Abstract
Myocardial infarction (MI) remains a leading cause of in-hospital mortality. Early identification of high-risk patients is essential for improving clinical outcomes and optimizing hospital resource allocation. This study presents a machine learning framework for predicting mortality following MI using a publicly available dataset of 1700 patient records, and after excluding records with over 20 missing values and features with more than 300 missing entries, the final dataset included 1547 patients and 113 variables, categorized as binary, categorical, integer, or continuous. Missing values were addressed using denoising autoencoders for continuous features and variational autoencoders for the remaining data. In contrast, feature selection was performed using Random Forest, and PowerTransformer scaling was applied, addressing class imbalance by using SMOTE. Twelve models were evaluated, including Focal-Loss Neural Networks, TabNet, XGBoost, LightGBM, CatBoost, Random Forest, SVM, Logistic Regression, and a voting ensemble. Performance was assessed using multiple metrics, with SVM achieving the highest F1 score (0.6905), ROC-AUC (0.8970), and MCC (0.6464), while Random Forest yielded perfect precision and specificity. To assess generalizability, a subpopulation external validation was conducted by training on male patients and testing on female patients. XGBoost and CatBoost reached the highest ROC-AUC (0.90), while Focal-Loss Neural Network achieved the best MCC (0.53). Overall, the proposed framework outperformed previous studies in key metrics and maintained better performance under demographic shift, supporting its potential for clinical decision-making in post-MI care.
1. Introduction
Cardiovascular disease (CVD) remains the largest cause of death worldwide and accounts for nearly a third of all deaths everywhere in the world each year [1]. In 2021, there were 20.5 million CVD deaths, with acute myocardial infarction (AMI) and stroke accounting for approximately 85% of such deaths, using statistics provided by the World Health Organization [2]. In this respect, acute coronary syndromes (ACS), both ST-elevation and non-ST-elevation myocardial infarction, continue to be a heavy clinical and economic load. Post-AMI mortality in the hospital is over 5%, while long-term mortality is up to 26%, depending on the patient’s characteristics and comorbidities [3].
Myocardial infarction (MI) incidence is also considerable. A recent meta-analysis has reported worldwide prevalence rates of 3.8% in those less than 60 years and 9.5% in those aged more than 60, in which men are affected approximately five times more than women [4]. Despite improvements achieved in preventive therapy as well as in improved treatment protocols in respective populations, MI remains a major cause of death and a persistent challenge for healthcare systems worldwide [5].
Accurate risk stratification at the time of admission plays a critical role in clinical decision-making—guiding triage, therapeutic strategies, and ICU prioritization—as early mortality risk estimation has been shown to improve outcomes by enabling patient-specific interventions; tools like the ACTION-ICU score exemplify this approach by leveraging admission data to identify NSTEMI patients likely to require intensive care [3,6].
Traditional risk prediction models, including the Global Registry of Acute Coronary Events (GRACE) and Thrombolysis in Myocardial Infarction (TIMI) scores, have been utilized in clinical practice for several years. However, it is questionable whether they remain applicable since they were developed from older patient groups and with fewer clinical variables. Most of these tools were developed before the introduction of drug-eluting stents and newer antiplatelet therapies, thus raising questions about their relevance to contemporary practice [7]. In addition, reliance on linear or logistic regression methods restricts their ability to model complex, nonlinear relationships [3,8]. GRACE, while extensive, is not practical in the acute setting due to its complexity, and TIMI has been shown to have poor discrimination within intermediate-risk groups. Other models, such as Primary Angioplasty in Myocardial Infarction (PAMI), are no longer in use, and most of the available scores only consider short-term outcomes such as in-hospital or 30-day mortality [9].
The latest studies have attempted to incorporate computational approaches. For instance, Newaz et al. employed conventional data mining techniques to predict MI complications at an early stage and reported moderate results, with F1 values approaching 0.54 and a ROC-AUC below 0.77 [10]. Their study, however, involved limited preprocessing and naive imputation techniques. On the other hand, the present study suggests a more integrated model that includes autoencoder-based imputation, enhanced feature selection, data balancing with SMOTE and Focal Loss, and a larger set of modern classifiers. This led to significantly improved performance on all the evaluation measures.
Prior studies have used the same publicly available myocardial infarction dataset for related prediction tasks but employed limited preprocessing and imbalance handling. Diakou et al. [11] applied multi-label classification (e.g., One-vs-Rest with XGBoost) to predict complications, but without deep imputation, advanced normalization, or structured imbalance control. Joshi et al. [12] achieved high F1 scores using classical algorithms such as Ridge Classifier and SVC, yet relied only on median imputation, mutual information feature selection, and no balancing or normalization. Reddy and Thangam [13] predicted relapse risk using resampling methods such as SMOTE and SVM-SMOTE, but they lacked deep preprocessing and model-specific optimization.
Machine learning (ML) provides data-driven and adaptive alternatives to traditional models, enabling the capture of nonlinear and high-dimensional patterns without relying on predefined assumptions. These methods are particularly well-suited to heterogeneous clinical datasets and dynamically evolving treatment environments . Prior studies have demonstrated the predictive superiority of ML algorithms. For example, Lee et al. reported that models like Random Forest and XGBoost outperformed logistic regression in predicting AMI-related mortality, achieving higher AUC scores [7]. Apart from statistical performance, nowadays, ML also begins to show clinical applicability. ML models processing ECG data could identify occlusion MI in NSTEMI patients with higher sensitivity than cardiologists, making them suitable for applications for real-time diagnoses, as revealed from a publication in Nature Medicine [14].
In conclusion, data imputation, class imbalance, and an extensive comparison of advanced algorithms for predicting mortality among patients with myocardial infarction have been employed. The method is tested on the same publicly accessible dataset employed by Newaz et al. [10], thereby facilitating direct comparison and revealing significant improvements on core performance measures.
