Abstract
To address the challenges of poor model generalization and suboptimal recognition accuracy stemming from limited and imbalanced sample sizes in tomato leaf disease identification, this study proposes a novel recognition strategy. This approach synergistically combines an enhanced image augmentation method based on generative adversarial networks with a lightweight deep learning model. Initially, an Atrous Spatial Pyramid Pooling (ASPP) module is integrated into the CycleGAN framework. This integration enhances the generator’s capacity to model multi-scale pathological lesion features, thereby significantly improving the diversity and realism of synthesized images. Subsequently, the Convolutional Block Attention Module (CBAM), incorporating both channel and spatial attention mechanisms, is embedded into the MobileNetV4 architecture. This enhancement boosts the model’s ability to focus on critical disease regions. Experimental results demonstrate that the proposed ASPP-CycleGAN significantly outperforms the original CycleGAN across multiple disease image generation tasks. Furthermore, the developed CBAM-MobileNetV4 model achieves a remarkable average recognition accuracy exceeding 97% for common tomato diseases, including early blight, late blight, and mosaic disease, representing a 1.86% improvement over the baseline MobileNetV4. The findings indicate that the proposed method offers exceptional data augmentation capabilities and classification performance under small-sample learning conditions, providing an effective technical foundation for the intelligent identification and control of tomato leaf diseases.
1. Introduction
Tomato is a globally cultivated and economically vital crop. However, its cultivation is frequently threatened by various diseases during its growth cycle, such as bacterial spot, early blight, late blight, and mosaic virus disease [1]. Given the prevalent intensive and concentrated farming practices employed for tomato production in China, disease outbreaks can spread rapidly, leading to widespread yield losses and significant economic impacts on farmers [2]. Therefore, developing accurate and rapid disease identification methods is crucial for effective disease management in tomato farming.
Although several publicly available tomato disease datasets exist, the small-sample problem remains a critical challenge in real-world applications. Large datasets have greatly facilitated model development, yet they are typically collected under controlled laboratory conditions or from specific tomato varieties. In practice, disease symptoms can vary considerably across different varieties, cultivation environments, and imaging conditions. Consequently, models trained on such datasets may not generalize well to local farms, where collecting a representative dataset becomes necessary. However, building farm-specific datasets often results in limited data volumes because the process of data collection and expert annotation is both labor-intensive and time-consuming. In this study, we define a “small dataset” as one containing approximately one hundred images per class, which is insufficient for training deep neural networks to achieve stable convergence and robust generalization. By contrast, datasets with thousands of images per class (e.g., over 5000) can generally be regarded as sufficient for training without a severe risk of overfitting. Therefore, despite the existence of larger public datasets, the small-data problem persists in practice and motivates the need for effective data augmentation and lightweight classification strategies, as investigated in this work.
Addressing crop disease identification under limited training samples, researchers have proposed methods focusing on small-sample leaf disease image recognition. These methods aim to learn classification models from scarce supervised samples. Current prevalent approaches for small-sample recognition encompass transfer learning, conventional data augmentation techniques, and generative adversarial networks (GANs) [3,4].
Among these, transfer learning represents a well-established strategy for small-sample recognition. It typically involves fine-tuning networks pre-trained on large-scale datasets like ImageNet [5,6]. For example, Astolfi et al. [7] applied transfer learning to adapt the LeNet5 network for rice blast disease recognition using a dataset provided by the Jiangsu Academy of Agricultural Sciences, achieving a top accuracy of 95.83%. Crucially, however, transfer learning does not fundamentally resolve the issue of insufficient data; models fine-tuned from pre-trained networks on very small datasets remain highly susceptible to overfitting [8].
Conventional data augmentation (CDA) offers another pathway. It artificially increases dataset size through geometric transformations (e.g., rotation, cropping, flipping) and photometric adjustments (e.g., contrast alteration) of original samples, followed by CNN-based recognition. While CDA can increase sample numbers and potentially enhance network adaptability and robustness to a degree, it primarily generates spatially or photometrically altered versions of the original pixels. It fails to synthesize truly novel, semantically relevant features, often resulting in samples with high similarity and low diversity. This limitation contributes to overfitting and inadequate generalization performance on unseen data.
