A Knowledge-Based Bidirectional Encoder Representation from Transformers to Predict a Paratope Position from a B Cell Receptor’s Amino Acid Sequence Alone
Abstract
1. Introduction
2. Methods
2.1. Data
2.2. Architecture
2.3. Analysis
- Model 1 (No Oversampling Common Knowledge Context): This model served as the baseline model. This baseline model used the original data with no oversampling. The original 20,679 cases were split into training and test sets with a 50:50 ratio (10,356 vs. 10,323 cases). The training and test sets consisted of 9381:975 and 9343:980 cases for labels 0:1, respectively. The knowledge contexts of this baseline model were common among all amino acids within each BCR chain as shown in Figure 1.
- Model 2 (No Oversampling Different Knowledge Context): This model was similar to Model 1 (baseline model) given that this model employed the original data with no oversampling. The original 20,679 cases were divided into training and test sets with a 50:50 ratio (10,356 vs. 10,323 cases). The training and test sets included 9381:975 and 9343:980 cases for labels 0:1, correspondingly. However, the knowledge contexts of this model were different for different amino acids within each BCR chain as presented in Figure 1.
- Model 3 (Weak Oversampling Different Knowledge Context): The original 18,724:1955 cases for labels 0:1 were oversampled with 18,724:3165 for labels 0:1. Weak oversampling was conducted, i.e., 1210 positive cases were created based on the Synthetic Minority Oversampling Technique. Then, these 21,889 cases were split into training and test sets with a 50:50 ratio (10,961 vs. 10,928 cases). The training and test sets consisted of 9381:1580 and 9343:1585 cases for labels 0:1, respectively. The knowledge contexts were different for different amino acids within each BCR chain.
- Model 4 (Strong Oversampling Different Knowledge Context): The original 18,724:1955 cases for labels 0:1 were oversampled with 18,724:14,055 for labels 0:1. Strong oversampling was performed, that is, 12,100 positive cases were created based on the Synthetic Minority Oversampling Technique. Then, these 32,779 cases were divided into training and test sets with a 50:50 ratio (16,406 vs. 16,373 cases). The training and test sets included 9381:7025 and 9343:7030 cases for labels 0:1, correspondingly. The knowledge contexts were different for different amino acids within each BCR chain.
