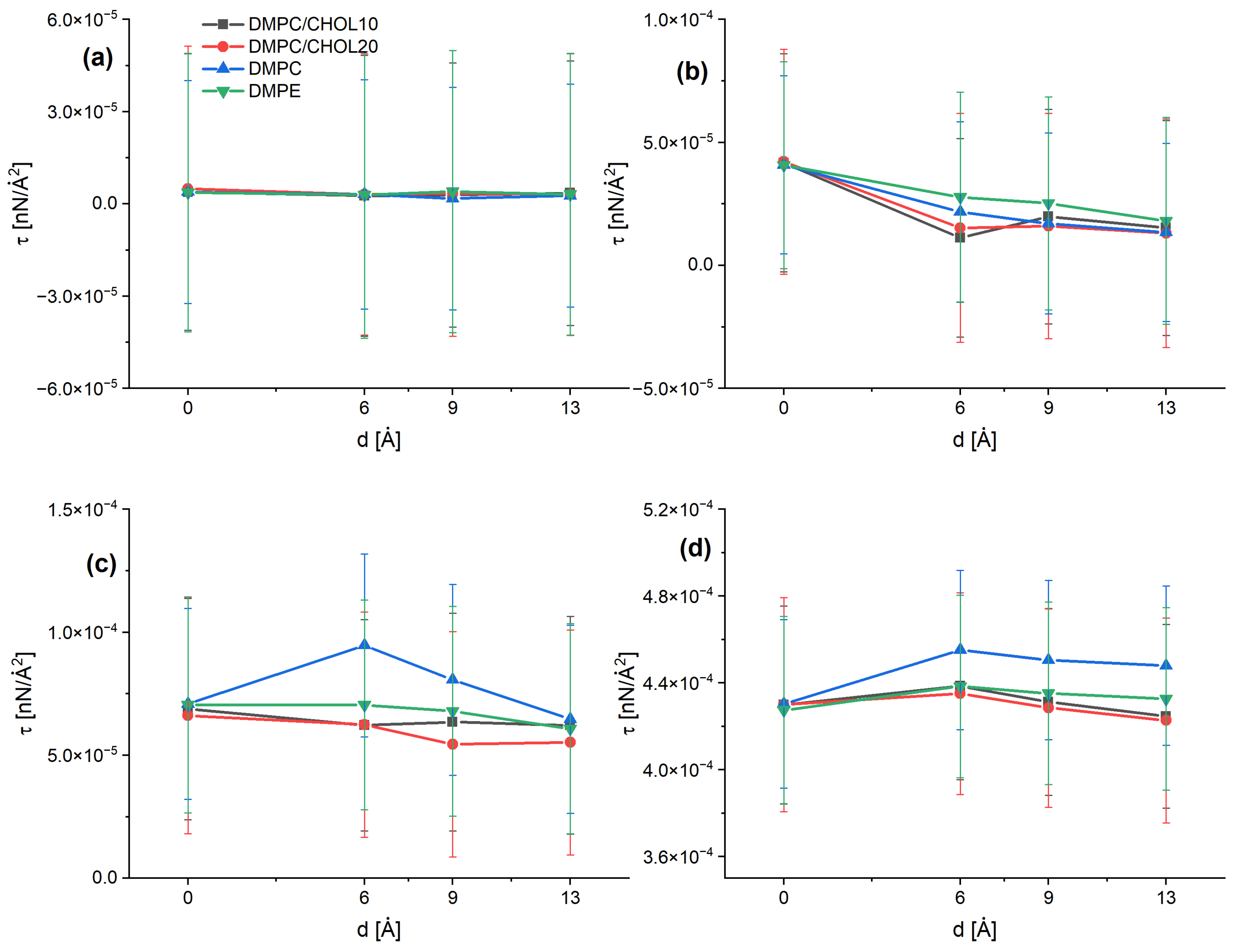
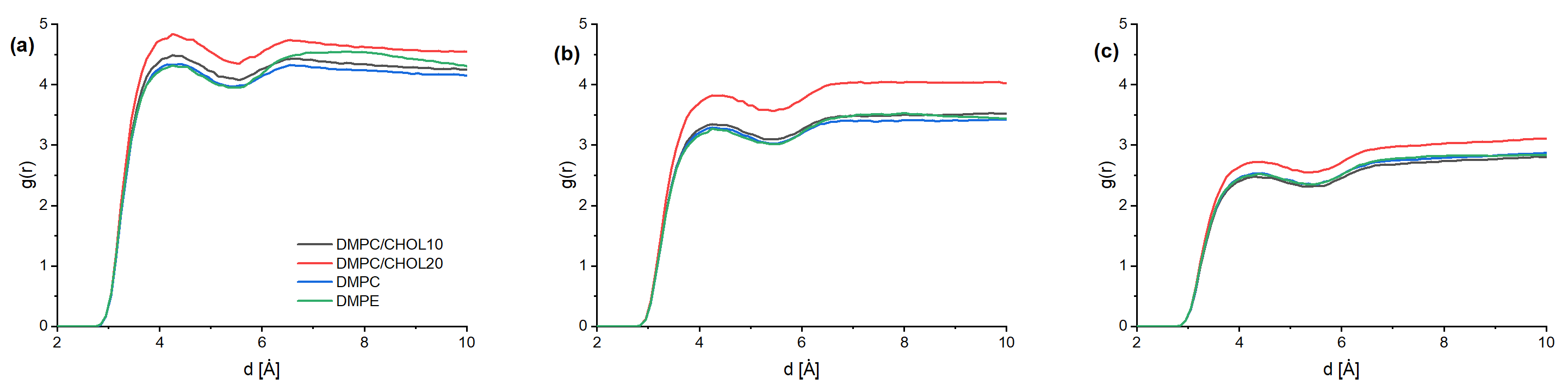
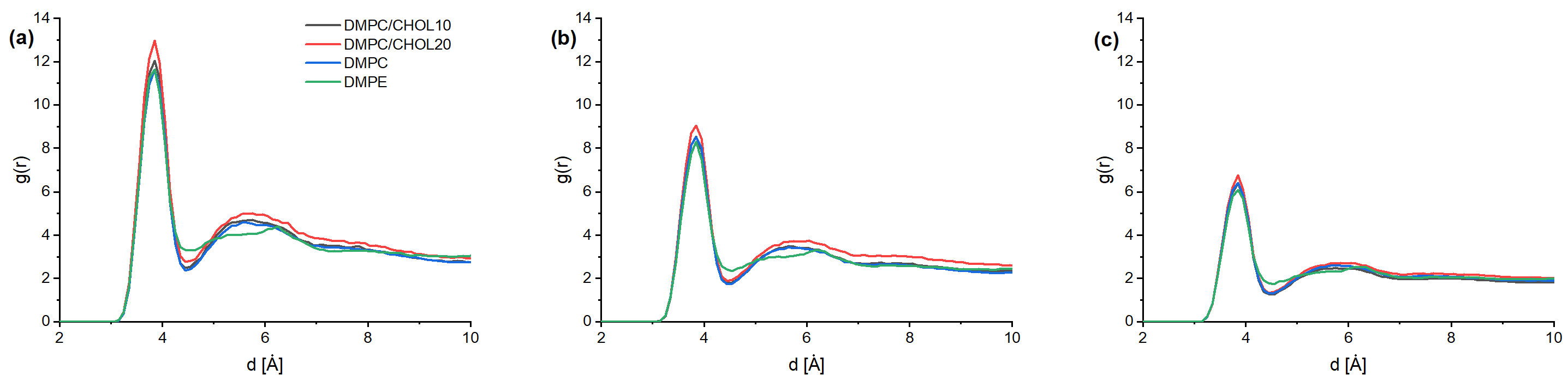
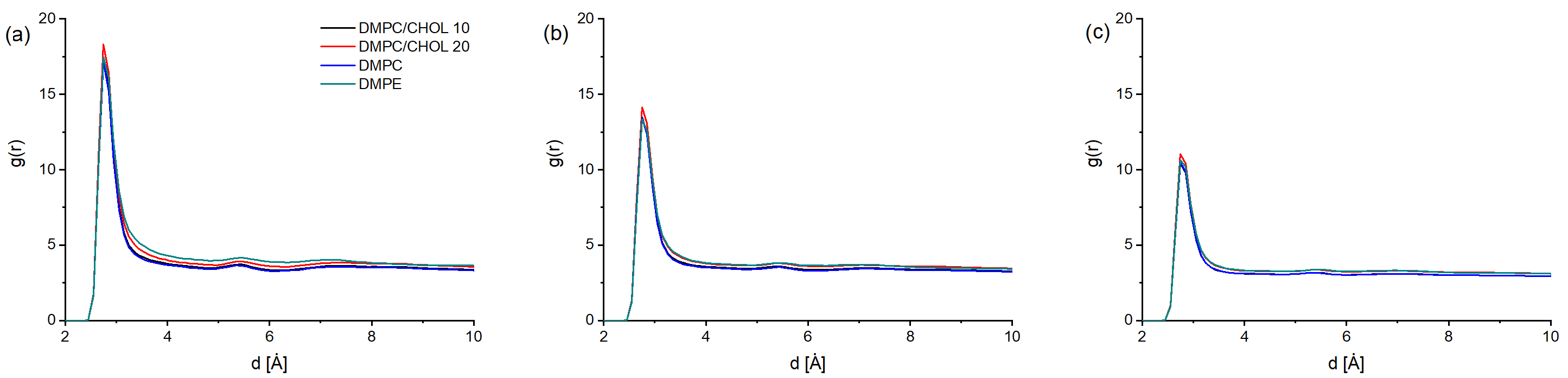
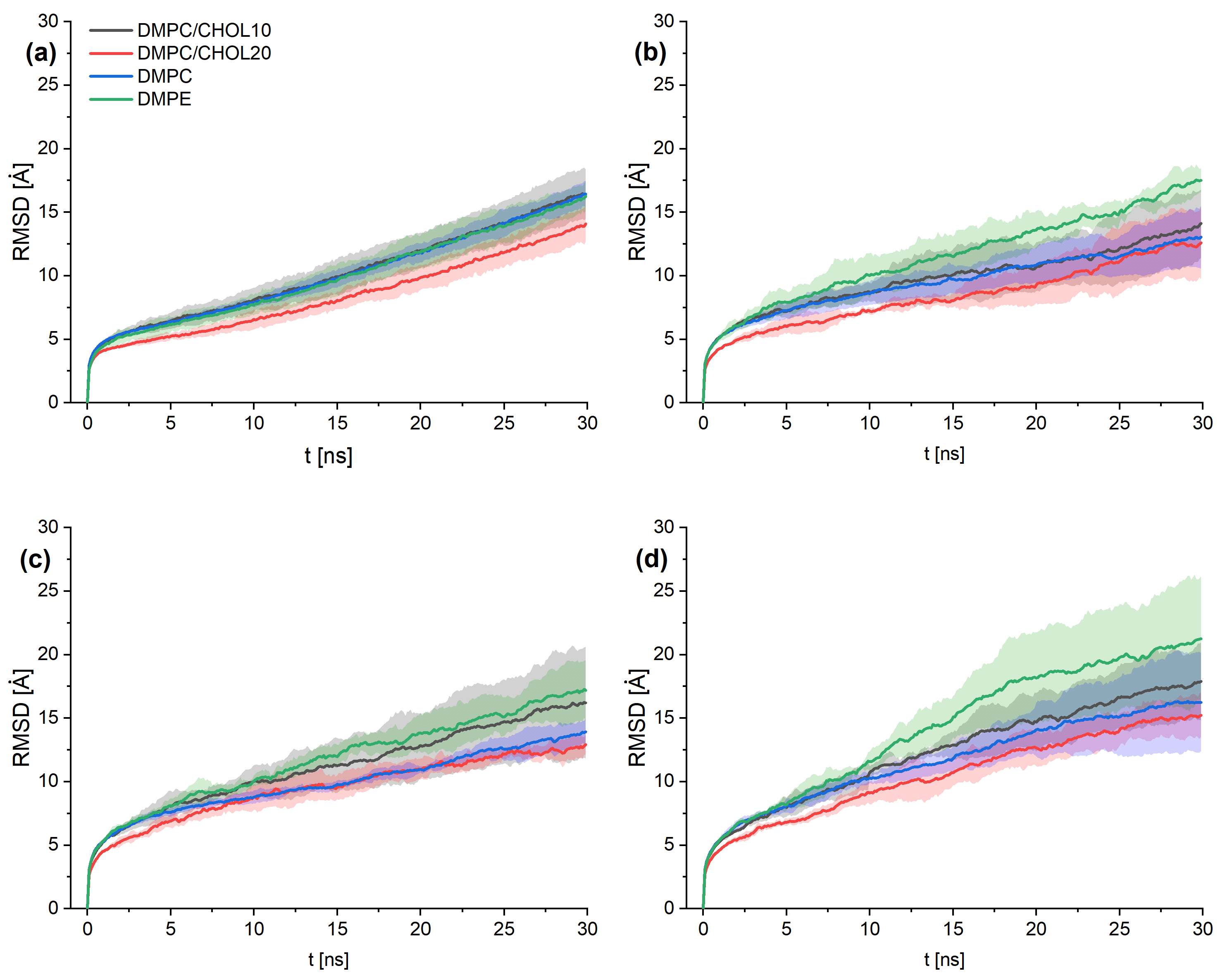
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, D.; Kaner, R.B. Graphene-based materials. Science 2008, 320, 1170–1171. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.R.; Kim, J.; Sim, S.; Jang, H.; Jang, W.S. Graphene oxide–based nanocomposite as an effective, multifunctional, antibiofilm coating. ACS Nano 2015, 9, 9097–9104. [Google Scholar]
- Lee, C.; Wei, X.; Kysar, J.W.; Hone, J. Measurement of the elastic properties and intrinsic strength of monolayer graphene. Science 2008, 321, 385–388. [Google Scholar] [CrossRef]
- Hu, Y.; Li, D.; Luo, Y.; Tang, Y.; Li, J. Graphene and graphene oxide: Two ideal choices for the enrichment and ionization of long-chain fatty acids by MALDI-TOF MS. Chem. Commun. 2015, 51, 3176–3179. [Google Scholar]
- Ali Olad, H.B.K.H. Graphene oxide and amin-modified graphene oxide incorporated chitosan-gelatin scaffolds as promising materials for tissue engineering. Compos. Part B Eng. 2019, 6, 692–702. [Google Scholar] [CrossRef]
- Chathuranga, H.; Wijesinghe, I.; Marriam, I.; Yan, C. Recent advances in graphene allotropes-based fire detection sensors. Crit. Rev. Solid State Mater. Sci. 2024, 50, 54–76. [Google Scholar] [CrossRef]
- Chang, H.; Ho, T.; Su, Y. Graphene-Enhanced Battery Components in Rechargeable Lithium-Ion and Lithium Metal Batteries. C—J. Carbon Res. 2021, 7, 65. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, M. Hydrogel-integrated graphene superstructures for tissue engineering. Carbon 2024, 215, 118000. [Google Scholar] [CrossRef]
