Multi-Class Segmentation and Classification of Intestinal Organoids: YOLO Stand-Alone vs. Hybrid Machine Learning Pipelines
Abstract
1. Introduction
- We evaluate YOLOv10 as a standalone model for organoid segmentation and classification, demonstrating its superior accuracy and efficiency over earlier YOLO architectures.
- We introduce a hybrid pipeline where features extracted from YOLO or ResNet50 are classified using state-of-the-art ML algorithms, including Logistic Regression, Random Forest, Naive Bayes, Multi-Layer Perceptrons (MLP), K-Nearest Neighbors (KNN), and eXtreme Gradient Boosting (XGBoost).
- To further enhance performance, we implement an AUC-weighted ensemble method that integrates predictions across multiple classifiers, achieving robust and scalable morphological classification.
2. Methods
2.1. Dataset
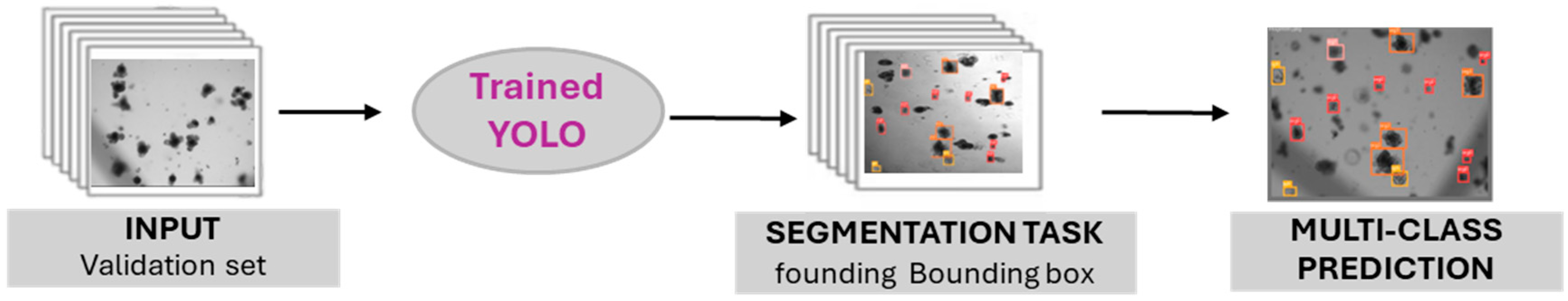
2.2. YOLO Standalone Model for Segmentation and Classification
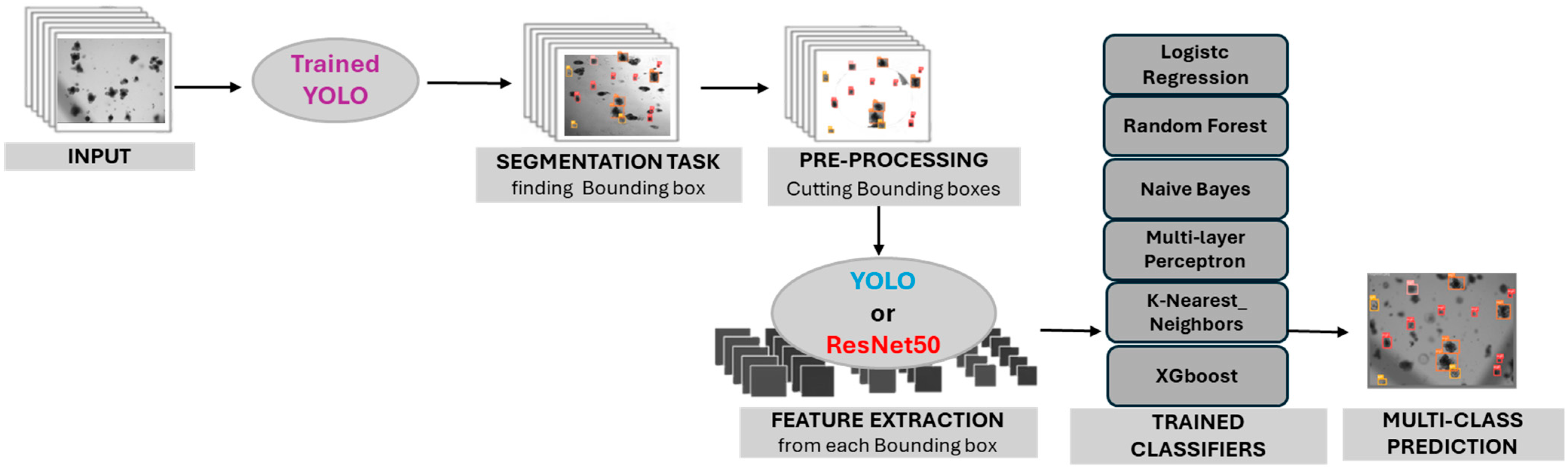
2.3. Hybrid Pipeline: YOLO Segmentation and Feature Extraction
2.4. Machine Learning Classifiers
2.5. AUC-Based Ensemble Method
2.6. Computing Environment and Runtime
3. Results
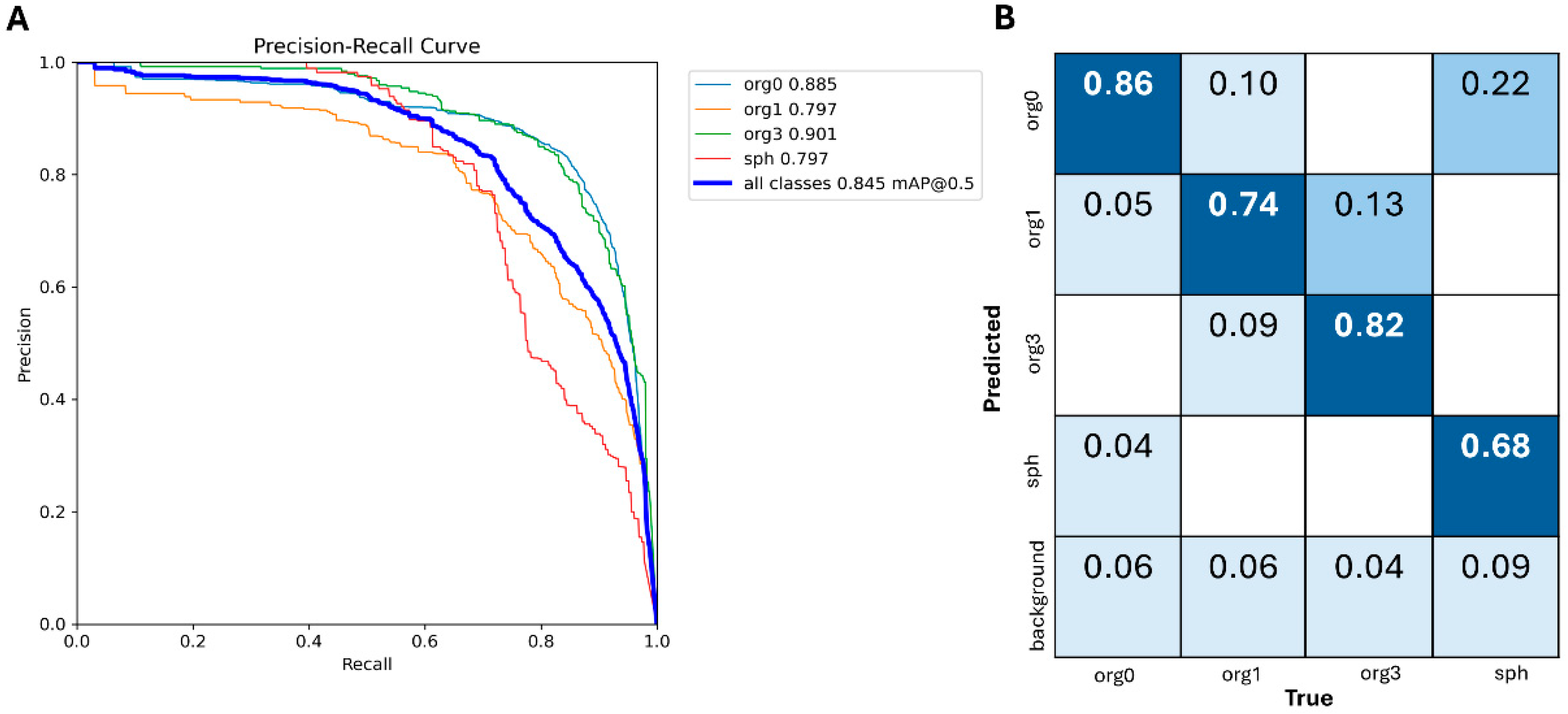
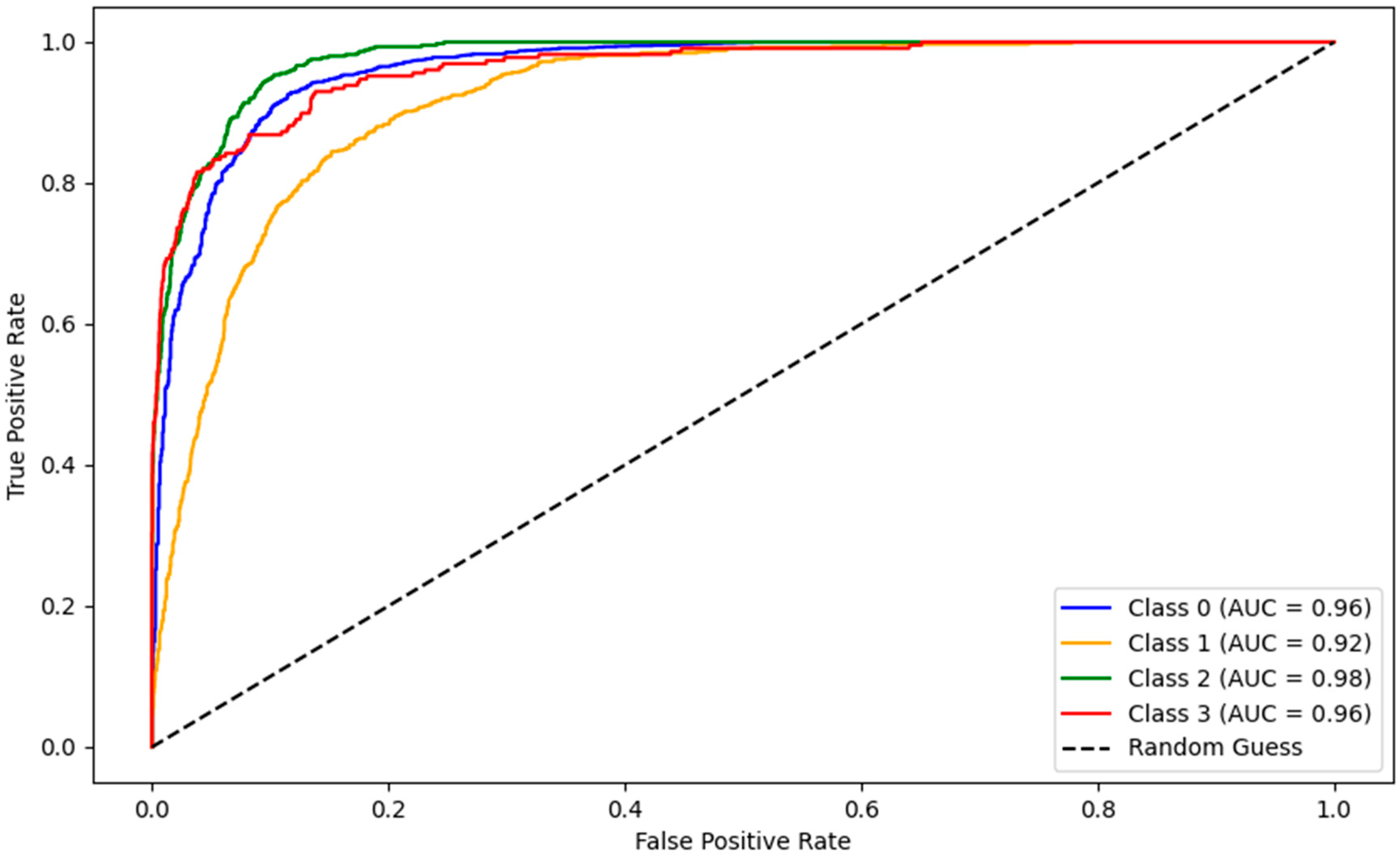
3.1. YOLOv10 Standalone Model Performance
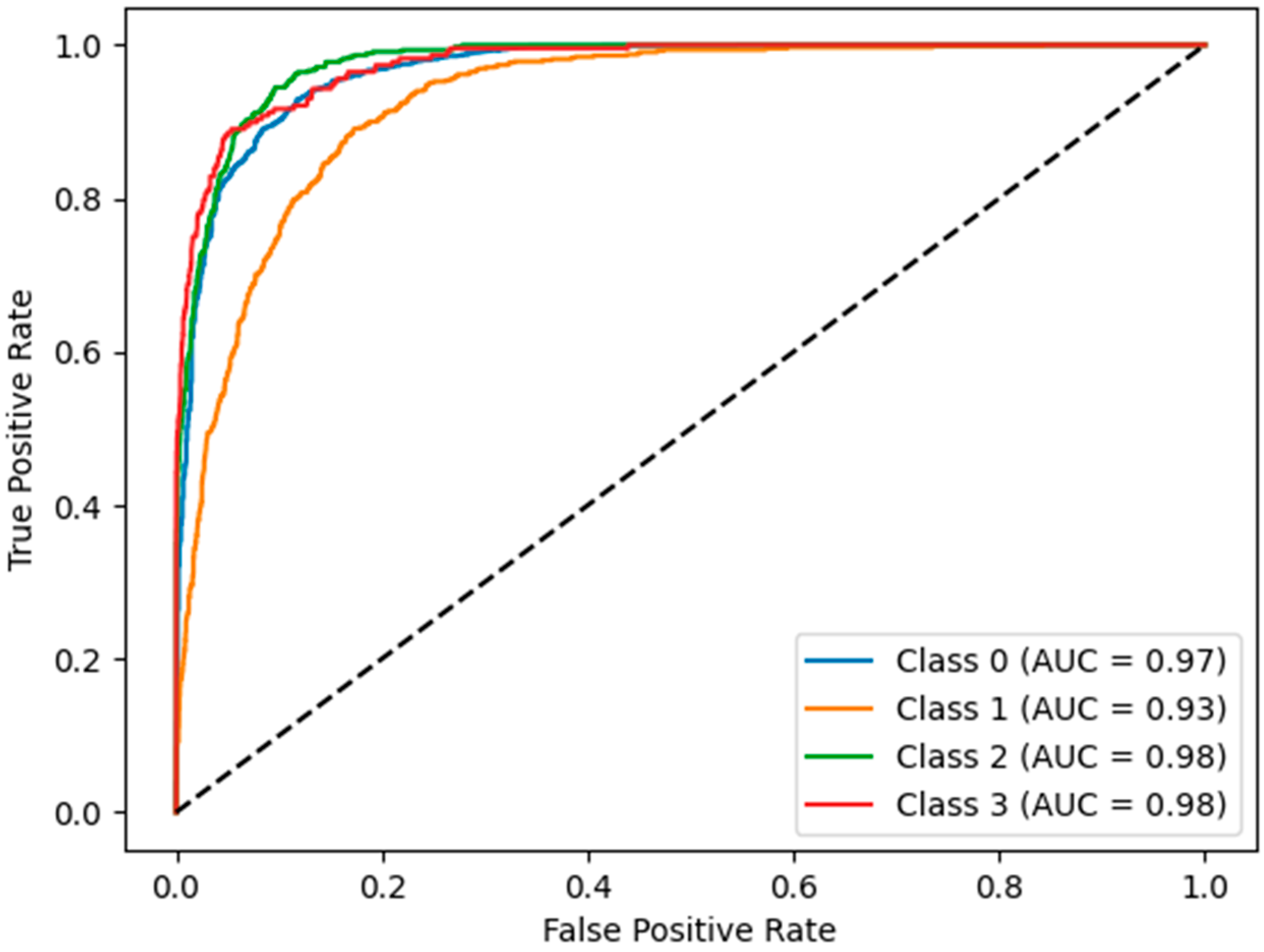
3.2. Hybrid Pipeline Performance
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 Stem Cells Build Crypt-Villus Structures in Vitro Without a Mesenchymal Niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Sato, T.; Stange, D.E.; Ferrante, M.; Vries, R.G.J.; Van Es, J.H.; Van den Brink, S.; Van Houdt, W.J.; Pronk, A.; Van Gorp, J.; Siersema, P.D.; et al. Long-Term Expansion of Epithelial Organoids from Human Colon, Adenoma, Adenocarcinoma, and Barrett’s Epithelium. Gastroenterology 2011, 141, 1762–1772. [Google Scholar] [CrossRef]