3. Results
3.1. Background Information
3.2. Model Performance
4. Discussion
4.1. Summary
4.2. Contributions
4.3. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hardy, R.R.; Hayakawa, K. B cell development pathways. Annu. Rev. Immunol. 2001, 19, 595–621. [Google Scholar] [CrossRef]
- Küppers, R. Mechanisms of B-cell lymphoma pathogenesis. Nat. Rev. Cancer 2005, 5, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Brezski, R.J.; Monroe, J.G. B-cell receptor. Adv. Exp. Med. Biol. 2008, 640, 12–21. [Google Scholar] [PubMed]
- Treanor, B. B-cell receptor: From resting state to activate. Immunology 2012, 136, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Hua, Z.; Hou, B. TLR signaling in B-cell development and activation. Cell. Mol. Immunol. 2013, 10, 103–106. [Google Scholar] [CrossRef]
- Rawlings, D.J.; Metzler, G.; Wray-Dutra, M.; Jackson, S.W. Altered B cell signalling in autoimmunity. Nat. Rev. Immunol. 2017, 17, 421–436. [Google Scholar] [CrossRef]
- Burger, J.A.; Wiestner, A. Targeting B cell receptor signalling in cancer: Preclinical and clinical advances. Nat. Rev. Cancer 2018, 18, 148–167. [Google Scholar] [CrossRef]
- Tanaka, S.; Baba, Y. B cell receptor signaling. Adv. Exp. Med. Biol. 2020, 1254, 23–36. [Google Scholar]
- Tkachenko, A.; Kupcova, K.; Havranek, O. B-cell receptor signaling and beyond: The role of Igalpha (CD79a)/Igbeta (CD79b) in normal and malignant B cells. Int. J. Mol. Sci. 2023, 25, 10. [Google Scholar] [CrossRef]
- Yuuki, H.; Itamiya, T.; Nagafuchi, Y.; Ota, M.; Fujio, K. B cell receptor repertoire abnormalities in autoimmune disease. Front. Immunol. 2024, 15, 1326823. [Google Scholar] [CrossRef]