To address the limitations of CDA in generating diverse and semantically rich data, researchers have explored using GANs for synthesizing crop disease samples [9]. GANs employ a generator-discriminator adversarial framework: the generator learns the distribution of the input data to produce new samples, while the discriminator tries to distinguish real from generated samples. GAN-generated images share the underlying data distribution of training samples but incorporate varied semantic information (e.g., altered backgrounds, textures, shapes), significantly enhancing dataset diversity. GANs have demonstrated success in diverse image generation tasks, including crop disease identification. Applications include Noise-to-Image GANs. Hu et al. utilized an improved Conditional Deep Convolutional GAN (C-DCGAN) to augment tea leaf images and combined it with a VGG-16 classifier, achieving 90% average accuracy [10]. Van Der Maaten et al. fused Deep Convolutional GAN (DCGAN) with GoogLeNet for tomato leaf disease recognition, reporting 94.33% average accuracy [11]. Their study also evaluated DCGAN outputs using t-distributed Stochastic Neighbor Embedding (t-SNE) and Turing tests, confirming the generated images possessed the same semantic information as real images.
Image-to-Image GANs is another important method. Tian et al. employed the cycle-consistent adversarial network (CycleGAN) to synthesize diseased apple leaf images from healthy ones and used YOLO (You Only Look Once) for tomato disease detection, attaining 95.57% detection accuracy [12]. Liu et al. proposed a Leaf GAN to generate novel grape leaf disease images for training an Xception model, which achieved a high average recognition accuracy of 98.70% [13].
Despite the promising applications of GANs for data augmentation, inherent challenges remain. GAN training is notoriously unstable and prone to mode collapse, which can drastically reduce the diversity and quality of generated samples. Tomato cultivation environments are complex, influenced by variable factors like lighting conditions, posing a significant challenge for small-sample disease identification against complex backgrounds. Furthermore, existing studies suggest that training deep convolutional neural networks (DCNNs) on segmented lesion images can effectively improve disease recognition accuracy [14,15]. This highlights the potential benefit of leveraging processed lesion images rather than or in conjunction with whole-leaf images for training DCNNs.
Motivated by the above challenges and opportunities, this paper proposes a novel tomato leaf disease recognition method integrating an enhanced generative adversarial network with an attention mechanism. During the image augmentation phase, we develop the ASPP-CycleGAN model, building upon CycleGAN by incorporating an atrous spatial pyramid pooling (ASPP) module. This integration strengthens the generator’s capacity to model multi-scale lesion features, thereby producing more diverse and detail-preserving synthetic disease images. In the recognition phase, we construct a lightweight CBAM-MobileNetV4 classification model. By embedding the convolutional block attention module (CBAM)—which combines channel and spatial attention—this model significantly enhances its focus on critical disease regions and improves its ability to discern fine-grained pathological features. The proposed framework demonstrates superior data augmentation effects and enhanced classification performance under small-sample conditions, offering a promising new solution for intelligent disease image recognition.
2. Materials and Methods
2.1. Data Source
The dataset utilized in this study is sourced from the publicly available PlantVillage dataset. The full dataset accessed on 1 September 2024, which can be accessed at https://github.com/spMohanty/PlantVillage-Dataset [16]. This comprehensive dataset contains 54,306 images spanning 14 plant species. The tomato leaf subset comprises images of 9 common diseases and healthy leaves, totaling 18,160 images. To simulate a small-sample learning scenario, we selected representative images from four prevalent tomato leaf diseases: bacterial spot, early blight, late blight, and mosaic virus disease, along with healthy leaves. For each class, 100 images were randomly selected. These were partitioned into a training set (70 images), a validation set (15 images), and a test set (15 images). All images were resized to a uniform resolution of 256 × 256 pixels.
The selected diseases exhibit distinct visual characteristics—bacterial spot: Typically manifests as small, water-soaked spots with irregular margins; early blight: Lesions appear as dark brown concentric rings; late blight: Characterized by water-soaked lesions that expand rapidly; mosaic virus disease: Presents as uneven leaf coloration with a mottled, chlorotic pattern; healthy leaves: Display uniform coloration without visible lesions. Various leaves are shown in Figure 1.

Figure 1.
Examples of tomato leaves from public datasets.
2.2. Image Data Augmentation Using Generative Adversarial Networks
Generative Adversarial Networks (GANs) represent an unsupervised approach to image data augmentation. By learning the underlying distribution of the original image data, GANs can synthesize novel images. Compared to conventional data augmentation techniques, GANs do not require predefined transformation rules (e.g., rotation, flipping). Crucially, they can generate images that do not exist in the original dataset, altering key features within the images and thereby significantly enhancing the diversity of samples in the augmented dataset.