- Fordham, R.P.; Yui, S.; Hannan, N.R.F.; Soendergaard, C.; Madgwick, A.; Schweiger, P.J.; Nielsen, O.H.; Vallier, L.; Pedersen, R.A.; Nakamura, T.; et al. Transplantation of Expanded Fetal Intestinal Progenitors Contributes to Colon Regeneration after Injury. Cell Stem Cell 2013, 13, 734–744. [Google Scholar] [CrossRef]
- Farin, H.F.; Van Es, J.H.; Clevers, H. Redundant Sources of Wnt Regulate Intestinal Stem Cells and Promote Formation of Paneth Cells. Gastroenterology 2012, 143, 1518–1529.e7. [Google Scholar] [CrossRef]
- Almeqdadi, M.; Mana, M.D.; Roper, J.; Yilmaz, Ö.H. Gut Organoids: Mini-Tissues in Culture to Study Intestinal Physiology and Disease. Am. J. Physiol.-Cell Physiol. 2019, 317, C405–C419. [Google Scholar] [CrossRef] [PubMed]
- Guiu, J.; Hannezo, E.; Yui, S.; Demharter, S.; Ulyanchenko, S.; Maimets, M.; Jørgensen, A.; Perlman, S.; Lundvall, L.; Mamsen, L.S.; et al. Tracing the Origin of Adult Intestinal Stem Cells. Nature 2019, 570, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Lupo, R.; Zaminga, M.; Carriero, M.C.; Santoro, P.; Artioli, G.; Calabrò, A.; Ilari, F.; Benedetto, A.; Caslini, M.; Clerici, M.; et al. Eating Disorders and Related Stigma: Analysis among a Population of Italian Nursing Students. Acta Biomed. 2020, 91, e2020011. [Google Scholar] [CrossRef]
- Ko, K.-P.; Zhang, S.; Huang, Y.; Kim, B.; Zou, G.; Jun, S.; Zhang, J.; Martin, C.; Dunbar, K.J.; Efe, G.; et al. Tumor Niche Network-Defined Subtypes Predict Immunotherapy Response of Esophageal Squamous Cell Cancer 2023. iScience 2023, 27, 109795. [Google Scholar] [CrossRef]
- Du, X.; Chen, Z.; Li, Q.; Yang, S.; Jiang, L.; Yang, Y.; Li, Y.; Gu, Z. Organoids Revealed: Morphological Analysis of the Profound next Generation in-Vitro Model with Artificial Intelligence. Bio-Des. Manuf. 2023, 6, 319–339. [Google Scholar] [CrossRef] [PubMed]