- Sun, X.; Liu, Z.; Wang, Y. PEGylated nanographene oxide for delivery of water-insoluble cancer drugs. J. Am. Chem. Soc. 2008, 130, 10876–10877. [Google Scholar] [CrossRef]
- Kim, K.S.; Zhao, Y.; Jang, H.; Lee, S.Y.; Kim, J.M.; Kim, K.S.; Ahn, J.H.; Kim, P.; Choi, J.Y.; Hong, B.H. Large-scale pattern growth of graphene films for stretchable transparent electrodes. Nature 2009, 457, 706–710. [Google Scholar] [CrossRef] [PubMed]
- Willems, N.; Urtizberea, A.; Verre, A.F.; Iliut, M.; Lelimousin, M.; Hirtz, M.; Vijayaraghavan, A.; Sansom, M.S.P. Biomimetic Phospholipid Membrane Organization on Graphene and Graphene Oxide Surfaces: A Molecular Dynamics Simulation Study. ACS Nano 2017, 11, 7171–7179. [Google Scholar] [CrossRef]
- Li, S.; Mulloor, J.J.; Wang, L.; Ji, Y.; Mulloor, C.J.; Micic, M.; Orbulescu, J.; Leblanc, R.M. Strong and Selective Adsorption of Lysozyme on Graphene Oxide. ACS Appl. Mater. Interfaces 2014, 6, 5704–5712. [Google Scholar] [CrossRef]
- Lipowsky, R. The conformation of membranes. Nature 1992, 349, 475–481. [Google Scholar] [CrossRef]
- Chen, Y.; Pandit, S.; Rahimi, S.; Mijakovic, I. Interactions Between Graphene-Based Materials and Biological Surfaces: A Review of Underlying Molecular Mechanisms. Adv. Mater. Interfaces 2021, 8, 2101132. [Google Scholar] [CrossRef]
- Nosonovsky, M.; Bhushan, B. Multiscale friction mechanisms and hierarchical surfaces in nano- and bio-tribology. Mater. Sci. Eng. R Rep. 2007, 58, 162–193. [Google Scholar] [CrossRef]