2. Materials and Methods
This materials and methods section outlines the dataset characteristics and machine learning models that have been employed in predicting in-hospital mortality from myocardial infarction. It lays emphasis on data cleaning, imputation techniques, feature engineering, and class imbalance correction. The paper then outlines the predictive models, hyperparameter tuning strategy, and performance metric adopted in gauging the difference between classifiers.
2.1. Data and Preprocessing
This study handled a dataset containing 1700 MI patients, with 123 clinical variables and a binary response of in-hospital mortality (0 = survival, 1 = death). The data consisted of several types of information, such as demographic data, clinical assessments, biochemical markers, and electrocardiographic (ECG) results. To support robust evaluation, the dataset was split into training (80%) and test (20%) sets using stratified sampling to preserve class balance. All preprocessing steps and SMOTE-based oversampling were confined to the training data within a 5-fold stratified cross-validation framework, ensuring that the test set remained entirely unseen until final evaluation. Missing values were processed using a combination of conventional statistical techniques (mean, median, mode) and deep learning-based techniques, including denoising autoencoders and variational autoencoders. Autoencoder models were demonstrated in earlier research to be better at imputing missing values in electronic health records (EHRs), with better accuracy and robustness than conventional techniques [15,16]. Notably, the AutoPopulus framework was recently proposed to evaluate the performance of autoencoders for large-scale clinical datasets [17], while variational autoencoders have proved especially useful in omics data for balancing computational efficiency and reconstruction accuracy [18]. Table 1 summarizes the dataset structure, types of variables, distribution of outcomes, degree of missingness, and imputation methods applied.

Table 1.
Dataset summary and outcome characteristics.
The preprocessing pipeline was designed to address key data quality issues, including missing values, measurement noise, redundancy, and class imbalance, by removing records with more than 20 missing fields and features with over 300 missing entries. Variables were categorized as binary, categorical, integer, or continuous based on their data type and clinical relevance, with numeric columns initially imputed using mean-based techniques and subsequently reconstructed through a denoising autoencoder to capture underlying patterns in the data. Before model imputation, binary and categorical variables were one-hot encoded to preserve structural integrity without information loss, while feature selection was performed using Random Forest-based importance ranking to identify the most informative variables. Remaining missing values were further imputed using a variational autoencoder, while skewed distributions of continuous variables were normalized with PowerTransformer to reduce skewness and approximate normality, thereby improving the convergence and stability of gradient-based learners and enhancing the reliability of distance-based metrics by ensuring comparability across feature scales [19,20]. Table 2 is a summary of the preprocessing steps, types of variables affected, and the techniques used.

Table 2.
Preprocessing pipeline overview.
A fully structured preprocessing architecture was established to enhance data integrity and model performance. The full machine learning pipeline is depicted in Figure 1.
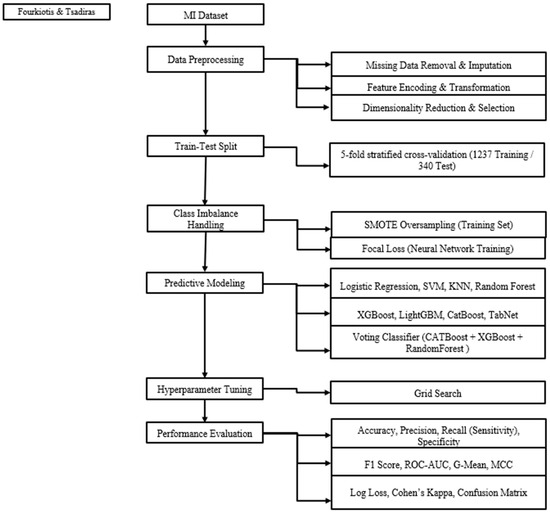
Figure 1.
Flowchart machine learning pipeline.
2.2. Handling Class Imbalance
The dataset exhibited substantial class imbalance, with deaths markedly underrepresented compared to survivals; to mitigate this, SMOTE was applied post-preprocessing to generate synthetic minority-class samples and enhance the model’s capacity to identify rare outcomes [21]. Alternative oversampling methods, including data augmentation with Generative Adversarial Networks (GANs) [22] and ensemble-based resampling via EasyEnsemble [23], were also evaluated but did not yield performance improvements under the given data conditions and were therefore excluded from the final modeling pipeline.
To further address class imbalance during training, Focal Loss was incorporated into the deep learning models, reducing the influence of well-classified majority-class instances while emphasizing harder, minority-class examples—thereby improving sensitivity without distorting the decision boundary [24].
2.3. Feature Scaling and Transformation
To enhance numerical stability and support effective model training, both continuous and integer variables underwent scaling utilizing PowerTransformer in conjunction with StandardScaler, after reconstruction through autoencoder-based imputation [25]. This method addressed skewness, minimized variance inconsistencies, and assisted in approximating normality within feature distributions. Normality approximation benefits several downstream algorithms. It improves the convergence of gradient-based learners. It preserves the validity of distance-based methods such as K-Nearest Neighbors and SVMs with RBF kernels, which assume isotropic feature distributions in Euclidean space [26]. Categorical and binary variables were imputed using the most frequent class and transformed through one-hot encoding. This processing approach guaranteed compatibility across various model architectures, maintained the integrity of discrete features, and mitigated distributional irregularities that could hinder training efficacy [27].
2.4. Dimensionality Reduction
Two complementary strategies were employed for dimensionality reduction. First, the SelectFromModel technique, utilizing a Random Forest estimator, was implemented to identify and preserve the most pertinent features post-preprocessing. Next, a Variational Autoencoder (VAE) was used to encode the data into nonlinear latent spaces, customized to suit the complexity and scale of high-dimensional biomedical inputs [28]. The integration of feature selection and representation learning played a crucial role in reducing redundancy and improving generalization in subsequent model training [18].