- Brémond Martin, C.; Simon Chane, C.; Clouchoux, C.; Histace, A. Recent Trends and Perspectives in Cerebral Organoids Imaging and Analysis. Front. Neurosci. 2021, 15, 629067. [Google Scholar] [CrossRef]
- Matthews, J.M.; Schuster, B.; Kashaf, S.S.; Liu, P.; Ben-Yishay, R.; Ishay-Ronen, D.; Izumchenko, E.; Shen, L.; Weber, C.R.; Bielski, M.; et al. OrganoID: A Versatile Deep Learning Platform for Tracking and Analysis of Single-Organoid Dynamics. PLoS Comput. Biol. 2022, 18, e1010584. [Google Scholar] [CrossRef]
- Park, T.; Kim, T.K.; Han, Y.D.; Kim, K.-A.; Kim, H.; Kim, H.S. Development of a Deep Learning Based Image Processing Tool for Enhanced Organoid Analysis. Sci. Rep. 2023, 13, 19841. [Google Scholar] [CrossRef]
- Lefferts, J.W.; Kroes, S.; Smith, M.B.; Niemöller, P.J.; Nieuwenhuijze, N.D.A.; Sonneveld van Kooten, H.N.; van der Ent, C.K.; Beekman, J.M.; van Beuningen, S.F.B. OrgaSegment: Deep-Learning Based Organoid Segmentation to Quantify CFTR Dependent Fluid Secretion. Commun. Biol. 2024, 7, 319. [Google Scholar] [CrossRef] [PubMed]
- Kok, R.N.U.; Hebert, L.; Huelsz-Prince, G.; Goos, Y.J.; Zheng, X.; Bozek, K.; Stephens, G.J.; Tans, S.J.; van Zon, J.S. OrganoidTracker: Efficient Cell Tracking Using Machine Learning and Manual Error Correction. PLoS ONE 2020, 15, e0240802. [Google Scholar] [CrossRef]
- Montes-Olivas, S.; Legge, D.; Lund, A.; Fletcher, A.G.; Williams, A.C.; Marucci, L.; Homer, M. In-Silico and in-Vitro Morphometric Analysis of Intestinal Organoids. PLoS Comput. Biol. 2023, 19, e1011386. [Google Scholar] [CrossRef] [PubMed]
- Powell, R.T.; Moussalli, M.J.; Guo, L.; Bae, G.; Singh, P.; Stephan, C.; Shureiqi, I.; Davies, P.J. DeepOrganoid: A Brightfield Cell Viability Model for Screening Matrix-Embedded Organoids. SLAS Discov. 2022, 27, 175–184. [Google Scholar] [CrossRef]
