Symmetric 3D Convolutional Network with Uncertainty Estimation for MRI-Based Striatal DaT-Uptake Assessment in Parkinson’s Disease
Abstract
1. Introduction
- We propose a symmetric regressor with a novel loss function for the striatal DaT-uptake prediction, which explicitlyutilizes the lateral symmetry and correlation of the right and left nigral patch inputs and the corresponding DaT-uptake outputs.
- We propose a symmetric MC dropout technique for improved uncertainty estimation, which enhances the reliability of the proposed method.
2. Related Works
2.1. Standard DaT-Uptake Assessment
2.2. DaT-Uptake Prediction Using MRI
2.3. Deep-Learning-Based Prediction
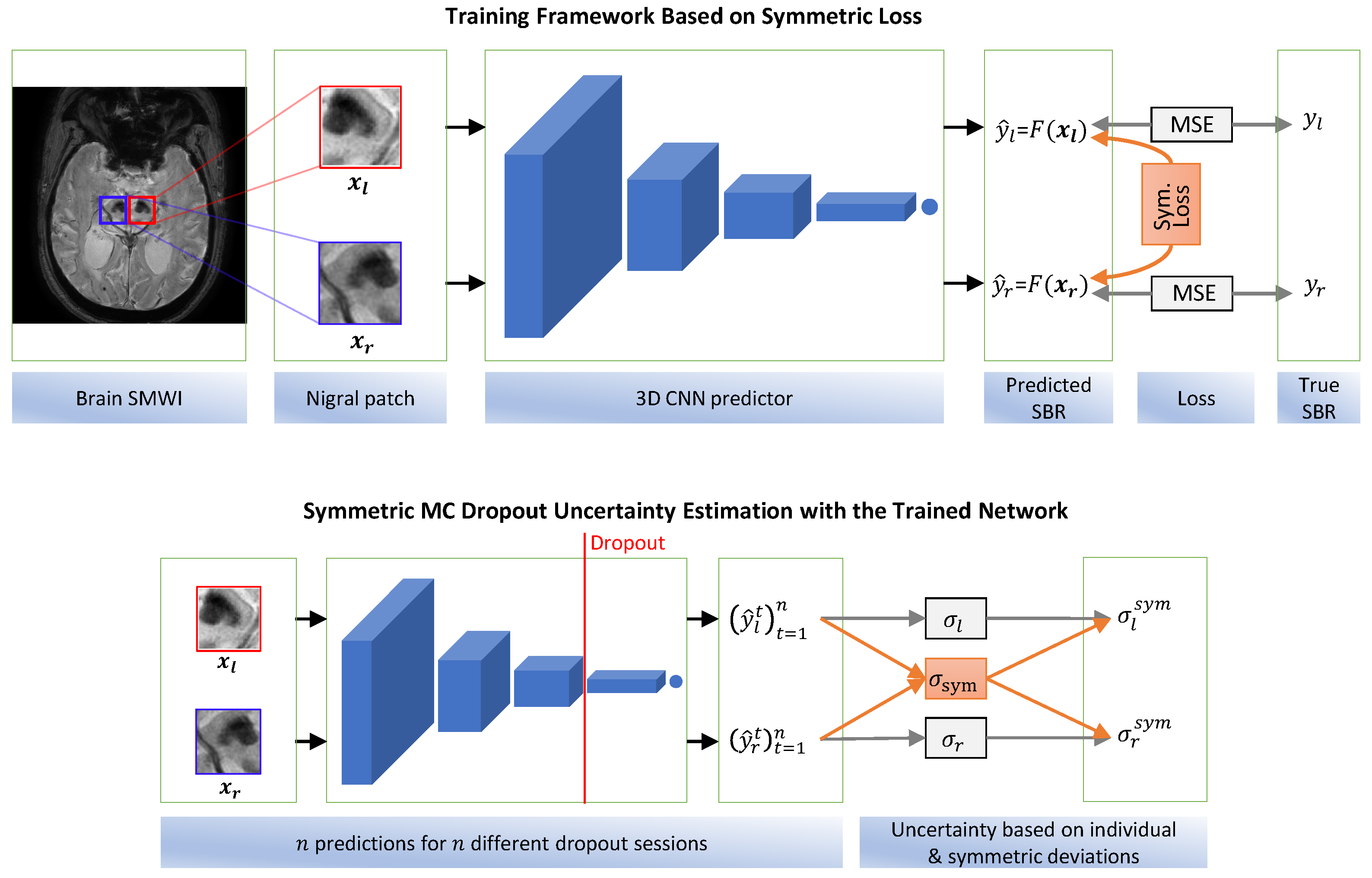
3. Materials and Methods
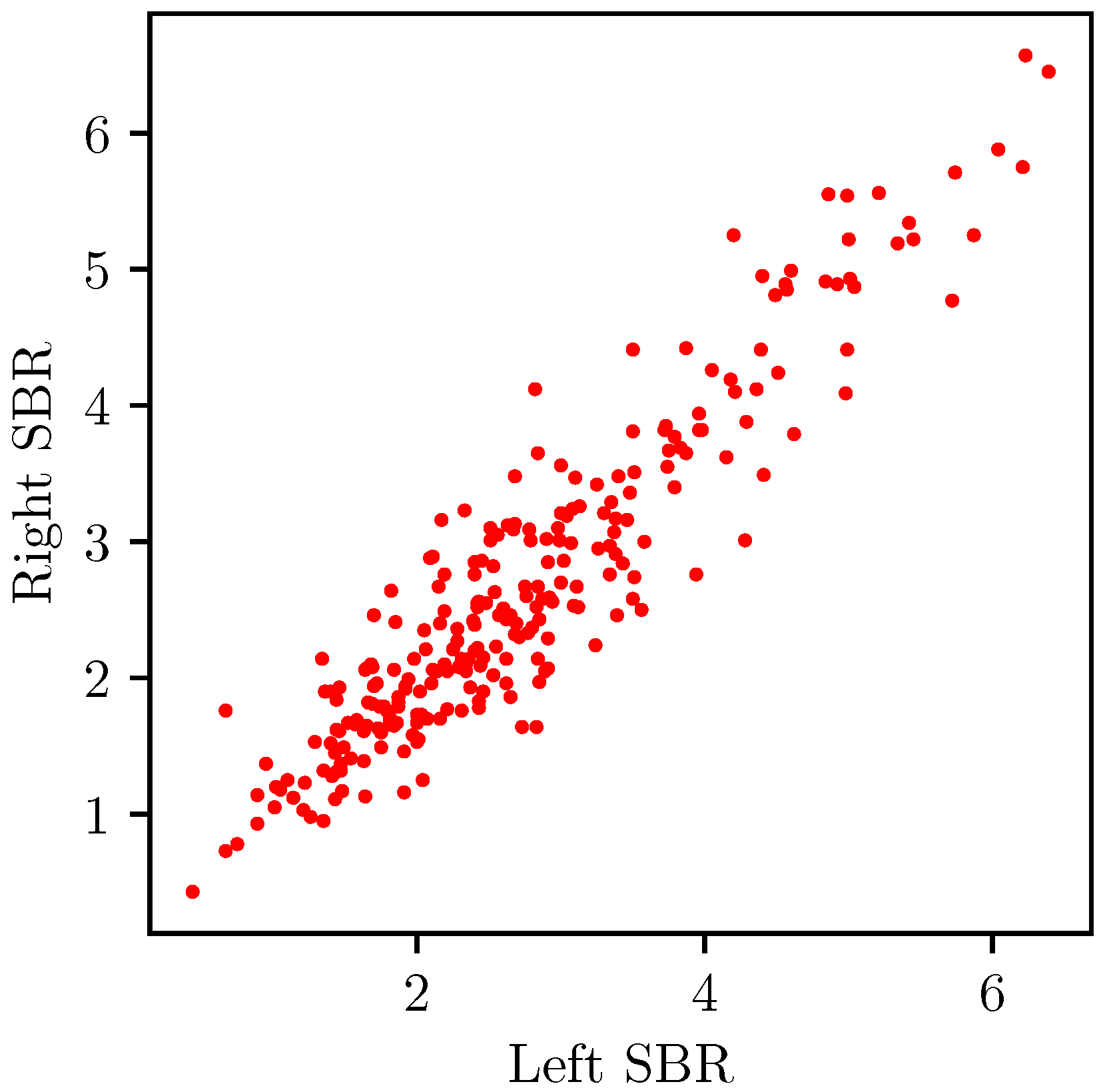
3.1. Data
3.2. Deep Regressor
3.3. Symmetric Regressor
3.3.1. Identical Input Type
3.3.2. Correlated Output
3.3.3. Proposed Model
3.4. Symmetric Uncertainty in Prediction
3.4.1. Monte Carlo Dropout
3.4.2. Symmetric MC Dropout
| Algorithm 1: Symmetric MC prediction interval |
|
4. Results
4.1. Experimental Setting
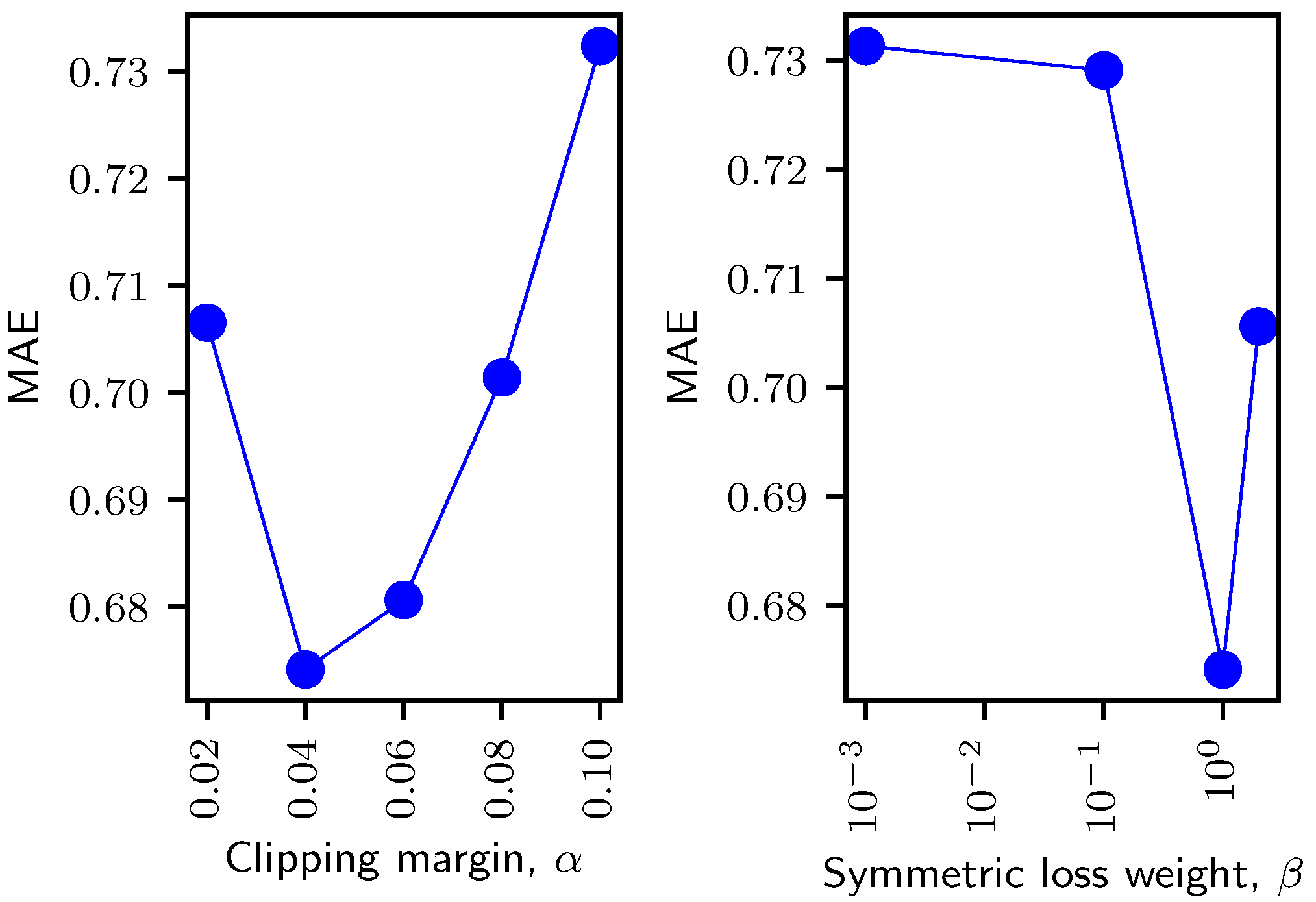
4.2. Hyperparameter Dependency
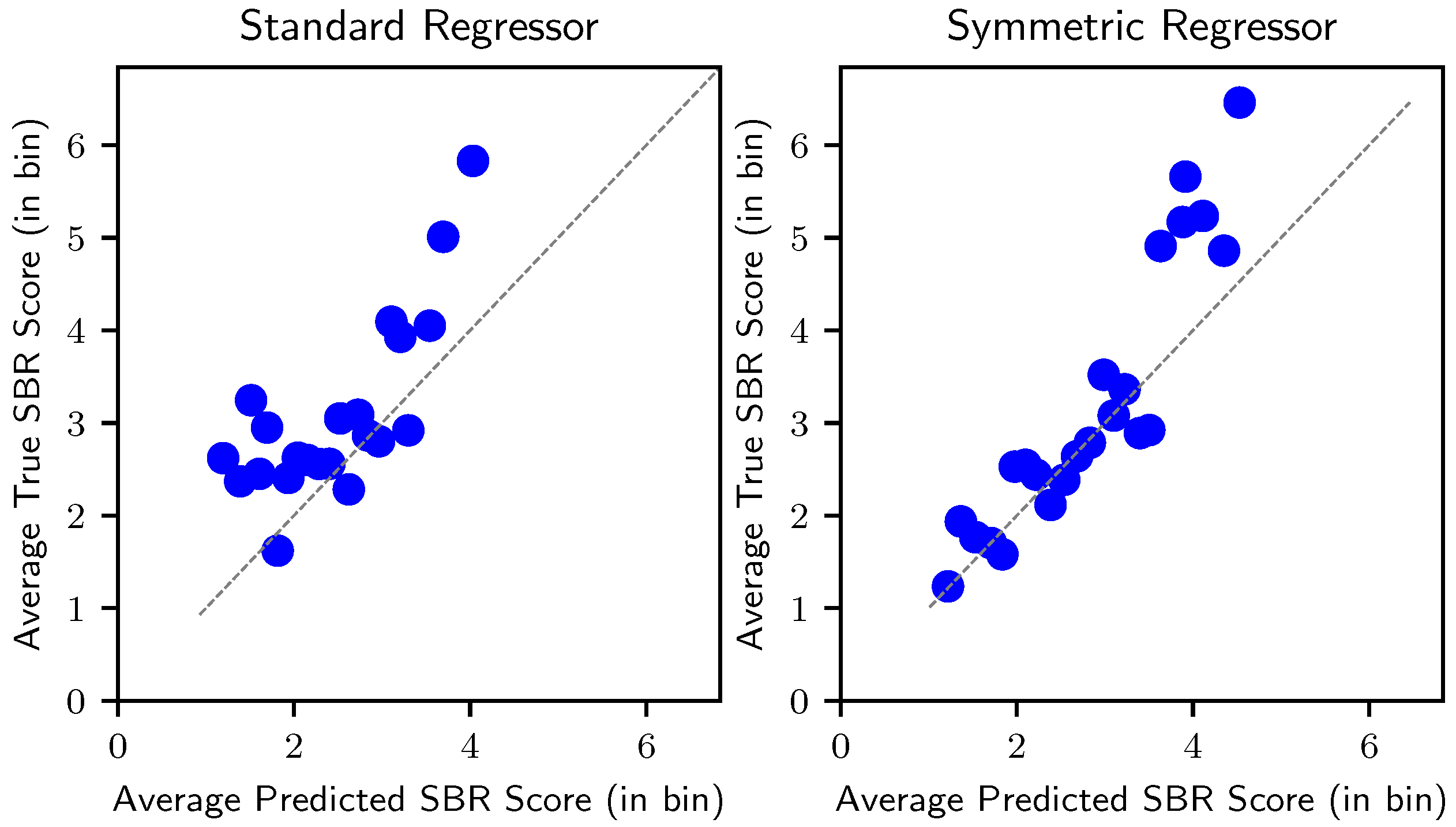
4.3. SBR Prediction Performance
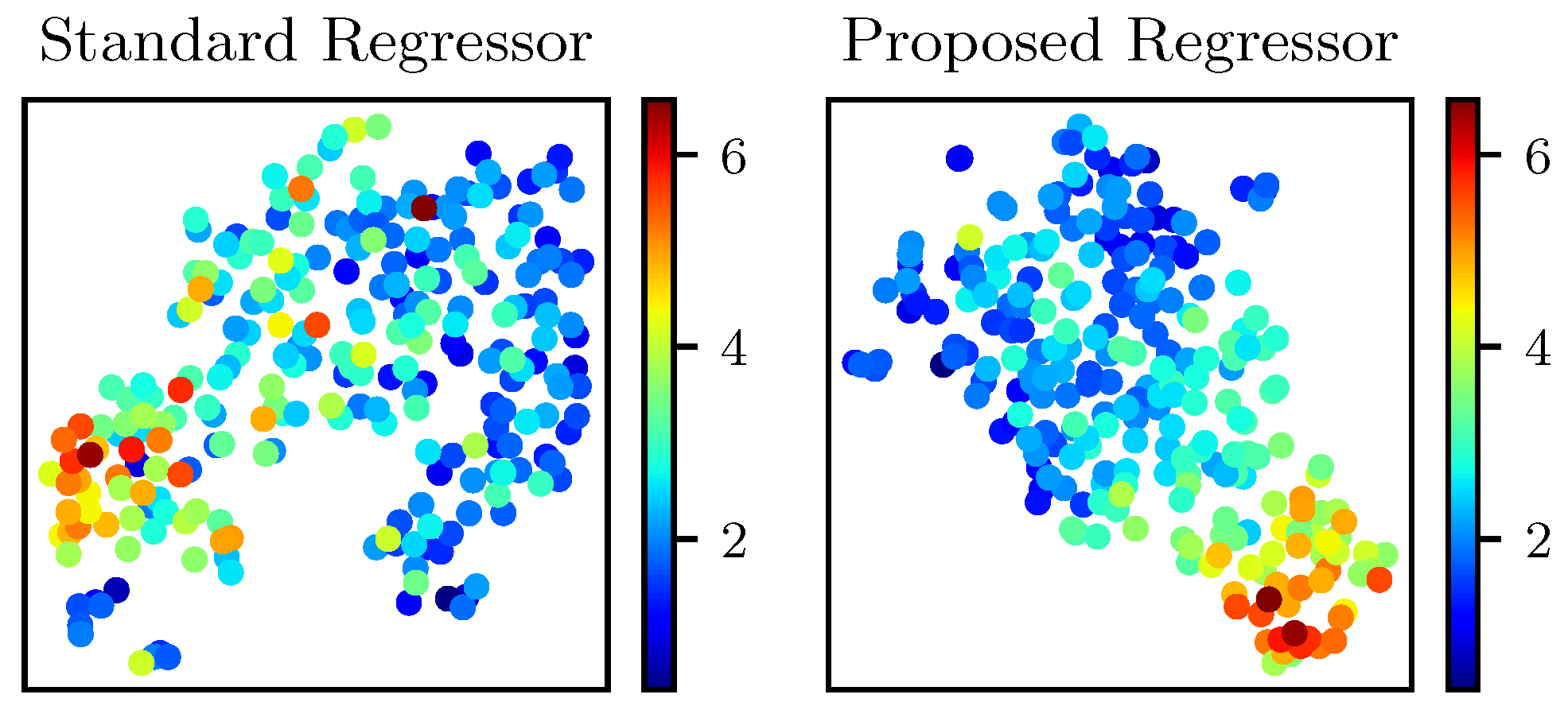
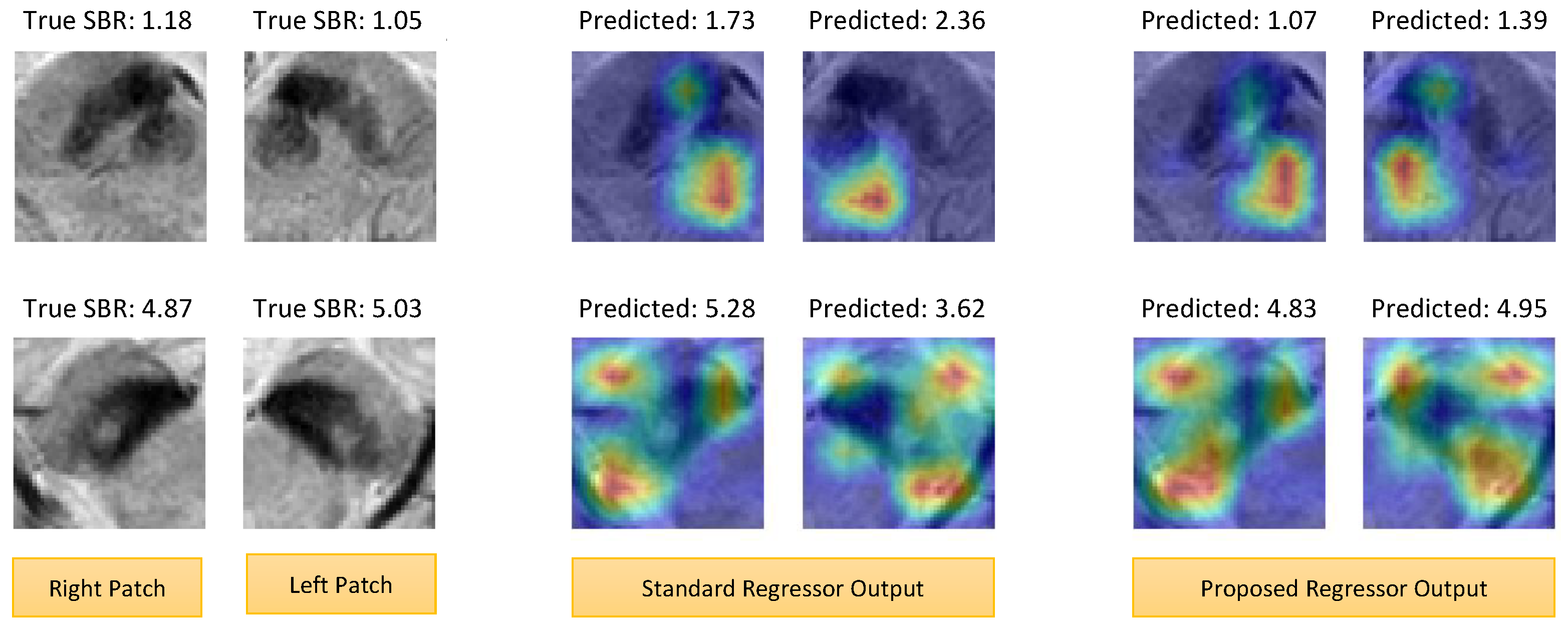
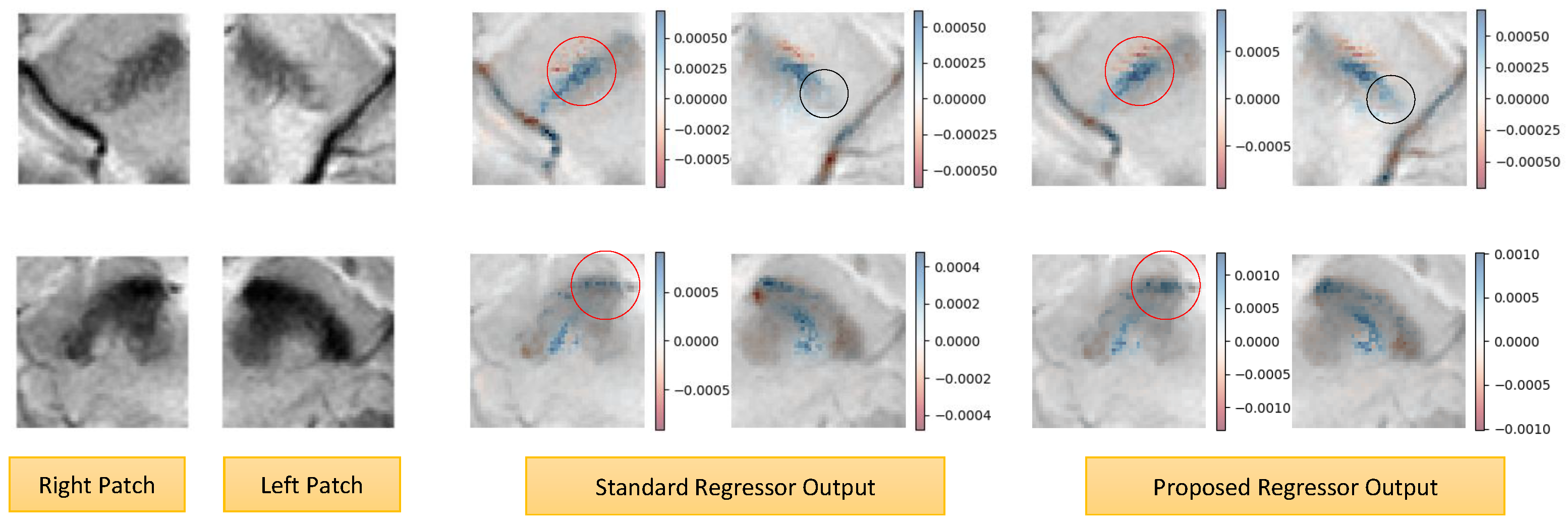
4.4. Explainability Analysis
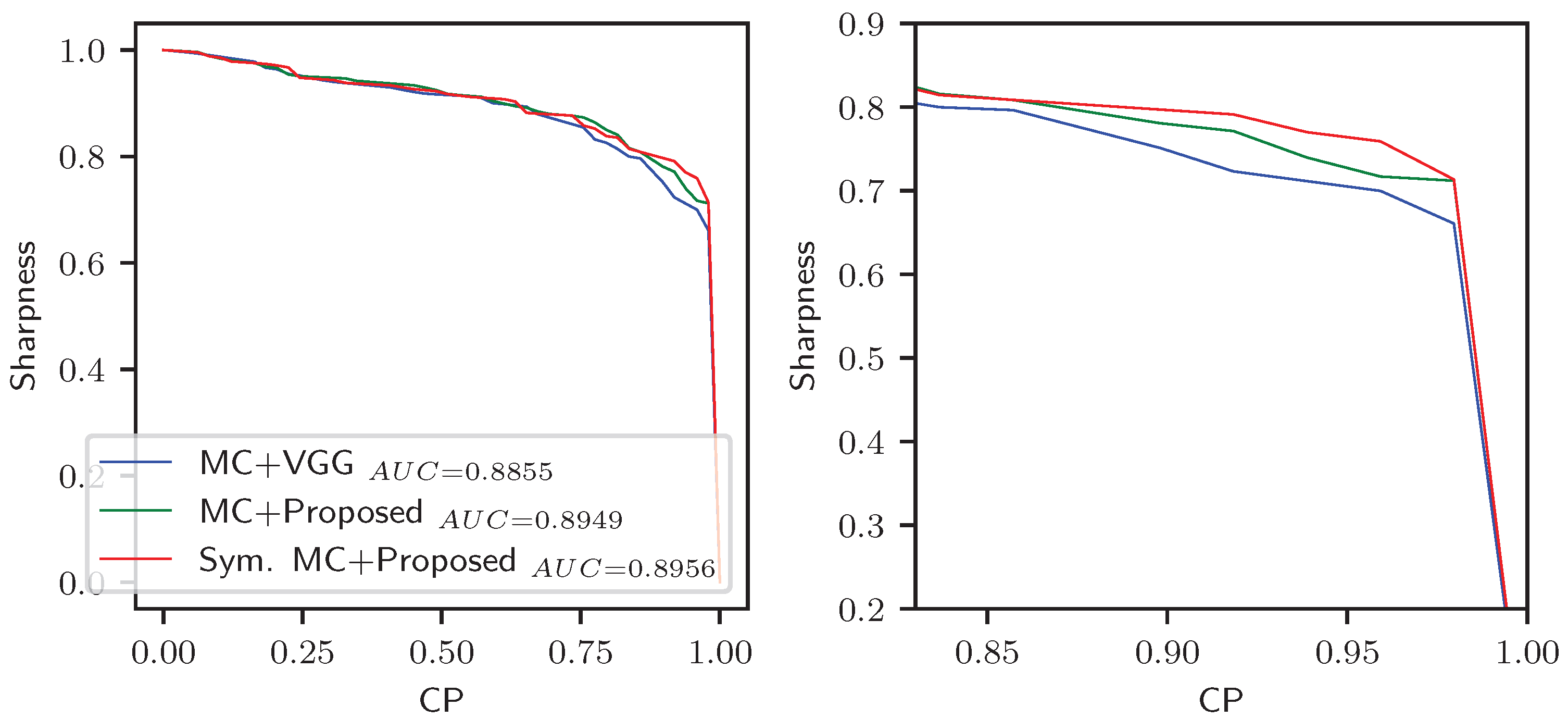
4.5. Quality of Uncertainty
4.6. Computation Time
5. Discussion
5.1. Validity of the Proposed Model
5.1.1. ConvNet Model Selection
5.1.2. Advantage of the Symmetric Model
5.2. Comparison with Previous Works
5.3. Uncertainty Analysis
5.4. Limitations and Future Directions
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bloem, B.R.; Okun, M.S.; Klein, C. Parkinson’s disease. Lancet 2021, 397, 2284–2303. [Google Scholar] [CrossRef]
- Damier, P.; Hirsch, E.; Agid, Y.; Graybiel, A. The substantia nigra of the human brain: II. Patterns of loss of dopamine-containing neurons in Parkinson’s disease. Brain 1999, 122, 1437–1448. [Google Scholar] [CrossRef] [PubMed]