All machine learning and deep learning models were trained on the same reduced feature set, obtained via a SelectFromModel step using a Random Forest solely for generic feature ranking. Features with importance above the mean threshold were retained, ensuring a consistent, noise-reduced input space for all models. This model-agnostic selection prevented bias toward Random Forest, improved convergence stability, and allowed each classifier to be fine-tuned independently on the optimized features [29].
2.5. Predictive Modeling
A varied array of supervised learning algorithms was applied to estimate the risk of in-hospital mortality, encompassing linear models, ensemble techniques, and deep learning architecture. Logistic Regression was included as a clinical benchmark due to its interpretability and historical application in binary outcome modeling [30]. Support Vector Machines (SVM) utilizing a radial basis function kernel were employed to capture nonlinear class boundaries and evaluate separation under conditions of imbalance [31]. The K-Nearest Neighbors (KNN) algorithm acted as a non-parametric reference to assessing local structures within the transformed feature space [32].
Among ensemble techniques, Random Forest functioned both as a predictive model and as a means of evaluating feature importance, appreciated for its robustness and transparency [33]. Gradient boosting methods encompass XGBoost, recognized for its regularization and generalization strengths [34]; LightGBM, optimized for computational efficiency and handling sparse inputs [35]; and CatBoost, which adeptly manages categorical variables without necessitating extensive preprocessing [36]. To evaluate ensemble performance, a voting classifier was developed by consolidating predictions from the leading boosting models, with the goal of enhancing predictive stability and minimizing variance [37]. Deep learning models featured a fully connected neural network trained with Focal Loss to emphasize misclassified minority (death) instances and alleviate the impact of class imbalance during training [38,39]. Moreover, TabNet, a sequential attention-based architecture specifically designed for tabular data, was integrated to improve both performance and interpretability through dynamic feature selection [40]. Table 3 provides the algorithms used and their predictive roles.

Table 3.
Algorithms and predictive roles.
2.6. Hyperparameter Optimization
For model performance optimization, a grid search strategy was adopted for systematic evaluation of hyperparameter combinations for each classifier [41]. The aim was to identify configurations that support generalization without sacrificing computational efficiency. Hyperparameter optimization was performed through five-fold stratified cross-validation on the training set to allow confident model selection and avoid overfitting. Ranges of hyperparameters were determined from the existing literature and best practices in biomedical classification problems. For TabNet, hyperparameter tuning was over architectural parameters such as decision and attention dimensions (n_d, n_a), the number of decision steps (n_steps), and the gamma parameter for feature reuse control. For boosting methods (XGBoost, LightGBM, CatBoost), the key parameters were n_estimators and learning_rate. Conventional models, such as Logistic Regression and SVM, were tuned using regularization strength, solver, and kernel parameters. K-Nearest Neighbors experimented with varying values of k, while the voting ensemble (XGBoost, CatBoost, and Random Forest) was hyperparameter-tuned for improved prediction stability. The summary of the baseline hyperparameter values determined before the grid search is presented in Table 4.

Table 4.
Hyperparameter search space per model.
2.7. Subpopulation-Based External Validation
Machine learning algorithms must perform well on held-out samples from the same population but also generalize new, demographically or institutionally distinct cohorts. Testing on outside datasets is the gold standard test for this, by testing on datasets from other hospitals, regions, or periods. Such an external dataset did not exist in this study. Lacking other hospitals’ data, an external validation strategy has been designed based on subpopulation by the most recent best practices in the literature [42].
Specifically, in this paper, males have been used to train models and performance was tested on females, inducing a biologically and clinically significant distributional shift. This has two applications. First, it allows one to test the performance of models under demographic shift. Second, it allows one to test whether model decisions are over-optimized for specific subgroups or reflect more generalizable patterns in the data.
During this research, the process of validation was a systematic and reproducible one. The identical same preprocessing pipeline of the baseline modeling experiment was employed, involving the following steps: (i) missing value imputation by a combination of basic statistical estimates (mean/mode) and sophisticated variational autoencoder-based methods, (ii) one-hot encoding of categorical variables, (iii) denoising autoencoder transformation with subsequent PowerTransformer scaling of continuous variables, (iv) feature selection by embedded methods (Random Forest with SelectFromModel), and (v) class imbalance adjustment by SMOTE oversampling. Interestingly, the entire pipeline was fitted on the male group only (n = 974), without ever being exposed to any female samples during training. After fitting, the pipeline that had been trained was then applied to the female group (n = 573), with transformation operations only being involved, thus ensuring a strict separation of training and test groups and not allowing for any information leakage.
Following preprocessing, all models were retrained on the preprocessed male data without further hyperparameter tuning. Final testing was then performed on the unpreprocessed, individually transformed female test set, mirroring the case of an external test set from a demographically distinct subpopulation. Testing employed the same set of metrics listed in Section 3, i.e., F1-score, ROC-AUC, and MCC, to adhere to the internal validation paradigm. The performance of each model under this setup is summarized in Table 5, with the best-performing values highlighted in bold.

Table 5.
Performance of ML models under subpopulation external validation.
The result shows that most of the models had good performance, even though they were trained on male patients alone and evaluated on female patients. The gradient boosting models (XGBoost, LightGBM, and CatBoost) worked best across all models with ROC-AUC scores in the range of 0.90 and decent performances in both F1 and MCC. The neural network, also trained on Focal Loss, performed well, especially on MCC, showing it is capable of handling class imbalance effectively.
More surprisingly, even more basic models like Logistic Regression and Random Forest also worked well. They are, therefore, good alternatives when interpretability and speed are essential. Models like TabNet and K-Nearest Neighbors, however, did not work well. It may be because they needed bigger datasets or were very sensitive to feature structure. Although this study had data from only one hospital, testing models on a different population of patients (men vs. women) gave valuable information. It showed that many models could still make good predictions on a patient set they had not yet seen during training. This gives credibility to their capacity to generalize to new patients. These performance results are supplemented by those of Section 4, which confirm that models with acceptable internal measures also perform well on test sets that have been demographically modified. These results support the suggestions made by Rockenschaub et al. [43], who state that testing on other groups is a good way of performing external validation where access to other hospitals’ data is not possible.