- Conte, L.; Lupo, R.; Lezzi, A.; Sciolti, S.; Rubbi, I.; Carvello, M.; Calabrò, A.; Botti, S.; Fanizzi, A.; Massafra, R.; et al. Breast Cancer Prevention Practices and Knowledge in Italian and Chinese Women in Italy: Clinical Checkups, Free NHS Screening Adherence, and Breast Self-Examination (BSE). J. Cancer Educ. 2024, 40, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Lupo, R.; Zacchino, S.; Caldararo, C.; Calabrò, A.; Carriero, M.C.; Santoro, P.; Carvello, M.; Conte, L. The Use of Electronical Devices and Relative Levels of Nomophobia within a Group of Italian Nurses: An Observational Study. Epidemiol. Biostat. Public Health 2020, 17, e13272. [Google Scholar] [CrossRef]
- Vitale, E.; Lupo, R.; Artioli, G.; Mea, R.; Lezzi, P.; Conte, L.; De Nunzio, G. How Shift Work Influences Anxiety, Depression, Stress and Insomnia Conditions in Italian Nurses: An Exploratory Study. Acta Biomed. 2023, 94, e2023102. [Google Scholar] [CrossRef]
- Conte, L.; Rizzo, E.; Civino, E.; Tarantino, P.; De Nunzio, G.; De Matteis, E. Enhancing Breast Cancer Risk Prediction with Machine Learning: Integrating BMI, Smoking Habits, Hormonal Dynamics, and BRCA Gene Mutations—A Game-Changer Compared to Traditional Statistical Models? Appl. Sci. 2024, 14, 8474. [Google Scholar] [CrossRef]
- Caldo, D.; Bologna, S.; Conte, L.; Amin, M.S.; Anselma, L.; Basile, V.; Hossain, M.M.; Mazzei, A.; Heritier, P.; Ferracini, R.; et al. Machine Learning Algorithms Distinguish Discrete Digital Emotional Fingerprints for Web Pages Related to Back Pain. Sci. Rep. 2023, 13, 4654. [Google Scholar] [CrossRef]
- Vitale, E.; Lupo, R.; Calabrò, A.; Cornacchia, M.; Conte, L.; Marchisio, D.; Caldararo, C.; Carvello, M.; Carriero, M.C. Mapping Potential Risk Factors in Developing Burnout Syndrome Between Physicians and Registered Nurses Suffering from an Aggression in Italian Emergency Departments. J. Psychopathol. 2021, 27, 148–155. [Google Scholar] [CrossRef]
- Kassis, T.; Hernandez-Gordillo, V.; Langer, R.; Griffith, L.G. OrgaQuant: Human Intestinal Organoid Localization and Quantification Using Deep Convolutional Neural Networks. Sci. Rep. 2019, 9, 12479. [Google Scholar] [CrossRef]