2.2.1. CycleGAN Network Architecture and Principles
To enhance data diversity under small-sample conditions, we employed the Cycle-Consistent Adversarial Network (CycleGAN) [17] for disease image synthesis. CycleGAN operates in an unsupervised manner, learning mappings between two image domains (e.g., healthy leaves ↔ diseased leaves) without requiring paired examples. This enables style transfer between healthy and diseased leaf appearances.
CycleGAN comprises two generators and two discriminators (Dx, Dy). The adversarial loss function for mapping images from domain X to domain Y is defined as:
Similarly, the adversarial loss for mapping from domain Y to domain X is:
The generators are responsible for mapping images from the source domain to the target domain, while the discriminators aim to distinguish real images from generated ones. To ensure that generated images not only transfer style successfully but also maintain content consistency (e.g., leaf structure), CycleGAN introduces a cycle consistency loss. This loss enforces that an image translated from domain X to Y and back to X should closely resemble the original image (and vice versa), thereby enhancing the fidelity and structural integrity of the generated images. This architecture is particularly suitable for leaf disease images, which exhibit significant style differences (healthy vs. diseased appearance) while retaining fundamental structural elements. CycleGAN can effectively generate synthetic images featuring lesion characteristics based on original images, mitigating the challenges posed by limited data availability.
2.2.2. Generator Structure and ASPP Enhancement
To improve the CycleGAN generator’s capability in modeling multi-scale texture variations inherent in disease lesions, we integrated the ASPP module [18] into its architecture. The original generator consists of three main components: an encoder (downsampling), a transformer (residual blocks), and a decoder (upsampling). In this study, the ASPP module was embedded between the transformer and the decoder to perform multi-scale context feature aggregation, as illustrated in Figure 2.
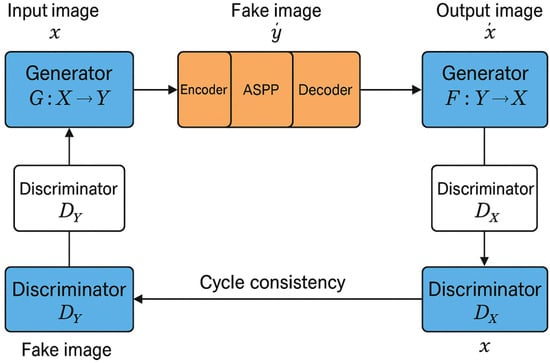
Figure 2.
CycleGAN combined with ASPP network structure diagram.
The ASPP module leverages multiple convolutional branches with different dilation rates to capture multi-scale contextual information without increasing the number of parameters [19]. It consists of four parallel branches: three 3 × 3 convolutions with distinct dilation rates (d1, d2, d3) and one global average pooling branch. Given an input feature map F, the output of the ASPP module is computed as:
where denote the dilation rates of the atrous convolutions (corresponding to r = 6, 12, 18 in Figure 3). denotes global average pooling applied to the input feature map F; denotes upscaling the result to the same spatial dimensions as the input feature map. The outputs from all branches are concatenated and then fused using a 1×1 convolution after appropriate dimension alignment. This process yields feature representations that effectively combine local details with global context [20]. This enhancement empowers the generator to better simulate the shape, size, and distribution patterns of various disease spots, thereby improving the realism and detail richness of the synthesized disease images. The ASPP network structure is depicted in Figure 3.
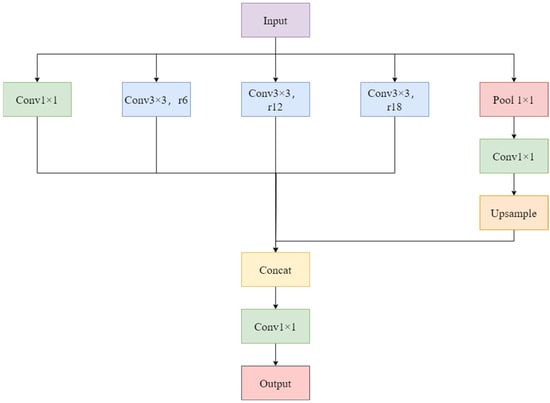
Figure 3.
ASPP network framework diagram.
2.3. MobileNetV4 Network Architecture and Enhancement
MobileNetV4, introduced by Google in 2024, represents the latest generation of lightweight convolutional neural networks [21]. Building upon the MobileNetV1–V3 series, its architecture is further optimized for both model compactness and inference efficiency. It integrates several key technologies, including Depthwise Separable Convolution [22], Inverted Residual Blocks, the Swish activation function, Squeeze-and-Excitation (SE) modules, and Hardware-Aware Neural Architecture Search (NAS), making it highly suitable for mobile deployment and edge computing scenarios.