3. Evaluation Metrics
Conventional metrics like accuracy frequently give a skewed picture of performance in clinical risk prediction tasks with a clear class imbalance, such as mortality modeling, where deaths are significantly lower than survivals. Even though accuracy is mentioned in more than half of the related studies [44], it does not represent the classifier’s capacity to recognize minority outcomes. A model that predicts all patients will survive, for example, may have a high accuracy rate but completely overlook fatal cases. As a result, when assessing unbalanced clinical data, accuracy by itself is not very useful.
Metrics that specifically measure minority-class performance are given priority in order to get around this restriction. As TP/(TP + FP), precision (positive predictive value) represents the percentage of predicted deaths that are accurate. The percentage of real deaths that the model correctly detects is known as recall (sensitivity), which is calculated as TP/(TP + FN) [45]. Recall is particularly important in clinical settings because neglecting high-risk patients can have dire repercussions. Recall, which is used to identify people who are ill or at high risk, was the most reported metric (~63%) in a review of medical AI models [46].
The trade-off between false positives and false negatives is balanced by the F1-score, which is the harmonic mean of precision and recall. In imbalanced scenarios, it provides a more informative evaluation than accuracy and is present in almost half of biomedical machine learning studies [47,48]. By measuring the accurate identification of non-fatal outcomes, specificity (TN/(TN + FP))—the true negative rate—enhances sensitivity [49]. Specificity is still crucial for assessing both outcome classes, even though it is not as stressful as recall in mortality studies.
Models that perform poorly in either class are penalized by some studies using composite indicators like the G-Mean, which is the geometric mean of sensitivity and specificity. Because it reflects performance symmetry without requiring a separate examination of recall and specificity, this metric is especially well-suited to imbalanced medical datasets [48].
The area under the Receiver Operating Characteristic curve (ROC-AUC) is commonly used to assess classifier performance independent of thresholds. It calculates the likelihood that a randomly chosen death will have a higher predicted risk than a randomly chosen survivor. Although ROC-AUC is frequently used in about half of predictive modeling studies, it can yield excessively optimistic results in datasets that are highly imbalanced, which could lead to a misleading interpretation [50].
For binary classification under imbalance, the Matthews Correlation Coefficient (MCC) is generally accepted as a fair and trustworthy metric. The confusion matrix’s four components, TP, TN, FP, and FN are all included, and it only yields high values when predictions are correct for both classes. MCC performs better than accuracy and F1-score in reflecting real-world reliability, according to comparative analyses, particularly in clinical datasets [51,52].
The confusion matrix, which records true positives, true negatives, false positives, and false negatives, is the source of all evaluation metrics. Direct examination of the confusion matrix, particularly in clinical applications, offers insight into model behavior beyond numerical summaries. It identifies misclassification trends, like unreported fatalities or inaccurate survivor labeling, which have an immediate impact on triage and care distribution. Therefore, in high-stakes healthcare settings, the confusion matrix is still crucial for assessing predictive systems [53]. An overview of each evaluation metric, along with its interpretation and applicability to this study, is provided in Table 6. Previous research on biomedical classification and medical AI performance evaluation informs interpretations and use cases [44,45,46,47,48,49,50,51,52,53].

Table 6.
Evaluation metrics used in imbalanced clinical risk prediction.
4. Results
To comprehensively assess the proposed machine learning framework for predicting in-hospital mortality after myocardial infarction, multiple classification algorithms were evaluated using the standardized preprocessing pipeline outlined in Section 2. Performance was examined under two scenarios, without SMOTE-based class imbalance correction and with SMOTE oversampling. Table 7 reports Accuracy, Precision, F1-score, ROC-AUC, MCC, Sensitivity, Specificity, and G-Mean for all models in the baseline (no SMOTE) configuration, with best-performing values highlighted in bold.

Table 7.
Comparative performance of machine learning models without SMOTE.
Without SMOTE, the Voting Classifier achieved the highest accuracy (0.9129), precision (0.9130), and MCC (0.6089), alongside near-perfect specificity (0.9924), indicating balanced performance and minimal false positives. Logistic Regression yielded the highest F1-score (0.6154), recall (0.5217), and G-Mean (0.7113), highlighting its effectiveness in identifying death cases despite class imbalance. LightGBM attained the top ROC-AUC (0.8929), confirming strong discriminative ability.
Table 8 reports model performance with SMOTE across all metrics. Due to the dataset’s class imbalance, most classifiers achieved high accuracy values. Following full preprocessing and hyperparameter tuning, each model was tested on a held-out set, with best scores highlighted in bold.

Table 8.
Comparative performance of machine learning models with SMOTE.
The Support Vector Machine achieved the most balanced performance, leading in F1-score (0.6905), ROC-AUC (0.8970), and MCC (0.6464), effectively managing false positive–false negative trade-offs. Random Forest reached perfect precision and specificity (1.0000) but had low recall (0.3913), reflecting a conservative approach that missed many true deaths. Logistic Regression attained the highest recall (0.6957) and G-Mean (0.7836), indicating superior sensitivity with acceptable specificity. These results underscore distinct classifier strengths in balancing sensitivity and specificity, both critical for clinically reliable mortality prediction.
To assess the effect of class imbalance correction, top-performing results with and without SMOTE were compared, revealing the potential benefits of synthetic oversampling for highly skewed clinical data. Table 9 presents the highest scores for each of the eight evaluation metrics alongside the corresponding best-performing model.

Table 9.
Comparative summary of the top-performing models across key evaluation metrics, with and without SMOTE oversampling.
As shown in Table 9, SMOTE consistently yielded the highest values across all evaluation metrics compared to the no-SMOTE configuration. It improved recall, F1-score, G-Mean, and in several cases, ROC-AUC. Performance gains were observed for most algorithms, underscoring the effectiveness of oversampling in enhancing minority-class detection in mortality prediction.