- Lee, K.S.; Kim, E.S. Generative artificial intelligence in the early diagnosis of gastrointestinal disease. Appl. Sci. 2024, 14, 11219. [Google Scholar] [CrossRef]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial networks. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention is all you need. arXiv 2017, arXiv:1706.03762. [Google Scholar]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. BERT, Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- OpenAI. GPT-4 technical report. arXiv 2023, arXiv:2303.08774. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16 x 16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Dewaker, V.; Morya, V.K.; Kim, Y.H.; Park, S.T.; Kim, H.S.; Koh, Y.H. Revolutionizing oncology: The role of Artificial Intelligence (AI) as an antibody design, and optimization tools. Biomark. Res. 2025, 13, 52. [Google Scholar] [CrossRef] [PubMed]
- Ruffolo, J.A.; Gray, J.J.; Sulam, J. Deciphering antibody affinity maturation with language models and weakly supervised learning. arXiv 2021, arXiv:2112.07782. [Google Scholar] [CrossRef]
- Kenlay, H.; Dreyer, F.A.; Kovaltsuk, A.; Miketa, D.; Pires, D.; Deane, C.M. Large scale paired antibody language models. arXiv 2024, arXiv:2403.17889. [Google Scholar] [CrossRef] [PubMed]
- Melnyk, I.; Chenthamarakshan, V.; Chen, P.Y.; Das, P.; Dhurandhar, A.; Padhi, I.; Das, D. Reprogramming pretrained language models for antibody sequence infilling. Proc. Mach. Learn. Res. 2023, 202, 24398–24419. [Google Scholar]
- Hadsund, J.T.; Satława, T.; Janusz, B.; Shan, L.; Zhou, L.; Röttger, R.; Krawczyk, K. nanoBERT: A deep learning model for gene agnostic navigation of the nanobody mutational space. Bioinform. Adv. 2024, 4, vbae033. [Google Scholar] [CrossRef]
- Elnaggar, A.; Heinzinger, M.; Dallago, C.; Rehawi, G.; Wang, Y.; Jones, L.; Gibbs, T.; Feher, T.; Angerer, C.; Steinegger, M.; et al. ProtTrans: Towards cracking the language of life’s code through self-supervised learning. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 44, 7112–7127. [Google Scholar] [CrossRef]
- Leem, J.; Mitchell, L.S.; Farmery, J.H.R.; Barton, J.; Galson, J.D. Deciphering the language of antibodies using self-supervised learning. Patterns 2022, 3, 100513. [Google Scholar] [CrossRef]