- Vincent, J.F.; Bogatyreva, O.A. Biomimetics: Its practice and theory. J. R. Soc. Interface 2007, 4, 637–656. [Google Scholar] [CrossRef] [PubMed]
- Veatch, S.L.; Soubias, O.; Keller, S.L.; Gawrisch, K. Critical fluctuations in domain-forming lipid mixtures. Proc. Natl. Acad. Sci. USA 2007, 104, 17650–17655. [Google Scholar] [CrossRef]
- Kučerka, N.; Tristram-Nagle, S.; Nagle, J.F. Structure of fully hydrated fluid phase DMPC and DLPC lipid bilayers using X-ray scattering from oriented multilamellar arrays and from unilamellar vesicles. Biophys. J. 2005, 88, 2626–2637. [Google Scholar] [CrossRef] [PubMed]
- Kučerka, N.; Tristram-Nagle, S.; Nagle, J.F. Cholesterol in bilayers: A combined X-ray and neutron scattering study. Biochim. Biophys. Acta-(BBA)-Biomembr. 2005, 1716, 77–88. [Google Scholar]
- Epand, R.M. Cholesterol and the interaction of proteins with membrane domains. Prog. Lipid Res. 2006, 45, 279–294. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Tristram-Nagle, S.; Nagle, J.F. Cholesterol perturbs lipid bilayers nonuniversally. Phys. Rev. E 2008, 77, 011901. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y.; Du, X.; Zhang, J.; Wang, Y.; Fan, J. Interaction of Graphene and Its Oxide with Lipid Membranes: A Molecular Dynamics Simulation Study. J. Phys. Chem. C 2016, 120, 9566–9573. [Google Scholar] [CrossRef]
- Yue, T.; Wang, X.; Zhang, X.; Huang, F. Molecular modeling of interaction between lipid monolayer and graphene nanosheets: Implications for pulmonary nanotoxicity and pulmonary drug delivery. RSC Adv. 2015, 5, 30092–30106. [Google Scholar] [CrossRef]