- Wakabayashi, Y.; Takahashi, R.; Kanda, T.; Zeng, F.; Nogami, M.; Ishii, K.; Murakami, T. Semi-quantitative dopamine transporter standardized uptake value in comparison with conventional specific binding ratio in [123I] FP-CIT single-photon emission computed tomography (DaTscan). Neurol. Sci. 2018, 39, 1401–1407. [Google Scholar] [CrossRef]
- Suwijn, S.R.; van Boheemen, C.J.; de Haan, R.J.; Tissingh, G.; Booij, J.; de Bie, R.M. The diagnostic accuracy of dopamine transporter SPECT imaging to detect nigrostriatal cell loss in patients with Parkinson’s disease or clinically uncertain parkinsonism: A systematic review. EJNMMI Res. 2015, 5, 12. [Google Scholar] [CrossRef]
- Akdemir, Ü.Ö.; BORA, H.A.T.; Atay, L.Ö. Dopamine transporter SPECT imaging in Parkinson’s disease and parkinsoniandisorders. Turk. J. Med. Sci. 2021, 51, 400–410. [Google Scholar] [CrossRef]
- Mahlknecht, P.; Krismer, F.; Poewe, W.; Seppi, K. Meta-analysis of dorsolateral nigral hyperintensity on magnetic resonance imaging as a marker for Parkinson’s disease. Mov. Disord. 2017, 32, 619–623. [Google Scholar] [CrossRef]
- Schwarz, S.T.; Afzal, M.; Morgan, P.S.; Bajaj, N.; Gowland, P.A.; Auer, D.P. The ‘swallow tail’appearance of the healthy nigrosome–a new accurate test of Parkinson’s disease: A case-control and retrospective cross-sectional MRI study at 3T. PLoS ONE 2014, 9, e93814. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Ashimgaliyev, M.; Matkarimov, B.; Barlybayev, A.; Li, R.Y.M.; Zhumadillayeva, A. Accurate MRI-based brain tumor diagnosis: Integrating segmentation and deep learning approaches. Appl. Sci. 2024, 14, 7281. [Google Scholar] [CrossRef]
- Güler, M.; Namlı, E. Brain tumor detection with deep learning methods’ classifier optimization using medical images. Appl. Sci. 2024, 14, 642. [Google Scholar] [CrossRef]
- Devindi, G.; Dissanayake, D.; Liyanage, S.; Francis, F.; Pavithya, M.; Piyarathne, N.; Hettiarachchi, P.; Rasnayaka, R.; Jayasinghe, R.D.; Ragel, R.G.; et al. Multimodal deep convolutional neural network pipeline for AI-assisted early detection of oral cancer. IEEE Access 2024, 12, 124375–124390. [Google Scholar] [CrossRef]
- Swathi, G.; Altalbe, A.; Kumar, R.P. QuCNet: Quantum-inspired convolutional neural networks for optimized thyroid nodule classification. IEEE Access 2024, 12, 27829–27842. [Google Scholar] [CrossRef]