The fundamental building block of MobileNetV4 is the Bottleneck module. Each module typically consists of three stages: a 1 × 1 convolution for channel expansion, a 3 × 3 depthwise separable convolution, and a 1 × 1 convolution for channel reduction. Skip connections form the inverted residual structure [23]. Lightweight attention mechanisms, such as SE modules, are embedded within some bottleneck blocks to enhance channel-wise feature modeling [24]. The overall network employs a hybrid design incorporating width diversity and non-linear activation functions, leading to improved performance in tasks like image classification and object detection.
Despite its significant advantages in model compression and computational efficiency, MobileNetV4 exhibits limitations in extracting fine-grained local features crucial for disease recognition tasks. Plant disease lesions are often small, irregularly distributed, and susceptible to interference from factors like varying illumination and occlusion, which can lead to misclassification. To address this, we incorporated the Convolutional Block Attention Module (CBAM) [25] into the MobileNetV4 backbone network, aiming to further enhance its discriminative power and localized focus capability. The network framework diagram is shown in Figure 4.
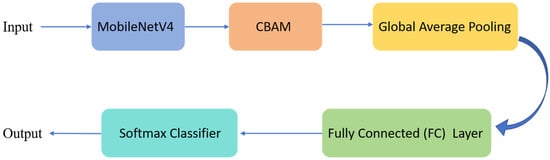
Figure 4.
MobileNetV4 Integrated with CBAM Network Overall Architecture Diagram.
2.4. Integration of CBAM Attention Mechanism
The Convolutional Block Attention Module (CBAM) is a lightweight attention mechanism comprising a Channel Attention Module (CAM) and a Spatial Attention Module (SAM) connected sequentially. CBAM enhances the model’s responsiveness to “what is important” (channels) and “where is important” (spatial locations) during feature extraction [26], as shown in Figure 5.
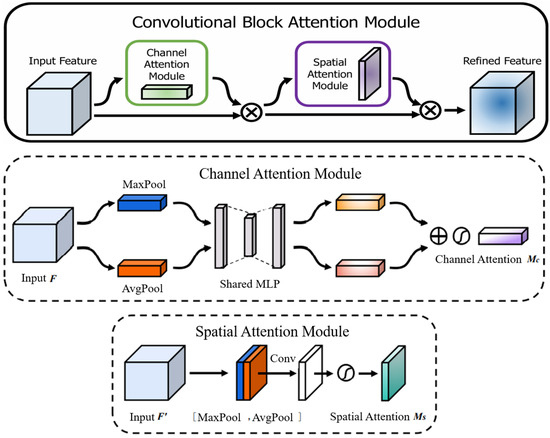
Figure 5.
CBAM attention mechanism structure diagram [25].
Channel Attention Module (CAM): This module first extracts channel-wise statistics using both global average pooling and global max pooling. These statistics are then processed by a shared Multi-Layer Perceptron (MLP) to model inter-channel dependencies. Finally, a Sigmoid activation function generates channel attention weights, amplifying the response of significant feature channels. The computation is expressed as:
where denotes the Sigmoid function, F is the input feature map, AvgPool is global average pooling, MaxPool is global max pooling, and MLP represents the shared multi-layer perceptron.
The intermediate output after applying channel attention is computed as:
Spatial Attention Module (SAM): This module computes the average and maximum values across the channel dimension for each spatial location in the feature map. The resulting two feature maps are concatenated and processed by a single 7 × 7 convolution to generate a spatial attention map, highlighting the most discriminative spatial regions. The computation is expressed as:
where denotes a 7 × 7 convolution operation, [ ; ] represents concatenation along the channel dimension, and is the Sigmoid function.
The final output after applying spatial attention is computed as:
In crop disease recognition tasks, lesion regions are typically localized, exhibit subtle features, and are often obscured by complex backgrounds. Integrating CBAM effectively guides the model to focus on disease-specific regions while suppressing irrelevant background information, thereby enhancing discriminative power and robustness under challenging conditions [27,28]. Due to its low computational overhead, CBAM is well-suited for lightweight architectures like MobileNetV4, offering an efficient means to boost its representational capacity.
In terms of network integration, the CBAM is inserted after each Bottleneck block output, acting as a supplementary attention mechanism along the residual pathway. CBAM sequentially applies CAM (enhancing “what” to focus on) and SAM (locating “where to focus on), achieving joint optimization. This design not only significantly improves the model’s perception of disease regions but also preserves the high efficiency of MobileNetV4 for deployment on edge devices.