- Chen, X.; Wang, Y.; Du, X.; Zhang, J.; Wang, Y.; Fan, J. Interaction of graphene oxide with artificial cell membranes: Role of anionic phospholipid and cholesterol in nanoparticle attachment and membrane disruption. Chemosphere 2021, 285, 131438. [Google Scholar] [CrossRef]
- Bonakala, S.; Lalitha, A.; Shin, J.E.; Moghadam, F.; Semino, R.; Park, H.B.; Maurin, G. Interaction of Graphene Nanoparticles and Lipid Membranes Displaying Different Liquid Orderings: A Molecular Dynamics Study. Langmuir 2019, 35, 16661–16668. [Google Scholar] [CrossRef]
- Li, Z.; Zhu, X.; Li, J.; Zhong, J.; Zhang, J.; Fan, J. Molecular insights into the resistance of phospholipid heads to the membrane penetration of graphene nanosheets. Nanoscale 2022, 14, 5384–5391. [Google Scholar] [CrossRef]
- Lee, H.; Ko, J.H.; Choi, J.S.; Hwang, J.H.; Kim, Y.H.; Salmeron, M.; Park, J.Y. Enhancement of Friction by Water Intercalated between Graphene and Mica. J. Phys. Chem. Lett. 2017, 8, 3482–3487. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Jeong, H.; Suh, J.; Doh, W.H.; Baik, J.; Shin, H.J.; Ko, J.H.; Wu, J.; Kim, Y.H.; Park, J.Y. Nanoscale Friction on Confined Water Layers Intercalated between MoS2 Flakes and Silica. J. Phys. Chem. C 2019, 123, 8827–8835. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VDM: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 27–28. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- Brünger, A.; Brooks, C.L., III; Karplus, M. Stochastic boundary conditions for molecular dynamics simulations of ST2 water. Chem. Phys. Lett. 1984, 105, 495–500. [Google Scholar] [CrossRef]