- Mohsen, H.; El-Dahshan, E.S.A.; El-Horbaty, E.S.M.; Salem, A.B.M. Classification using deep learning neural networks for brain tumors. Future Comput. Inform. J. 2018, 3, 68–71. [Google Scholar]
- Yadav, S.S.; Jadhav, S.M. Deep convolutional neural network based medical image classification for disease diagnosis. J. Big Data 2019, 6, 113. [Google Scholar] [CrossRef]
- Aboutalebi, H.; Pavlova, M.; Shafiee, M.J.; Sabri, A.; Alaref, A.; Wong, A. Covid-net cxr-s: Deep convolutional neural network for severity assessment of COVID-19 cases from chest x-ray images. Diagnostics 2021, 12, 25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Petitjean, C.; Lopez, P.; Ainouz, S. Direct estimation of fetal head circumference from ultrasound images based on regression CNN. In Proceedings of the Medical Imaging with Deep Learning, Montreal, QC, Canada, 6–8 July 2020; pp. 914–922. [Google Scholar]
- Bae, Y.J.; Choi, B.S.; Kim, J.M.; Ai, W.A.; Yun, I.; Song, Y.S.; Nam, Y.; Cho, S.J.; Kim, J.H. Deep learning regressor model based on nigrosome MRI in Parkinson syndrome effectively predicts striatal dopamine transporter-SPECT uptake. Neuroradiology 2023, 65, 1101–1109. [Google Scholar] [CrossRef]
- Gal, Y.; Ghahramani, Z. Dropout as a bayesian approximation: Representing model uncertainty in deep learning. In Proceedings of the International Conference on Machine Learning, New York, NY, USA, 19–24 June 2016; pp. 1050–1059. [Google Scholar]
- Rahman, M.G.; Islam, M.M.; Tsujikawa, T.; Kiyono, Y.; Okazawa, H. Count-based method for specific binding ratio calculation in [I-123] FP-CIT SPECT analysis. Ann. Nucl. Med. 2019, 33, 14–21. [Google Scholar] [CrossRef]
- Kuo, P.H.; Eshghi, N.; Tinaz, S.; Blumenfeld, H.; Louis, E.D.; Zubal, G. Optimization of parameters for quantitative analysis of 123I-ioflupane SPECT images for monitoring progression of Parkinson disease. J. Nucl. Med. Technol. 2019, 47, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Filippi, L.; Manni, C.; Pierantozzi, M.; Brusa, L.; Danieli, R.; Stanzione, P.; Schillaci, O. 123I-FP-CIT semi-quantitative SPECT detects preclinical bilateral dopaminergic deficit in early Parkinson’s disease with unilateral symptoms. Nucl. Med. Commun. 2005, 26, 421–426. [Google Scholar] [CrossRef]