- Ostrop, J.; Zwiggelaar, R.T.; Terndrup Pedersen, M.; Gerbe, F.; Bösl, K.; Lindholm, H.T.; Díez-Sánchez, A.; Parmar, N.; Radetzki, S.; von Kries, J.P.; et al. A Semi-Automated Organoid Screening Method Demonstrates Epigenetic Control of Intestinal Epithelial Differentiation. Front. Cell Dev. Biol. 2021, 8, 618552. [Google Scholar] [CrossRef]
- Domènech-Moreno, E.; Brandt, A.; Lemmetyinen, T.T.; Wartiovaara, L.; Mäkelä, T.P.; Ollila, S. Tell—An Object-Detector Algorithm for Automatic Classification of Intestinal Organoids. Dis. Model. Mech. 2023, 16, dmm049756. [Google Scholar] [CrossRef] [PubMed]
- Deben, C.; De La Hoz, E.C.; Compte, M.L.; Van Schil, P.; Hendriks, J.M.H.; Lauwers, P.; Yogeswaran, S.K.; Lardon, F.; Pauwels, P.; Van Laere, S.; et al. OrBITS: Label-Free and Time-Lapse Monitoring of Patient Derived Organoids for Advanced Drug Screening. Cell. Oncol. 2023, 46, 299–314. [Google Scholar] [CrossRef]
- Xiang, T.; Wang, J.; Li, H. Current Applications of Intestinal Organoids: A Review. Stem Cell Res. Ther. 2024, 15, 155. [Google Scholar] [CrossRef] [PubMed]
- Laudadio, I.; Carissimi, C.; Scafa, N.; Bastianelli, A.; Fulci, V.; Renzini, A.; Russo, G.; Oliva, S.; Vitali, R.; Palone, F.; et al. Characterization of Patient-Derived Intestinal Organoids for Modelling Fibrosis in Inflammatory Bowel Disease. Inflamm. Res. 2024, 73, 1359–1370. [Google Scholar] [CrossRef] [PubMed]
- Abdul, L.; Xu, J.; Sotra, A.; Chaudary, A.; Gao, J.; Rajasekar, S.; Anvari, N.; Mahyar, H.; Zhang, B. D-CryptO: Deep Learning-Based Analysis of Colon Organoid Morphology from Brightfield Images. Lab Chip 2022, 22, 4118–4128. [Google Scholar] [CrossRef] [PubMed]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Wang, A.; Chen, H.; Liu, L.; Chen, K.; Lin, Z.; Han, J.; Ding, G. YOLOv10: Real-Time End-to-End Object Detection. In Proceedings of the 2024 Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 17–21 June 2024. [Google Scholar]