- Kale, L.; Skeel, R.; Bhandarkar, M.; Brunner, R.; Gursoy, A.; Krawetz, N.; Phillips, J.; Shinozaki, A.; Varadarajan, K.; Schulten, K. NAMD2: Greater Scalability for Parallel Molecular Dynamics. J. Comput. Phys. 1999, 151, 283–312. [Google Scholar] [CrossRef]
- Raczyński, P.; Górny, K.; Pabiszczak, M.; Gburski, Z. Nanoindentation of biomembrane by carbon nanotubes–MD simulation. Comput. Mater. Sci. 2013, 70, 13–18. [Google Scholar] [CrossRef]
- Raczynski, P.; Gorny, K.; Raczynska, V.; Pabiszczak, M.; Dendzik, Z.; Gburski, Z. On the impact of nanotube diameter on biomembrane indentation-Computer simulations study. Biochim. Biophys.-Acta-Biomembr. 2018, 1860, 310–318. [Google Scholar] [CrossRef]
- Raczynski, P.; Raczynska, V.; Gorny, K.; Gburski, Z. Properties of ultrathin cholesterol and phospholipid layers surrounding silicon-carbide nanotube: MD simulations. Arch. Biochem. Biophys. 2015, 580, 22–30. [Google Scholar] [CrossRef]
- Martyna, G.J.; Tobias, D.J.; Klein, M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994, 101, 4177. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926. [Google Scholar] [CrossRef]
- MacKerell, A.D., Jr.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef] [PubMed]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.W.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; MacKerell, A.D.; Pastor, R.W. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J. Phys. Chem. B 2010, 114, 7830–7843. [Google Scholar] [CrossRef] [PubMed]


| Graphene Rate [m/s] | Distance—Lipids [Å] | Distance—Graphene [Å] | Distance Percentage |