- Booij, J.; Tissingh, G.; Boer, G.; Speelman, J.; Stoof, J.; Janssen, A.; Wolters, E.C.; Van Royen, E. [123I] FP-CIT SPECT shows a pronounced decline of striatal dopamine transporter labelling in early and advanced Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry 1997, 62, 133. [Google Scholar] [CrossRef]
- Prashanth, R.; Roy, S.D.; Mandal, P.K.; Ghosh, S. Automatic classification and prediction models for early Parkinson’s disease diagnosis from SPECT imaging. Expert Syst. Appl. 2014, 41, 3333–3342. [Google Scholar] [CrossRef]
- Lehericy, S.; Vaillancourt, D.E.; Seppi, K.; Monchi, O.; Rektorova, I.; Antonini, A.; McKeown, M.J.; Masellis, M.; Berg, D.; Rowe, J.B.; et al. The role of high-field magnetic resonance imaging in parkinsonian disorders: Pushing the boundaries forward. Mov. Disord. 2017, 32, 510–525. [Google Scholar]
- Gao, P.; Zhou, P.Y.; Li, G.; Zhang, G.B.; Wang, P.Q.; Liu, J.Z.; Xu, F.; Yang, F.; Wu, X.X. Visualization of nigrosomes-1 in 3T MR susceptibility weighted imaging and its absence in diagnosing Parkinson’s disease. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 4603–4609. [Google Scholar] [PubMed]
- Reiter, E.; Mueller, C.; Pinter, B.; Krismer, F.; Scherfler, C.; Esterhammer, R.; Kremser, C.; Schocke, M.; Wenning, G.K.; Poewe, W.; et al. Dorsolateral nigral hyperintensity on 3.0 T susceptibility-weighted imaging in neurodegenerative Parkinsonism. Mov. Disord. 2015, 30, 1068–1076. [Google Scholar] [PubMed]
- Cosottini, M.; Frosini, D.; Pesaresi, I.; Donatelli, G.; Cecchi, P.; Costagli, M.; Biagi, L.; Ceravolo, R.; Bonuccelli, U.; Tosetti, M. Comparison of 3T and 7T susceptibility-weighted angiography of the substantia nigra in diagnosing Parkinson disease. Am. J. Neuroradiol. 2015, 36, 461–466. [Google Scholar] [CrossRef]
- Bae, Y.J.; Kim, J.M.; Kim, E.; Lee, K.M.; Kang, S.Y.; Park, H.S.; Kim, K.J.; Kim, Y.E.; Oh, E.S.; Yun, J.Y.; et al. Loss of nigral hyperintensity on 3 Tesla MRI of parkinsonism: Comparison with 123I-FP-CIT SPECT. Mov. Disord. 2016, 31, 684–692. [Google Scholar] [CrossRef] [PubMed]
- Bae, Y.J.; Kim, J.M.; Kim, K.J.; Kim, E.; Park, H.S.; Kang, S.Y.; Yoon, I.Y.; Lee, J.Y.; Jeon, B.; Kim, S.E. Loss of substantia nigra hyperintensity at 3.0-T MR imaging in idiopathic REM sleep behavior disorder: Comparison with 123I-FP-CIT SPECT. Radiology 2018, 287, 285–293. [Google Scholar]