- Greiff, V.; Weber, C.R.; Palme, J.; Bodenhofer, U.; Miho, E.; Menzel, U.; Reddy, S.T. Learning the high-dimensional immunogenomic features that predict public and private antibody repertoires. J. Immunol. 2017, 199, 2985–2997. [Google Scholar] [CrossRef] [PubMed]
- Petroni, F.; Rocktäschel, T.; Lewis, P.; Bakhtin, A. Language models as knowledge bases? arXiv 2019, arXiv:1909.01066. [Google Scholar] [CrossRef]
- Safavi, T.; Koutra, D. Relational world knowledge representation in contextual language models: A review. arXiv 2021, arXiv:2104.05837. [Google Scholar] [CrossRef]
- Yu, W.; Iter, D.; Wang, S.; Xu, Y.; Ju, M.; Sanyal, S.; Zhu, C.; Zeng, M.; Jiang, M. Generate rather than retrieve: Large language models are strong context generators. arXiv 2022, arXiv:2209.10063. [Google Scholar]
- Singhal, K.; Azizi, S.; Tu, T.; Mahdavi, S.S.; Wei, J.; Chung, H.W.; Scales, N.; Tanwani, A.; Cole-Lewis, H.; Pfohl, S.; et al. Large language models encode clinical knowledge. Nature 2023, 620, 172–180. [Google Scholar] [CrossRef]
- Ju, T.; Sun, W.; Du, W.; Yuan, X.; Ren, Z.; Liu, G. How large language models encode context knowledge? A layer-wise probing study. arXiv 2024, arXiv:2402.16061. [Google Scholar] [CrossRef]
- Karim, M.R.; Islam, T.; Shajalal, M.; Beyan, O.; Lange, C.; Cochez, M.; Rebholz-Schuhmann, D.; Decker, S. Explainable AI for bioinformatics: Methods, tools and applications. Brief. Bioinform. 2023, 24, bbad236. [Google Scholar] [CrossRef]
- Vilhekar, R.S.; Rawekar, A. Artificial Intelligence in Genetics. Cureus 2024, 16, e52035. [Google Scholar] [CrossRef]
- Claverie, J.M.; Cedric Notredame, C. Bioinformatics for Dummies, 2nd ed.; Wiley: Indianapolis, IN, USA, 2007. [Google Scholar]
- Pevsner, J. Bioinformatics and Functional Genomics, 3rd ed.; Wiley: Oxford, UK, 2015. [Google Scholar]
- European Molecular Biology Laboratory-European Bioinformatics Institute. UniProt. Available online: https://www.uniprot.org/ (accessed on 1 June 2025).
- Weizmann Institute of Science. GeneCards. Available online: https://www.genecards.org/ (accessed on 1 June 2025).
- National Center for Biotechnology Information. GenBank. Available online: https://www.ncbi.nlm.nih.gov/genbank/about/ (accessed on 1 June 2025).