|---|---|---|---|
| 0.1 | 9.8 (2.3) | 15.1 | 64.9 |
| 1 | 61.6 (7.5) | 152.2 | 40.4 |
| 10 | 185.1 (7.6) | 1502.0 | 12.3 |
| 100 | 242.3 (3.9) | 15,020.0 | 1.6 |
| Graphene Rate [m/s] | Distance—Lipids [Å] | Distance—Graphene [Å] | Distance Percentage |
|---|---|---|---|
| 0.1 | 8.3 (4.0) | 15.1 | 55.0 |
| 1 | 34.7 (11.7) | 152.2 | 22.8 |
| 10 | 76.1 (3.3) | 1502.0 | 5.1 |
| 100 | 163.5 (3.4) | 15,020.0 | 1.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raczyński, P.; Górny, K.; Bełdowski, P.; Lutowski, Z.; Kloska, S.; Dendzik, Z. Graphene-Mediated Lubrication of Phospholipid Membranes: Insights from Molecular Dynamics Simulations. Appl. Sci. 2025, 15, 12369. https://doi.org/10.3390/app152312369
Raczyński P, Górny K, Bełdowski P, Lutowski Z, Kloska S, Dendzik Z. Graphene-Mediated Lubrication of Phospholipid Membranes: Insights from Molecular Dynamics Simulations. Applied Sciences. 2025; 15(23):12369. https://doi.org/10.3390/app152312369
Chicago/Turabian StyleRaczyński, Przemysław, Krzysztof Górny, Piotr Bełdowski, Zbigniew Lutowski, Sylwester Kloska, and Zbigniew Dendzik. 2025. "Graphene-Mediated Lubrication of Phospholipid Membranes: Insights from Molecular Dynamics Simulations" Applied Sciences 15, no. 23: 12369. https://doi.org/10.3390/app152312369
APA StyleRaczyński, P., Górny, K., Bełdowski, P., Lutowski, Z., Kloska, S., & Dendzik, Z. (2025). Graphene-Mediated Lubrication of Phospholipid Membranes: Insights from Molecular Dynamics Simulations. Applied Sciences, 15(23), 12369. https://doi.org/10.3390/app152312369