- Uchida, Y.; Kan, H.; Sakurai, K.; Inui, S.; Kobayashi, S.; Akagawa, Y.; Shibuya, K.; Ueki, Y.; Matsukawa, N. Magnetic susceptibility associates with dopaminergic deficits and cognition in Parkinson’s disease. Mov. Disord. 2020, 35, 1396–1405. [Google Scholar] [CrossRef]
- Gho, S.M.; Liu, C.; Li, W.; Jang, U.; Kim, E.Y.; Hwang, D.; Kim, D.H. Susceptibility map-weighted imaging (SMWI) for neuroimaging. Magn. Reson. Med. 2014, 72, 337–346. [Google Scholar] [CrossRef]
- Nam, Y.; Gho, S.M.; Kim, D.H.; Kim, E.Y.; Lee, J. Imaging of nigrosome 1 in substantia nigra at 3T using multiecho susceptibility map-weighted imaging (SMWI). J. Magn. Reson. Imaging 2017, 46, 528–536. [Google Scholar] [CrossRef]
- Bae, Y.J.; Kim, J.M.; Sohn, C.H.; Choi, J.H.; Choi, B.S.; Song, Y.S.; Nam, Y.; Cho, S.J.; Jeon, B.; Kim, J.H. Imaging the substantia nigra in Parkinson disease and other Parkinsonian syndromes. Radiology 2021, 300, 260–278. [Google Scholar] [CrossRef]
- Bae, Y.; Song, Y.; Kim, J.M.; Choi, B.; Nam, Y.; Choi, J.H.; Lee, W.; Kim, J. Determining the degree of dopaminergic denervation based on the loss of nigral hyperintensity on SMWI in parkinsonism. Am. J. Neuroradiol. 2021, 42, 681–687. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar]
- Raj, A.; Shah, N.A.; Tiwari, A.K.; Martini, M.G. Multivariate regression-based convolutional neural network model for fundus image quality assessment. IEEE Access 2020, 8, 57810–57821. [Google Scholar] [CrossRef]
- Rehman, S.U.; Gruhn, V. A sequential VGG16+ CNN-based automated approach with adaptive input for efficient detection of knee osteoarthritis stages. IEEE Access 2024, 12, 62407–62415. [Google Scholar] [CrossRef]
- Rodriguez-Oroz, M.C.; Rodriguez, M.; Leiva, C.; Rodriguez-Palmero, M.; Nieto, J.; Garcia-Garcia, D.; Luis Zubieta, J.; Cardiel, C.; Obeso, J.A. Neuronal activity of the red nucleus in Parkinson’s disease. Mov. Disord. 2008, 23, 908–911. [Google Scholar] [PubMed]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16 × 16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Koonce, B. MobileNetV3. In Convolutional Neural Networks with Swift for Tensorflow: Image Recognition and Dataset Categorization; Springer: Berlin/Heidelberg, Germany, 2021; pp. 125–144. [Google Scholar]