- Sun, Y.; Zhang, H.; Huang, F.; Gao, Q.; Li, P.; Li, D.; Luo, G. Deliod a Lightweight Detection Model for Intestinal Organoids Based on Deep Learning. Sci. Rep. 2025, 15, 5040. [Google Scholar] [CrossRef]
- Bukas, C.; Subramanian, H.; See, F.; Steinchen, C.; Ezhov, I.; Boosarpu, G.; Asgharpour, S.; Burgstaller, G.; Lehmann, M.; Kofler, F.; et al. MultiOrg: A Multi-Rater Organoid-Detection Dataset. In Proceedings of the NeurIPS 2025, San Diego, CA, USA, 2–7 December 2024. [Google Scholar]
- Sureshkumar, V.; Prasad, R.S.N.; Balasubramaniam, S.; Jagannathan, D.; Daniel, J.; Dhanasekaran, S. Breast Cancer Detection and Analytics Using Hybrid CNN and Extreme Learning Machine. J. Pers. Med. 2024, 14, 792. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Shrimali, V.; Kumar, M. Detection and Classification of Brain Tumor Using Hybrid Feature Extraction Technique. Multimed. Tools Appl. 2023, 82, 21483–21507. [Google Scholar] [CrossRef]
- Iqball, T.; Wani, M.A. Weighted Ensemble Model for Image Classification. Int. J. Inf. Technol. 2023, 15, 557–564. [Google Scholar] [CrossRef]
- Musa, A.B. Comparative Study on Classification Performance Between Support Vector Machine and Logistic Regression. Int. J. Mach. Learn. Cybern. 2013, 4, 13–24. [Google Scholar] [CrossRef]
- Couronné, R.; Probst, P.; Boulesteix, A.-L. Random Forest Versus Logistic Regression: A Large-Scale Benchmark Experiment. BMC Bioinform. 2018, 19, 270. [Google Scholar] [CrossRef]
- Leng, B.; Jiang, H.; Wang, B.; Wang, J.; Luo, G. Deep-Orga: An Improved Deep Learning-Based Lightweight Model for Intestinal Organoid Detection. Comput. Biol. Med. 2024, 169, 107847. [Google Scholar] [CrossRef]