- Li, D.; Pucci, F.; Rooman, M. Prediction of paratope-epitope pairs using convolutional neural networks. Int. J. Mol. Sci. 2024, 25, 5434. [Google Scholar] [CrossRef]
- Silver, D.; Huang, A.; Maddison, C.J.; Guez, A.; Sifre, L.; van den Driessche, G.; Schrittwieser, J.; Antonoglou, I.; Panneershelvam, V.; Lanctot, M.; et al. Mastering the game of Go with deep neural networks and tree search. Nature 2016, 529, 484–489. [Google Scholar] [CrossRef]
- Hambly, B.; Xu, R.; Yang, H. Recent advances in reinforcement learning in finance. arXiv 2022, arXiv:2112.04553. [Google Scholar]
- Yu, C.; Liu, J.; Nemati, S. Reinforcement learning in healthcare: A survey. arXiv 2020, arXiv:1908.08796. [Google Scholar] [CrossRef]
- Dognin, P.; Padhi, I.; Melnyk, I.; Das, P. ReGen: Reinforcement learning for text and knowledge base generation using pretrained language models. In Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing, Punta Cana, Dominican Republic, 7–11 November 2021; pp. 1084–1099. [Google Scholar]
- Pang, J.C.; Yang, S.H.; Li, K.; Zhang, J.; Chen, X.H.; Tang, N.; Yu, Y. Knowledgeable agents by offline reinforcement learning from large language model rollouts. arXiv 2024, arXiv:2404.09248. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, Q.; Zeng, Z.; Ren, L.; Liu, L.; Peng, B.; Cheng, H.; He, X.; Wang, K.; Gao, J.; et al. Reinforcement learning for reasoning in large language models with one training example. arXiv 2025, arXiv:2504.20571. [Google Scholar] [CrossRef]

| Amino Acid | RNA Codons | DNA Codons |
|---|---|---|
| Ala A | GCU, GCC, GCA, GCG | GCT, GCC, GCA, GCG |
| Arg R | CGU, CGC, CGA, CGG; AGA, AGG | CGT, CGC, CGA, CGG; AGA, AGG |
| Asn N | AAU, AAC | AAT, AAC |
| Asp D | GAU, GAC | GAT, GAC |
| Asn/Asp B | AAU, AAC; GAU, GAC | AAT, AAC; GAT, GAC |
| Cys C | UGU, UGC | TGT, TGC |
| Gln Q | CAA, CAG | CAA, CAG |
| Glu E | GAA, GAG | GAA, GAG |
| Gln/Glu Z | CAA, CAG; GAA, GAG | CAA, CAG; GAA, GAG |
| Gly G | GGU, GGC, GGA, GGG | GGT, GGC, GGA, GGG |
| His H | CAU, CAC | CAT, CAC |
| Ile I | AUU, AUC, AUA | ATT, ATC, ATA |