- Kaur, P.; Stoltzfus, J.C. Bland–Altman plot: A brief overview. Int. J. Acad. Med. 2017, 3, 110–111. [Google Scholar] [CrossRef]
- Van der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Draelos, R.L.; Carin, L. Use HiResCAM instead of Grad-CAM for faithful explanations of convolutional neural networks. arXiv 2020, arXiv:2011.08891. [Google Scholar]
- Nohara, Y.; Matsumoto, K.; Soejima, H.; Nakashima, N. Explanation of machine learning models using improved shapley additive explanation. In Proceedings of the 10th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, Niagara Falls, NY, USA, 7–10 September 2019; p. 546. [Google Scholar]
- Psaros, A.F.; Meng, X.; Zou, Z.; Guo, L.; Karniadakis, G.E. Uncertainty quantification in scientific machine learning: Methods, metrics, and comparisons. J. Comput. Phys. 2023, 477, 111902. [Google Scholar] [CrossRef]

| Reference/Method | Key Finding | Limitation | Our Contribution |
|---|---|---|---|
| Gao et al., 2015 [26] Nigral hyperintensity-based decision using 3T MRI | Hyperintensity absence indicates PD | Manual observation; PD identification only | - Deep learning approach, rather than manual observation |
| Reiter et al., 2015 [27] Nigral hyperintensity-based decision using 3T SWI | Hyperintensity in the dorsolateral nigra is a useful PD indicator | Manual observation; PD identification only | - Exact SBR prediction, rather than binary identification |
| Bae et al., 2016 [29] Nigral hyperintensity-based DaT-uptake prediction using 3T SWI | Good agreement btn. nigral hyperintensity in SWI and DaT Uptake in SPECT | Manual observation; Binary (high/low SBR) prediction only | |
| Nam et al., 2017 [33] Nigral hyperintensity-based PD diagnosis using 3T SMWI | Improved visualization of nigrosome; improved diagnosis | Manual observation; PD identification only | |
| Bae et al., 2021 [35] Nigral hyperintensity-based DaT-uptake prediction using 3T SMWI | Improved agreement to DaT uptake in SPECT | Manual observation; Binary (high/low SBR) prediction only | |