3. Experimental Setup and Results Analysis
3.1. Experimental Environment and Parameter Settings
All model training and testing were conducted on a local computing platform. The hardware configuration comprised an Intel Core i9-14900HX CPU (Intel, Santa Clara, CA, USA), 32 GB of RAM, an NVIDIA GeForce RTX 4060 GPU (NVIDIA, Santa Clara, CA, USA) with 8 GB of VRAM, and 1 TB of HDD/SSD storage, running under the Windows 11 operating system. The software environment included Python 3.10, the PyTorch version 2.3.0 deep learning framework, and CUDA version 12.6 to leverage GPU acceleration.
During model training, the hyperparameters were set as follows: the number of training epochs was 200, the batch size was 16, and the initial learning rate was 0.0001. Both the generative adversarial networks (CycleGAN and ASPP-CycleGAN) and the disease classification model (CBAM-MobileNetV4) were trained and evaluated under this identical configuration to ensure the fairness and reproducibility of the experimental results.
3.2. Evaluation of Image Generation Quality
To quantitatively assess the quality of the images generated by the data augmentation methods, two widely adopted metrics were employed: Fréchet Inception Distance (FID) [29] and Kernel Inception Distance (KID) [30]. These metrics were used to compare the augmented images produced by the original CycleGAN and the proposed ASPP-CycleGAN.
Fréchet Inception Distance (FID): FID measures the dissimilarity between the distributions of real images and generated images within the feature space of an Inception-v3 model pre-trained on ImageNet. A lower FID score indicates greater similarity and higher perceptual quality of the generated images relative to the real ones. Formally, FID is calculated as:
where and are the mean feature vectors of the real and generated image sets, respectively, and and are their covariance matrices. The optimal FID score is 0.0, signifying identical image distributions.
Kernel Inception Distance (KID): KID provides an unbiased estimate of the Maximum Mean Discrepancy (MMD) using a polynomial kernel (typically cubic) within the Inception feature space. Like FID, a lower KID score signifies better image quality. KID is considered more robust to some biases and aligns well with human perception.
As shown in the corresponding Table 1, ASPP-CycleGAN achieved significantly lower FID and KID scores across all disease classes compared to the original CycleGAN. These results demonstrate that incorporating the ASPP module effectively enhanced both the realism and diversity of the synthetic images, improving the generator’s ability to model lesions across multiple scales.

Table 1.
Comparison of FID/KID scores for four types of disease images using different methods.
3.3. Disease Recognition Accuracy Experiment
To evaluate the impact of different data augmentation strategies on classification performance, three distinct training datasets were constructed: (1) the Original Dataset (baseline), (2) a CycleGAN-Augmented Dataset (augmented using the original CycleGAN), and (3) an ASPP-CycleGAN-Augmented Dataset (augmented using the proposed ASPP-CycleGAN). After applying ASPP-CycleGAN, we expanded the dataset by generating 70 additional images per class. As a result, the final training set contained 140 images per class, while the validation and test sets remained unchanged. This ensured that only the training data were augmented and that the evaluation was conducted on unseen original images. The dataset section is shown in Figure 6.
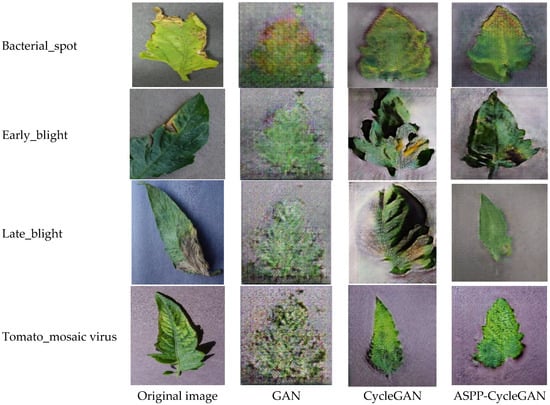
Figure 6.
Comparison of Data Visualizations Generated by Different Algorithms.
The CBAM-MobileNetV4 classification model was trained separately on each of these datasets. Model performance was evaluated using standard classification metrics:
The model trained on the ASPP-CycleGAN-Augmented Dataset exhibited substantially superior overall performance compared to models trained on the other datasets. Specifically, it achieved an average recognition accuracy nearly 5 percentage points higher than the model trained only on the original data. This significant improvement underscores the stronger discriminative power and generalization capability imparted by the images synthesized using ASPP-CycleGAN. The comparison results of the effectiveness between different Generative Adversarial Networks (GANs) and CBAM-MobileNetV4 models are shown in Table 2.