- Cicceri, G.; Di Bella, S.; Di Franco, S.; Stassi, G.; Todaro, M.; Vitabile, S. Deep Learning Approaches for Morphological Classification of Intestinal Organoids. IEEE Access 2025, 13, 62267–62287. [Google Scholar] [CrossRef]
- Huang, K.; Li, M.; Li, Q.; Chen, Z.; Zhang, Y.; Gu, Z. Image-Based Profiling and Deep Learning Reveal Morphological Heterogeneity of Colorectal Cancer Organoids. Comput. Biol. Med. 2024, 173, 108322. [Google Scholar] [CrossRef]
- Otsu, N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. Man. Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Vincent, L.; Soille, P. Watersheds in Digital Spaces: An Efficient Algorithm Based on Immersion Simulations. IEEE Trans. Pattern Anal. Mach. Intell. 1991, 13, 583–598. [Google Scholar] [CrossRef]
- Chan, T.F.; Vese, L.A. Active Contours Without Edges. IEEE Trans. Image Process. 2001, 10, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man. Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Ojala, T.; Pietikainen, M.; Maenpaa, T. Multiresolution Gray-Scale and Rotation Invariant Texture Classification with Local Binary Patterns. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 971–987. [Google Scholar] [CrossRef]
- Dalal, N.; Triggs, B. Histograms of Oriented Gradients for Human Detection. In Proceedings of the 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05), San Diego, CA, USA, 20–25 June 2005; IEEE: New York, NY, USA, 2005; Volume 1, pp. 886–893. [Google Scholar]
| Classifier | Feature Extraction | Enseble ResNet50 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YOLO | ResNet50 | |||||||||||
| AUC | AUC | AUC | ||||||||||
| Org0 | Org1 | Org3 | Sph | Org0 | Org1 | Org3 | Sph | Org0 | Org1 | Org3 | Sph | |
| LR | 0.89 | 0.83 | 0.91 | 0.80 | 0.97 | 0.93 | 0.98 | 0.98 | 0.96 | 0.92 | 0.98 | 0.96 |
| Random Forest | 0.89 | 0.82 | 0.92 | 0.71 | 0.96 | 0.90 | 0.97 | 0.94 | ||||
| Naïve Bayes | 0.85 | 0.77 | 0.89 | 0.74 | 0.91 | 0.84 | 0.96 | 0.87 | ||||
| KNN | 0.80 | 0.75 | 0.69 | 0.71 | 0.92 | 0.83 | 0.93 | 0.87 | ||||