To enable a clinically relevant and transparent comparison between models trained with and without imbalance correction, a Composite Performance Index (CPI) was calculated for each setup [54]. The CPI, in Table 10, consolidates eight metrics into a weighted score reflecting the priorities of in-hospital mortality prediction, assigning the greatest weight to recall (0.25), followed by F1-score (0.20), MCC (0.20), ROC-AUC (0.15), and smaller weights to specificity (0.10) and G-Mean (0.10). Models are ranked in descending CPI, with bolded entries marking the top performer, allowing direct evaluation of SMOTE’s impact on predictive reliability.

Table 10.
CPI ranking of machine learning models with and without SMOTE.
Under SMOTE, SVM achieved the highest Composite Performance Index (0.7342), benefiting from the balanced class distribution to boost recall and overall discrimination while retaining high specificity. Without SMOTE, Logistic Regression ranked first (0.6688) due to its strong recall and G-Mean, essential for identifying high-risk patients. SMOTE improved CPI for most models, especially imbalance-sensitive ones like SVM and CatBoost, while tree-based ensembles such as Random Forest showed smaller gains, reflecting their inherent robustness. Overall, SMOTE enhanced the sensitivity–specificity trade-off, strengthening clinical applicability in skewed datasets.
To further assess model reliability, we calculated Type I (false positive) and Type II (false negative) error rates for both SMOTE and non-SMOTE experiments. In this context, Type I errors occur when low-risk patients are incorrectly classified as high risk, potentially leading to unnecessary interventions, while Type II errors occur when high-risk patients are incorrectly classified as low risk, posing a greater clinical threat due to missed opportunities for timely care [55,56]. Error rates were derived from the underlying confusion matrices used to derive the metrics in Table 7 and Table 8, with Type I defined as the proportion of false positives among actual negatives, Type II as the proportion of false negatives among actual positives, and total error as their sum. Since Type II errors are generally considered more critical in healthcare, comparing these rates provides a deeper understanding of model performance beyond aggregate accuracy or discrimination metrics.
Table 11 reports the Type I, Type II, and total error rates for all classifiers, ranked separately for SMOTE and non-SMOTE setups, with bold values indicating the lowest total error in each configuration.

Table 11.
Comparative Type I and Type II error rates with and without SMOTE.
While the CPI provides a weighted aggregate measure of model performance, Type I and Type II error rates offer a complementary view by isolating specific failure modes, particularly the clinically critical Type II errors associated with missed diagnoses of high-risk patients. As shown in Table 11, under SMOTE, SVM achieved the lowest total error rate (0.4037) through a balanced trade-off between the two error types, demonstrating strong reliability in both detecting high-risk patients and avoiding false alarms. Logistic Regression closely followed (0.4217), attaining the lowest Type II error among SMOTE models. In the absence of SMOTE, Logistic Regression ranked highest (0.5086), outperforming more complex algorithms, with XGBoost in second place (0.5227). The results indicate that synthetic oversampling particularly benefits algorithms sensitive to class imbalance, such as SVM and Logistic Regression, by reducing Type II errors without notably increasing Type I errors. Conversely, Random Forest maintained low Type I errors in both scenarios but exhibited higher Type II errors, reflecting a conservative decision boundary. These observations underscore the need to evaluate models not only through aggregate performance scores but also via clinically meaningful error types directly linked to patient outcomes.
To enhance the interpretability of the models, a post hoc feature importance analysis was performed using the Random Forest classifier. After training the models on the reduced feature set determined by the preprocessing pipeline, the fitted Random Forest model was used to extract impurity-based importance scores for each variable. The top 20 most influential features are shown in Figure 2. Myocardial rupture indicator RAZRIV was the most highly ranked variable, followed by serum potassium (R_AB_3_n) and sodium (NA_R_3_n) levels on day three, which are biomarkers known to affect post-infarction outcomes. The NOT_NA_3_n flag, indicating abnormal sodium levels, reinforced the clinical relevance of electrolyte disturbance in mortality risk. The ZSN_A angiographic stenosis score, reflecting residual coronary obstruction, was also ranked highly. Collectively, these features represent major clinical dimensions such as structural integrity, fluid–electrolyte balance, and ischemic burden. This post hoc analysis not only improved the interpretability of the pipeline but also highlighted clinically meaningful predictors that could inform risk stratification and decision support in real-world settings.
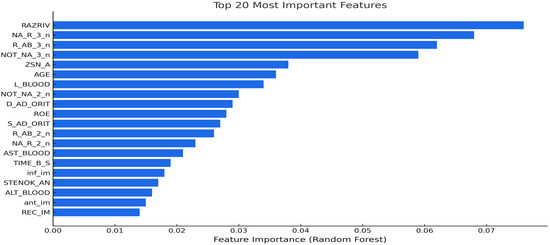
Figure 2.
Top 20 predictive features identified by Random Forest post hoc analysis.
Model performance was compared to the findings of Newaz et al. [10], who examined the same myocardial infarction dataset, to allow for direct comparison with earlier research. A more sophisticated preprocessing pipeline, including deep imputation, variance stabilizing transformations, class balancing through SMOTE, and an expanded model suite, was used in this work in contrast to the previous study. The top-performing models for each metric in the two studies are compared side by side in Table 12.

Table 12.
Comparative performance of this study model versus prior work.
The present approach performs greater than the prior results in most metrics, as indicated in Table 8. The SVM classifier produced better MCC (0.6464 vs. 0.6222), ROC-AUC (0.8970 vs. 0.7606), and F1-score (0.6905 vs. 0.5425). Additionally, this study’s G-Mean for Logistic Regression was higher (0.7836 vs. 0.7270), indicating better balance between the outcome classes. These improvements are ascribed to the more thorough preprocessing pipeline, specifically the application of SMOTE, variational autoencoders, and denoising autoencoders—all of which were not present in the previous framework. The results highlight how crucial sophisticated preprocessing and algorithm selection are to improving model performance for clinical risk stratification.