Table 2.
Comparison of the effectiveness of different GAN and CBAM-MobileNetV4 models.
3.4. Performance Comparison of Different Deep Learning Models
To further validate the effectiveness of the proposed CBAM-MobileNetV4 classifier for disease recognition, it was compared against four established image classification networks: VGG16, ResNet50, MobileNetV3, and the original MobileNetV4 (without CBAM). For a fair comparison, all models were trained and tested using the same ASPP-CycleGAN-Augmented Dataset. Their performance was evaluated on the task of recognizing five classes: four tomato leaf diseases (Bacterial spot, Early blight, Late blight, Mosaic virus disease) and Healthy leaves.
As detailed in Table 3, Table 4 and Table 5, CBAM-MobileNetV4 consistently outperformed all competing models. It achieved average Precision, Recall, and F1-Scores exceeding 97% across all four disease classes. The superior performance of CBAM-MobileNetV4 relative to VGG16, ResNet50, MobileNetV3, and the original MobileNetV4 can be primarily attributed to the integrated CBAM attention mechanism. CBAM effectively directs the model’s focus towards salient lesion regions while suppressing irrelevant background noise. This enhanced localization capability significantly boosts the model’s accuracy and robustness. Furthermore, the inherent multi-scale feature extraction capability of the model contributes to capturing richer and more diverse pathological features. The comparison chart of the results of the confusion matrices generated by the five different algorithms is shown in Figure 7.

Table 3.
Comparison of the accuracy of disease recognition using different deep learning models on expanded data.

Table 4.
Comparison of the recall rate of disease recognition using different deep learning models on expanded data.

Table 5.
Comparison of the F1 score of disease recognition using different deep learning models on expanded data.
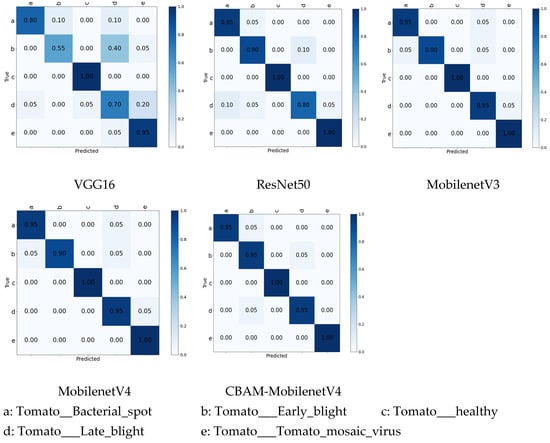
Figure 7.
Confusion matrix results for different algorithms.
4. Discussion
This study proposes a novel method that integrates enhancements at both the data level and the model level. At the data level, ASPP-CycleGAN substantially enriched the training set. Starting from only 70 original images per class, we generated an additional 400 synthetic images per class, increasing the dataset size to 140 images per class. This fivefold expansion significantly alleviated overfitting and improved model generalization. Compared with the original CycleGAN, ASPP-CycleGAN achieved consistently lower FID and KID scores. For instance, in bacterial spot disease, the FID decreased from 356.68 (CycleGAN) to 345.15 (ASPP-CycleGAN), and the KID decreased from 0.3349 to 0.3204. Similar improvements were observed across all disease types (Table 1). These results indicate that the proposed ASPP module improves the diversity and realism of generated samples, thereby enhancing the quality of data augmentation.
At the model level, CBAM-MobileNetV4 achieved an average accuracy of 97.10%, exceeding the standard MobileNetV4 (95.24%) by +1.86%, and outperforming classical models such as VGG16 (81.83%) and ResNet50 (92.92%) (Table 3, Table 4 and Table 5). Precision, recall, and F1-scores consistently improved across all categories; for example, in bacterial spot disease, CBAM-MobileNetV4 achieved 100% precision, 95% recall, and 100% F1, compared to MobileNetV4’s 90.48% precision, 95% recall, and 100% F1. Importantly, although the CBAM block slightly increases the number of parameters compared with MobileNetV4, the trade-off is justified by the observed performance gain. The model remains lightweight and feasible for deployment on edge devices.
Taken together, these results highlight the synergistic advantage of the “augmentation–recognition” framework: ASPP-CycleGAN mitigates the small-sample problem by expanding intra-class diversity, while CBAM-MobileNetV4 effectively exploits these enriched samples by focusing on critical lesion regions. Compared with recent tomato disease recognition studies, our approach achieves state-of-the-art accuracy under a small-sample setting, confirming its potential for practical agricultural applications.