| XGBoost | 0.92 | 0.87 | 0.94 | 0.82 | 0.96 | 0.92 | 0.98 | 0.97 | ||||
| MLP | 0.92 | 0.87 | 0.94 | 0.84 | 0.96 | 0.91 | 0.97 | 0.97 | ||||
| Classifier | Feature Extraction | Ensemble ResNet50 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YOLO | ResNet50 | |||||||||||
| Acc | P | R | F1 | Acc | P | R | F1 | Acc | P | R | F1 | |
| LR | 0.71 | 069 | 0.70 | 0.69 | 0.84 | 0.84 | 0.84 | 0.84 | 0.84 | 0.83 | 0.84 | 0.83 |
| Random Forest | 0.69 | 0.69 | 0.69 | 0.65 | 0.82 | 0.81 | 0.81 | 0.81 | ||||
| Naïve Bayes | 0.57 | 0.66 | 0.57 | 0.60 | 0.72 | 0.75 | 0.73 | 0.73 | ||||
| KNN | 0.60 | 0.60 | 0.60 | 0.54 | 0.79 | 0.77 | 0.78 | 0.77 | ||||
| XGBoost | 0.74 | 0.74 | 0.74 | 0.72 | 0.83 | 0.83 | 0.83 | 0.83 | ||||
| MLP | 0.74 | 0.74 | 0.74 | 0.73 | 0.82 | 0.81 | 0.82 | 0.82 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Conte, L.; De Nunzio, G.; Raso, G.; Cascio, D. Multi-Class Segmentation and Classification of Intestinal Organoids: YOLO Stand-Alone vs. Hybrid Machine Learning Pipelines. Appl. Sci. 2025, 15, 11311. https://doi.org/10.3390/app152111311
Conte L, De Nunzio G, Raso G, Cascio D. Multi-Class Segmentation and Classification of Intestinal Organoids: YOLO Stand-Alone vs. Hybrid Machine Learning Pipelines. Applied Sciences. 2025; 15(21):11311. https://doi.org/10.3390/app152111311
Chicago/Turabian StyleConte, Luana, Giorgio De Nunzio, Giuseppe Raso, and Donato Cascio. 2025. "Multi-Class Segmentation and Classification of Intestinal Organoids: YOLO Stand-Alone vs. Hybrid Machine Learning Pipelines" Applied Sciences 15, no. 21: 11311. https://doi.org/10.3390/app152111311
APA StyleConte, L., De Nunzio, G., Raso, G., & Cascio, D. (2025). Multi-Class Segmentation and Classification of Intestinal Organoids: YOLO Stand-Alone vs. Hybrid Machine Learning Pipelines. Applied Sciences, 15(21), 11311. https://doi.org/10.3390/app152111311