5. Discussion
This section covers the methodological advances of the proposed framework over earlier frameworks, its potential impact on intensive care and triage workflows, broader implications for healthcare economics and bioinformatics, and the study’s limitations and future research directions.
5.1. Methodological Advancements over Prior Frameworks
The novelty of this study lies in its focus on the clinically critical endpoint of in-hospital mortality and the integration of multiple methodological innovations into a single, reproducible pipeline. In contrast to previous work using the Myocardial Infarction Complications Database, the framework combines deep learning-based imputation (denoising autoencoders for continuous variables and variational autoencoders for categorical and binary variables), advanced normalization with the PowerTransformer, dual-level class imbalance handling through SMOTE oversampling and Focal Loss, and a heterogeneous ensemble incorporating both conventional and deep tabular learners such as TabNet. It further introduces a subpopulation-based external validation protocol, training on male patients and testing on female patients, to evaluate generalization under demographic shift, an assessment absent from earlier studies. Collectively, these innovations address methodological and clinical gaps, enhancing predictive reliability, interpretability, and applicability in intensive care and triage workflows.
Relative to prior studies using the same dataset [10,11,12,13], the present framework delivers substantive advances in preprocessing, imbalance handling, and evaluation design, while also achieving superior predictive performance. Diakou et al. addressed multi-label complication prediction using conventional multivariate imputation without normalization and evaluated models primarily via Hamming loss, whereas this study targets a single binary mortality endpoint, employing deep learning-based imputation and PowerTransformer normalization to improve numerical stability. Joshi et al. applied median imputation, mutual information-based feature selection, and limited evaluation to the F1-score, while our approach adopts model-agnostic Random Forest-based feature selection combined with a broader multi-metric evaluation (F1, ROC-AUC, MCC, Recall, Specificity, G-Mean) to capture clinically relevant trade-offs. Lokeswar Reddy K. & Thangam S. predicted relapse events using mean/median imputation, min–max scaling, and recursive feature elimination without any imbalance handling, whereas our pipeline incorporates dual-level class imbalance correction (SMOTE at the data level, Focal Loss at the algorithmic level) to enhance minority-class sensitivity without degrading specificity. Compared to Newaz et al., who used mean/mode imputation, min–max normalization, chi-square/correlation-based feature selection, and single-level SMOTE balancing, our framework ensures a more robust feature space, integrates advanced deep tabular learners, and implements a heterogeneous voting ensemble for improved generalization.
Importantly, this study introduces the Composite Performance Index (CPI) and Type I/II error rate analysis, absent in all compared works, providing a more clinically grounded assessment of predictive reliability. In direct benchmarking, our models consistently outperformed Newaz et al.’s results.
All methodological enhancements are unified within a modular, leakage-resistant pipeline that enforces strict separation between training and testing phases, supporting reproducibility. Post hoc feature importance analysis further identifies clinically coherent predictors, enhancing interpretability.
Table 13 presents a detailed methodological and performance comparison between the present study and the four prior works, highlighting differences in preprocessing strategies, class imbalance handling, model portfolios, evaluation metrics, and predictive outcomes.

Table 13.
Methodological comparison between the present study and recent works.
5.2. Implications for Triage and ICU Management
Machine learning models based on early triage data, such as vitals and initial clinical history, have demonstrated good performance in ICU admission and short-term mortality prediction, and have overall outperformed traditional scoring systems with ROC-AUC above 0.82–0.92 [57]. In the context of myocardial infarction, models trained on structured triage data can assist emergency departments in triaging high-risk patients at the time of admission. This allows for intervention earlier in time, more efficient use of ICU beds, and improved patient flow. Incorporation of mortality prediction into triage workflows can help reduce delays in the delivery of critical care and hospital resource use in the context of constrained availability.
5.3. Bioinformatics and Economic Perspectives
The application of denoising and variational autoencoders allows for the elusion of clinically meaningful patterns from complex, high-dimensional patient data, an approach that is in line with conventional bioinformatics practice. From an economic perspective, clinical decision support systems that utilize machine learning have shown promising results. Pacmed Critical, for example, which is a decision support system for the planning of ICU discharge, achieved an ICER of €18,507 per QALY by decreasing ICU length of stay through improved risk stratification [50]. The use of similar models for post-MI management can support timely decision-making while ensuring sustainable healthcare resource use.
5.4. Limitations
While the framework achieved superior predictive performance, SMOTE may introduce synthetic patterns that differ from real-world clinical data, though its stability and consistent validation justified its use. External validation on independent cohorts remains necessary to ensure generalizability. The subpopulation-based validation employed here, however, provides supporting evidence of robustness to demographic shifts, reinforcing the framework’s suitability for early mortality risk stratification.
5.5. Future Directions
To evaluate the robustness of the model in a variety of clinical settings, future research should give priority to external validation across multi-center datasets. Enhancing transparency and encouraging adoption by medical professionals may be achieved by incorporating interpretability strategies like attention-based visualization or SHAP values. To gauge the practical impact and viability of implementation, prospective clinical assessments and systematic health economic analyses will also be required.
6. Conclusions
This study presents a reproducible, end-to-end machine learning framework for predicting in-hospital mortality following myocardial infarction, integrating deep learning-based imputation, advanced normalization, dual-level class imbalance handling, and a diverse portfolio of state-of-the-art classifiers. The proposed pipeline operates within a strict, leakage-resistant evaluation design and incorporates a clinically meaningful subpopulation validation protocol, enhancing its robustness to demographic variation. This combination of methodological rigor and clinical awareness positions the framework as a practical tool for decision support in acute cardiac care.