| Uchida et al., 2020 [31] Mean nigral intensity-based PD diagnosis using 3T MRI | Mean intensity inversely correlates to DaT uptake | Low correlation () | - Leverage the entire intensity space of the 3D nigral patch, as opposed to a single summary statistic like the mean intensity |
| Bae et al., 2023 [18] Deep regressor-based DaT -uptake prediction using 3T SMWI | Improved correlation to DaT uptake in SPECT | Left SBR prediction only; right-to-left symmetry not utilized | - Simultaneous right and left SBR prediction - A symmetric regressor that leverages right- to-left anatomical symmetry |
| Layer | No. of Kernels, | Output Size |
|---|---|---|
| Kernel Size\Stride | ||
| Input | − | |
| Conv1-1 | \1 | |
| Conv1-2 | \1 | |
| Pool1 | \2 | |
| Conv2-1 | \1 | |
| Conv2-2 | \1 | |
| Pool2 | \2 | |
| Conv3-1 | \1 | |
| Conv3-2 | \1 | |
| Pool3 | \2 | |
| Conv4-1 | \1 | |
| Conv4-2 | \1 | |
| Pool4 | \2 | |
| Flatten | − | 4608 |
| FC | − | 1 |
| Attribute | Description |
|---|---|
| Total cases | 734 (367 right and 367 left |
| nigral patches) | |
| Train:validation:test | 512:112:110 (70%:15%:15%) |
| Data acquisition period | February 2017–December 2018 |
| MRI (SMWI) device | Ingenia and Ingenia CX, |
| Philips, The Netherlands. Detail | |
| protocol in [18] | |
| Nigral patch size | 50 × 50 × 20 voxels |
| Voxel size | 0.5 × 0.5 × 1 mm3 |
| SPECT device | DATrace-123™, Samyoung |
| Unitech, Korea with Trionix | |
| XLT, Trionix Research Lab, | |
| USA. Detail protocol in [18] | |
| SBR analysis program | DATquant, Xeleris 3.1, GE |
| Healthcare, USA | |
| SBR range | Right: [0.44, 6.84]; |
| left: [0.43, 6.80] | |
| MRI/SPECT time diff. | 42 ± 60 days |
| (median: 18 h) |
| RMSE | MAE | R | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Methods | Right | Left | Avg. | Right | Left | Avg. | Right | Left | Avg. |
| ViT [40] | 1.5487 | 1.4287 | 1.4887 | 1.1279 | 1.0192 | 1.0735 | −0.1431 | −0.0895 | −0.1163 |
| MobileNetV3 [41] | 2.1324 | 2.3530 | 2.2422 | 1.7173 | 1.9679 | 1.8426 | 0.2190 | 0.3172 | 0.2681 |
| ResNet-18 [9] | 1.3955 | 1.6411 | 1.5183 | 0.8836 | 0.9459 | 0.9147 | 0.4458 | 0.3255 | 0.3856 |
| VGG-16 [8] | 1.5515 | 1.6699 | 1.6107 | 0.9829 | 1.0236 | 1.0032 | 0.3978 | 0.3112 | 0.3545 |