| Leu L | CUU, CUC, CUA, CUG; UUA, UUG | CTT, CTC, CTA, CTG; TTA, TTG |
| Lys K | AAA, AAG | AAA, AAG |
| Met M | AUG | ATG |
| Phe F | UUU, UUC | TTT, TTC |
| Pro P | CCU, CCC, CCA, CCG | CCT, CCC, CCA, CCG |
| Ser S | UCU, UCC, UCA, UCG; AGU, AGC | TCT, TCC, TCA, TCG; AGT, AGC |
| Thr T | ACU, ACC, ACA, ACG | ACT, ACC, ACA, ACG |
| Trp W | UGG | TGG |
| Tyr Y | UAU, UAC | TAT, TAC |
| Val V | GUU, GUC, GUA, GUG | GTT, GTC, GTA, GTG |
| Start | AUG, CUG, UUG | ATG, TTG, GTG, CTG |
| Stop | UAA, UGA, UAG | TAA, TGA, TAG |
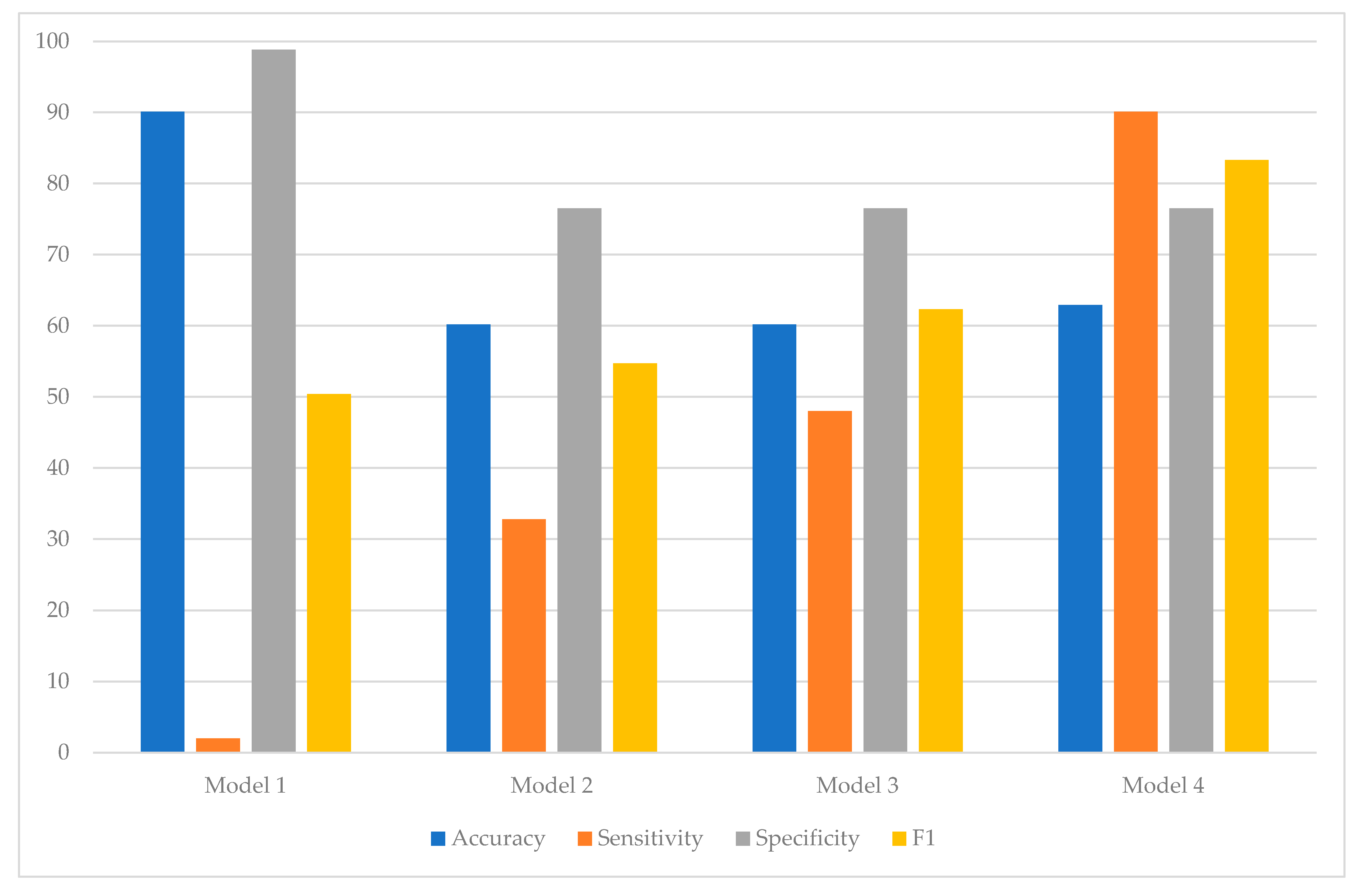
| Data | Metric | Model 1 | Model 2 | Model 3 | Model 4 |
|---|---|---|---|---|---|
| Training | Accuracy | 90.1 | 60.2 | 60.2 | 62.9 |
| Sensitivity | 2.0 | 32.8 | 48.0 | 90.1 | |
| Specificity | 98.8 | 76.5 | 76.5 | 76.5 | |
| F1 | 50.4 | 54.7 | 62.3 | 83.3 | |
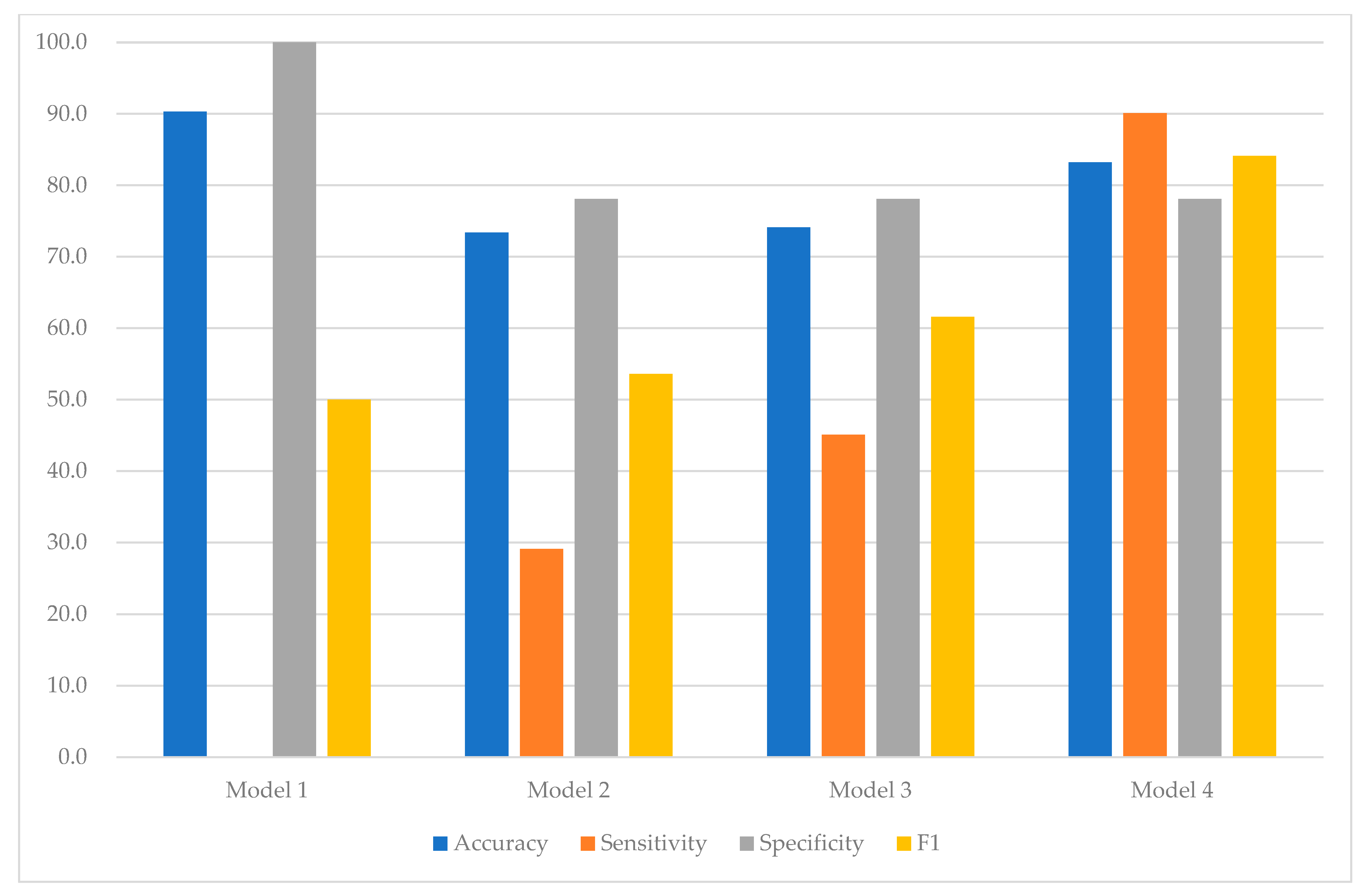
| Test | Accuracy | 90.3 | 73.4 | 74.1 | 83.2 |
| Sensitivity | 0.0 | 29.1 | 45.1 | 90.1 | |
| Specificity | 100.0 | 78.1 | 78.1 | 78.1 | |
| F1 | 50.0 | 53.6 | 61.6 | 84.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, H.; Lee, K.-S. A Knowledge-Based Bidirectional Encoder Representation from Transformers to Predict a Paratope Position from a B Cell Receptor’s Amino Acid Sequence Alone. Appl. Sci. 2025, 15, 10115. https://doi.org/10.3390/app151810115
Park H, Lee K-S. A Knowledge-Based Bidirectional Encoder Representation from Transformers to Predict a Paratope Position from a B Cell Receptor’s Amino Acid Sequence Alone. Applied Sciences. 2025; 15(18):10115. https://doi.org/10.3390/app151810115
Chicago/Turabian StylePark, Hyuntae, and Kwang-Sig Lee. 2025. "A Knowledge-Based Bidirectional Encoder Representation from Transformers to Predict a Paratope Position from a B Cell Receptor’s Amino Acid Sequence Alone" Applied Sciences 15, no. 18: 10115. https://doi.org/10.3390/app151810115
APA StylePark, H., & Lee, K.-S. (2025). A Knowledge-Based Bidirectional Encoder Representation from Transformers to Predict a Paratope Position from a B Cell Receptor’s Amino Acid Sequence Alone. Applied Sciences, 15(18), 10115. https://doi.org/10.3390/app151810115