The findings indicate that the integration of deep imputation methods, PowerTransformer-based normalization, and structured class imbalance correction enables consistent improvements in predictive performance and reliability across a range of models. Complementary evaluation using composite indices and error-type analysis highlights the framework’s capacity to balance sensitivity and specificity, addressing critical clinical priorities such as minimizing missed diagnoses while avoiding unnecessary interventions. Compared to prior work on the same dataset, the framework demonstrates substantive advances in preprocessing, evaluation breadth, and generalization capacity, particularly over studies that lacked one or more of these methodological components.
Beyond methodological contributions, the framework produces interpretable outputs, with feature importance analyses aligning with established pathophysiological mechanisms. This interpretability fosters clinician trust, facilitates targeted patient monitoring, and supports integration into triage and intensive care workflows. Its modular structure further allows adaptation to institutional priorities, enabling flexible tuning to emphasize either sensitivity or specificity depending on clinical and operational needs.
Nonetheless, limitations remain. The use of data from a single center constrains the diversity of patient populations. Future research should also explore health-economic implications to ensure that predictive gains translate into tangible benefits for patient outcomes and resource allocation.
In conclusion, this work advances the state of the art in post-MI mortality prediction by unifying methodological innovation, rigorous evaluation, and clinical interpretability into a deployable support framework, offering a foundation for both improved patient care and optimized healthcare operations.
Author Contributions
Conceptualization, K.P.F.; Methodology, K.P.F.; Software, K.P.F.; Formal analysis, K.P.F.; Data curation, K.P.F.; Writing—original draft, K.P.F.; Writing—review & editing, A.T.; Visualization, K.P.F.; Supervision, A.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The dataset and code used in this study are publicly available. Access information and relevant links are provided within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Heidari-Foroozan, M.; Farshbafnadi, M.; Golestani, A.; Younesian, S.; Jafary, H.; Rashidi, M.M.; Tabatabaei-Malazy, O.; Rezaei, N.; Kheirabady, M.M.; Ghotbi, A.B.; et al. National and Subnational Burden of Cardiovascular Diseases in Iran from 1990 to 2021: Results from Global Burden of Diseases 2021 Study. Glob. Heart 2025, 20, 43. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Cardiovascular Diseases (CVDs). Available online: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 28 June 2025).
- Zhang, X.; Wang, X.; Xu, L.; Ren, P.; Wu, H. The Predictive Value of Machine Learning for Mortality Risk in Patients with Acute Coronary Syndromes: A Systematic Review and Meta-Analysis. Eur. J. Med. Res. 2023, 28, 451. [Google Scholar] [CrossRef]
- Salari, N.; Morddarvanjoghi, F.; Abdolmaleki, A.; Rasoulpoor, S.; Khaleghi, A.A.; Hezarkhani, L.A.; Shohaimi, S.; Mohammadi, M. The Global Prevalence of Myocardial Infarction: A Systematic Review and Meta-Analysis. BMC Cardiovasc. Disord. 2023, 23, 206. [Google Scholar] [CrossRef]
- Yang, Y.; Tang, J.; Ma, L.; Wu, F.; Guan, X. A Systematic Comparison of Short-Term and Long-Term Mortality Prediction in Acute Myocardial Infarction Using Machine Learning Models. BMC Med. Inform. Decis. Mak. 2025, 25, 208. [Google Scholar] [CrossRef]
- Fanaroff, A.C.; Chen, A.Y.; Thomas, L.E.; Pieper, K.S.; Garratt, K.N.; Peterson, E.D.; Newby, L.K.; de Lemos, J.A.; Kosiborod, M.N.; Amsterdam, E.A.; et al. Risk Score to Predict Need for Intensive Care in Initially Hemodynamically Stable Adults With Non–ST-Segment–Elevation Myocardial Infarction. J. Am. Heart Assoc. 2018, 7, e008894. [Google Scholar] [CrossRef]
- Lee, W.; Lee, J.; Woo, S.I.; Choi, S.H.; Bae, J.W.; Jung, S.; Jeong, M.H.; Lee, W.K. Machine Learning Enhances the Performance of Short- and Long-Term Mortality Prediction Model in Non-ST-Segment Elevation Myocardial Infarction. Sci. Rep. 2021, 11, 12886. [Google Scholar] [CrossRef]
- Gupta, A.K.; Mustafiz, C.; Mutahar, D.; Zaka, A.; Parvez, R.; Mridha, N.; Stretton, B.; Kovoor, J.G.; Bacchi, S.; Ramponi, F.; et al. Machine Learning vs Traditional Approaches to Predict All-Cause Mortality for Acute Coronary Syndrome: A Systematic Review and Meta-Analysis. Can. J. Cardiol. 2025, in press. [CrossRef]
- Kumar, R.; Safdar, U.; Yaqoob, N.; Khan, S.F.; Matani, K.; Khan, N.; Jalil, B.; Yousufzai, E.; Shahid, M.O.; Khan, S. Assessment of the Prognostic Performance of TIMI, PAMI, CADILLAC and GRACE Scores for Short-Term Major Adverse Cardiovascular Events in Patients Undergoing Emergent Percutaneous Revascularisation: A Prospective Observational Study. BMJ Open 2025, 15, e091028. [Google Scholar] [CrossRef] [PubMed]