5. Conclusions
This study addressed the challenge of tomato disease recognition under small-sample conditions by proposing a two-level strategy. At the data level, ASPP-CycleGAN improved the quality and diversity of synthetic images, enabling a fivefold increase in training samples per class (from 70 to 140). At the model level, CBAM-MobileNetV4 combined lightweight design with attention mechanisms, boosting recognition accuracy to 97.10% while maintaining low computational complexity.
The findings demonstrate that generative augmentation can effectively alleviate data scarcity, and that lightweight models enhanced with attention are well-suited for practical deployment in intelligent agriculture.
For future work, we will extend our framework in three directions:
- (1)
- Real-world validation: Test the approach on tomato images collected from farms under natural growing conditions, where small-sample problems are more severe. A stepwise training strategy—first pretraining on public datasets, then fine-tuning on farm-specific images—will be explored.
- (2)
- Cross-crop generalization: Apply the framework to other crops (e.g., cucumbers, peppers, rice) to verify its universality.
- (3)
- Extreme small-sample scenarios: Investigate optimization strategies under one-shot or few-shot conditions to further reduce data dependency and enhance robustness.
Author Contributions
Conceptualization, T.J. and Y.J.; methodology, Y.J.; software, T.J.; validation, T.J. and Y.J.; formal analysis, T.J.; investigation, Y.J.; resources, J.W.; data curation, Y.J.; writing—original draft preparation, T.J.; writing—review and editing, Y.J.; visualization, T.J.; supervision, J.W. and C.S.; project administration, C.S.; funding acquisition, Y.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research is supported by Central University Projects (2572022BH03).
Data Availability Statement
The data the conclusions of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Liu, W.; Zhai, Y.; Xia, Y. Tomato leaf disease identification method based on improved YOLOX. Agronomy 2023, 13, 1455. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, Y.; Jiang, P.; Peng, J.; Chen, H. IBSA-Net: A network for tomato leaf disease identification based on transfer learning with small samples. Appl. Sci. 2023, 13, 4348. [Google Scholar] [CrossRef]
- Xie, S.; Wang, C.; Wang, C.; Lin, Y.; Dong, X. Online identification method of tea diseases in complex natural environments. IEEEO Pen J. Comput. Soc. 2023, 4, 62–71. [Google Scholar] [CrossRef]
- Chakraborty, K.K.; Mukherjee, R.; Chakroborty, C.; Bora, K. Automated recognition of optical image based potato leaf blight diseases using deep learning. Physiol. Mol. Plant Pathol. 2022, 117, 101781. [Google Scholar] [CrossRef]
- Lu, Z.; Reitschuster, S.; Tobie, T.; Stahl, K.; Liu, H.; Hu, X. Contact fatigue life prediction of PEEK gears based on CTAB-GAN data augmentation. Eng. Fract. Mech. 2024, 312, 110639. [Google Scholar] [CrossRef]
- Kugelman, J.; Alonso-Caneiro, D.; Read, S.A.; Vincent, S.J.; Collins, M.J. Enhancing OCT patch-based segmentation with improved GAN data augmentation and semi-supervised learning. Neural Comput. Appl. 2024, 36, 18087–18105. [Google Scholar] [CrossRef]
- Astolfi, P.; Casanova, A.; Verbeek, J.; Vincent, P.; Romero-Soriano, A.; Drozdzal, M. Instance-conditioned gan data augmentation for representation learning. arXiv 2023, arXiv:2303.09677. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, Z.; Luo, X.; Li, P.; Min, G.; Li, H. SlaugFL: Efficient edge federated learning with selective GAN-based data augmentation. IEEE Trans. Mob. Comput. 2024, 23, 11191–11208. [Google Scholar] [CrossRef]
- Cap, Q.H.; Uga, H.; Kagiwada, S.; Iyatomi, H. LeafGAN: An effective data augmentation method for practical plant disease diagnosis. IEEE Trans. Autom. Sci. Eng. 2022, 19, 1258–1267. [Google Scholar] [CrossRef]
- Hu, G.; Wu, H.; Zhang, Y.; Wan, M. A low shot learning method for tea leaf's disease identification. Comput. Electron. Agric. 2019, 163, 104852. [Google Scholar] [CrossRef]