| VGG-small [18] | 0.9410 | 1.0553 | 0.9982 | 0.7687 | 0.7663 | 0.7675 | 0.6281 | 0.7067 | 0.6674 |
| Sym. VGG * | 0.8477 | 0.6929 | 0.7703 | 0.7238 | 0.6738 | 0.6988 | 0.6745 | 0.7405 | 0.7075 |
| Sym. VGG + * | 0.8180 | 0.7081 | 0.7630 | 0.6774 | 0.6709 | 0.6741 | 0.7157 | 0.7426 | 0.7291 |
| Optimal (Sharpness, CP) | Sharpness @ CP | |||||
|---|---|---|---|---|---|---|
| Methods | Right | Left | Avg. | Right | Left | Avg. |
| MC + VGG | (0.815, 0.809) | (0.802, 0.822) | (0.809, 0.816) | 0.693 | 0.705 | 0.699 |
| MC + Proposed | (0.811, 0.845) | (0.815, 0.840) | (0.813, 0.843) | 0.710 | 0.728 | 0.719 |
| Symmetric MC + Proposed | (0.806, 0.867) | (0.819, 0.831) | (0.813, 0.849) | 0.733 | 0.765 | 0.749 |
| Prediction Correlation | Binary Agreement (Acc., Sen., Spe.) | |||
|---|---|---|---|---|
| Method; Goal; Data Size | Right | Left | Right | Left |
| Mean intensity [31]; classification; 61 | - | - | ||
| Observation [35]; classification; 138 | - | - | (0.652, 0.587, 0.897) | (0.667, 0.596, 0.932) |
| VGG regressor [18]; prediction; 367 | (0.764, 0.721, 0.750) | (0.855, 0.786, 0.756) | ||
| This work; prediction; 734 | (0.818, 0.860, 0.833) | (0.873, 0.854, 0.786) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdullah Al, W.; Yun, I.D.; Bae, Y.J. Symmetric 3D Convolutional Network with Uncertainty Estimation for MRI-Based Striatal DaT-Uptake Assessment in Parkinson’s Disease. Appl. Sci. 2025, 15, 10977. https://doi.org/10.3390/app152010977
Abdullah Al W, Yun ID, Bae YJ. Symmetric 3D Convolutional Network with Uncertainty Estimation for MRI-Based Striatal DaT-Uptake Assessment in Parkinson’s Disease. Applied Sciences. 2025; 15(20):10977. https://doi.org/10.3390/app152010977
Chicago/Turabian StyleAbdullah Al, Walid, Il Dong Yun, and Yun Jung Bae. 2025. "Symmetric 3D Convolutional Network with Uncertainty Estimation for MRI-Based Striatal DaT-Uptake Assessment in Parkinson’s Disease" Applied Sciences 15, no. 20: 10977. https://doi.org/10.3390/app152010977
APA StyleAbdullah Al, W., Yun, I. D., & Bae, Y. J. (2025). Symmetric 3D Convolutional Network with Uncertainty Estimation for MRI-Based Striatal DaT-Uptake Assessment in Parkinson’s Disease. Applied Sciences, 15(20), 10977. https://doi.org/10.3390/app152010977