- Van Der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2023, 9, 2579–2605. [Google Scholar]
- Tian, Y.; Yang, G.; Wang, Z.; Li, E.; Liang, Z. Detection of apple lesions in orchards based on deep learning methods of cycle-GAN and yolov3 -dense. J. Sens. 2019, 2019, 7630926. [Google Scholar] [CrossRef]
- Liu, B.; Tan, C.; Li, S.; He, J.; Wang, H. A data augmentation method based on generative adversarial networks for grape leaf disease identification. IEEE Access 2020, 8, 102188–102198. [Google Scholar] [CrossRef]
- Sharma, P.; Berwal, Y.P.S.; Ghai, W. Performance analysis of deep learning CNN models for disease detection in plants us- ing image segmentation. Inf. Process. Agric. 2020, 7, 566–574. [Google Scholar]
- Ma, J.; Du, K.; Zheng, F.; Zhang, L.; Gong, Z.; Sun, Z. A recognition method for cucumber diseases using leaf symptom images based on deep con- culture. Comput. Electron. Agric. 2018, 154, 18–24. [Google Scholar] [CrossRef]
- Hughes, D.; Salathé, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv 2015, arXiv:1511.08060. [Google Scholar]
- Chu, C.; Zhmoginov, A.; Sandler, M. Cyclegan, a master of steganography. arXiv 2017, arXiv:1712.02950. [Google Scholar] [CrossRef]
- Chen, L.-C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef]
- Nawaz, M.; Nazir, T.; Javed, A.; Masood, M.; Rashid, J.; Kim, J.; Hussain, A. A robust deep learning approach for tomato plant leaf disease localization and classification. Sci. Rep. 2022, 12, 18568. [Google Scholar] [CrossRef]
- Tian, K.; Zeng, J.; Song, T.; Li, Z.; Evans, A.; Li, J. Tomato leaf diseases recognition based on deep convolutional neural networks. J. Agric. Eng. 2023, 54. [Google Scholar] [CrossRef]
- Qin, D.; Leichner, C.; Delakis, M.; Fornoni, M.; Luo, S.; Yang, F.; Wang, W.; Banbury, C.; Ye, C.; Akin, B.; et al. MobileNetV4, universal models for the mobile ecosystem. In European Conference on Computer Vision; Springer Nature: Cham, Switzerland, 2024; pp. 78–96. [Google Scholar]
- Wang, X.; Liu, J.; Zhu, X. Early real-time detection algorithm of tomato diseases and pests in the natural environment. Plant Methods 2021, 17, 43. [Google Scholar] [CrossRef]
- Djimeli-Tsajio, A.B.; Thierry, N.; Jean-Pierre, L.T.; Kapche, T.F.; Nagabhushan, P. Improved detection and identification approach in tomato leaf disease using transformation and combination of transfer learning features. J. Plant Dis. Prot. 2022, 129, 665–674. [Google Scholar] [CrossRef]
- Thangaraj, R.; Anandamurugan, S.; Pandiyan, P.; Kaliappan, V.K. Artificial intelligence in tomato leaf disease detection: A comprehensive review and discussion. J. Plant Dis. Prot. 2022, 129, 469–488. [Google Scholar] [CrossRef]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Hoque, M.D.J.; Islam, M.S.; Khaliluzzaman, M. AI-Powered Precision in Diagnosing Tomato Leaf Diseases. Complexity 2025, 2025, 7838841. [Google Scholar] [CrossRef]
- Cheng, H.H.; Dai, Y.L.; Lin, Y.; Hsu, H.C.; Lin, C.P.; Huang, J.H.; Chen, S.F.; Kuo, Y.F. Identifying tomato leaf diseases under real field conditions using convolutional neural networks and a chatbot. Comput. Electron. Agric. 2022, 202, 107365. [Google Scholar] [CrossRef]
- Kaur, P.; Harnal, S.; Gautam, V.; Singh, M.P.; Singh, S.P. An approach for characterization of infected area in tomato leaf disease based on deep learning and object detection technique. Eng. Appl. Artif. Intell. 2022, 115, 105210. [Google Scholar] [CrossRef]
- Heusel, M.; Ramsauer, H.; Unterthiner, T.; Nessler, B.; Hochreiter, S. Gans trained by a two time-scale update rule converge to a local nash equilibrium. arXiv 2017, arXiv:1706.08500. [Google Scholar]
- Bińkowski, M.; Sutherland, D.J.; Arbel, M.; Gretton, A. Demystifying mmd gans. arXiv 2018, arXiv:1801.01401. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Howard, A.; Sandler, M.; Chu, G.; Chen, L.C.; Chen, B.; Tan, M.; Wang, W.; Zhu, Y.; Pang, R.; Vasudevan, V.; et al. Searching for mobilenetv3. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 1314–1324. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).