- Newaz, A.; Mohosheu, M.S.; Noman, M.A.A. Predicting Complications of Myocardial Infarction Within Several Hours of Hospitalization Using Data Mining Techniques. Inform. Med. Unlocked 2023, 42, 101361. [Google Scholar] [CrossRef]
- Diakou, I.; Iliopoulos, E.; Papakonstantinou, E.; Dragoumani, K.; Yapijakis, C.; Iliopoulos, C.; Spandidos, D.A.; Chrousos, G.P.; Eliopoulos, E.; Vlachakis, D. Multi-Label Classification of Biomedical Data. Med. Int. 2024, 4, 68. [Google Scholar] [CrossRef]
- Joshi, A.; Gunwant, H.; Sharma, M.; Chaudhary, V. Early Prognosis of Acute Myocardial Infarction Using Machine Learning Techniques. In Lecture Notes on Data Engineering and Communications Technologies, Proceedings of Data Analytics and Management, Polkowice, Poland, 26 June 2021; Springer: Singapore, 2022; Volume 91, pp. 815–829. [Google Scholar] [CrossRef]
- Lokeswar Reddy, K.; Thangam, S. Predicting Relapse of the Myocardial Infarction in Hospitalized Patients. In Proceedings of the 2022 3rd International Conference for Emerging Technology (INCET), Belgaum, India, 20–22 May 2022; pp. 1–6. [Google Scholar] [CrossRef]
- Al-Zaiti, S.S.; Martin-Gill, C.; Zègre-Hemsey, J.K.; Bouzid, Z.; Faramand, Z.; Alrawashdeh, M.O.; Gregg, R.E.; Helman, S.; Riek, N.T.; Kraevsky-Phillips, K.; et al. Machine Learning for ECG Diagnosis and Risk Stratification of Occluded Myocardial Infarction. Nat. Med. 2023, 29, 1804–1813. [Google Scholar] [CrossRef]
- Beaulieu-Jones, B.K.; Moore, J.H. Missing Data Imputation in the Electronic Health Record Using Deeply Learned Autoencoders. Pac. Symp. Biocomput. 2017, 22, 207–218. [Google Scholar] [CrossRef]
- Liu, M.; Li, S.; Yuan, H.; Ong, M.E.H.; Ning, Y.; Xie, F.; Saffari, S.E.; Shang, Y.; Volovici, V.; Chakraborty, B.; et al. Handling Missing Values in Healthcare Data: A Systematic Review of Deep Learning-Based Imputation Techniques. Artif. Intell. Med. 2023, 142, 102587. [Google Scholar] [CrossRef]
- Zamanzadeh, D.J.; Petousis, P.; Davis, T.A.; Nicholas, S.B.; Norris, K.C.; Tuttle, K.R.; Bui, A.A.T.; Sarrafzadeh, M. AutoPopulus: A Novel Framework for Autoencoder Imputation on Large Clinical Datasets. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2021, 2021, 2303–2309. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.L.; Zheng, H.; Gevaert, O. Genomic Data Imputation with Variational Autoencoders. GigaScience 2020, 9, giaa082. [Google Scholar] [CrossRef] [PubMed]
- Yeo, I.-K.; Johnson, R.A. A New Family of Power Transformations to Improve Normality or Symmetry. Biometrika 2000, 87, 954–959. [Google Scholar] [CrossRef]
- Kuhn, M.; Johnson, K. Applied Predictive Modeling; Springer: New York, NY, USA, 2013. [Google Scholar] [CrossRef]
- Zhu, J.; Pu, S.; He, J.; Su, D.; Cai, W.; Xu, X.; Liu, H. Processing Imbalanced Medical Data at the Data Level with Assisted-Reproduction Data as an Example. BioData Min. 2024, 17, 29. [Google Scholar] [CrossRef]
- Cusworth, S.; Gkoutos, G.V.; Acharjee, A. A Novel Generative Adversarial Networks Modelling for the Class Imbalance Problem in High-Dimensional Omics Data. BMC Med. Inform. Decis. Mak. 2024, 24, 90. [Google Scholar] [CrossRef]
- Liu, R.; Wang, M.; Zheng, T.; Zhang, R.; Li, N.; Chen, Z.; Yan, H.; Shi, Q. An Artificial Intelligence-Based Risk Prediction Model of Myocardial Infarction. BMC Bioinformatics 2022, 23, 217. [Google Scholar] [CrossRef]
- Singh, J.; Beeche, C.; Shi, Z.; Beale, O.; Rosin, B.; Leader, J.; Pu, J. Batch-Balanced Focal Loss: A Hybrid Solution to Class Imbalance in Deep Learning. J. Med. Imaging 2023, 10, 051809. [Google Scholar] [CrossRef]
- Vincent, P.; Larochelle, H.; Lajoie, I.; Bengio, Y.; Manzagol, P.A. Stacked Denoising Autoencoders: Learning Useful Representations in a Deep Network with a Local Denoising Criterion. J. Mach. Learn. Res. 2010, 11, 3371–3408. [Google Scholar]
- Ditzler, G.; Roveri, M.; Alippi, C.; Polikar, R. Learning in Nonstationary Environments: A Survey. IEEE Comput. Intell. Mag. 2015, 10, 12–25. [Google Scholar] [CrossRef]
- Ahsan, M.M.; Mahmud, M.A.P.; Saha, P.K.; Gupta, K.D.; Siddique, Z. Effect of Data Scaling Methods on Machine Learning Algorithms and Model Performance. Technologies 2021, 9, 52. [Google Scholar] [CrossRef]
- Yang, Y.; Sun, H.; Zhang, Y.; Zhang, T.; Gong, J.; Wei, Y.; Duan, Y.G.; Shu, M.; Yang, Y.; Wu, D.; et al. Dimensionality Reduction by UMAP Reinforces Sample Heterogeneity Analysis in Bulk Transcriptomic Data. Cell Rep. 2021, 36, 109442. [Google Scholar] [CrossRef]
- Bolón-Canedo, V.; Sánchez-Maroño, N.; Alonso-Betanzos, A. A Review of Feature Selection Methods on Synthetic Data. Knowl. Inf. Syst. 2013, 34, 483–519. [Google Scholar] [CrossRef]
- Hosmer, D.W.; Lemeshow, S.; Sturdivant, R.X. Applied Logistic Regression, 3rd ed.; Wiley: Hoboken, NJ, USA, 2013. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Cover, T.M.; Hart, P.E. Nearest Neighbor Pattern Classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.Y. LightGBM: A Highly Efficient Gradient Boosting Decision Tree. In Proceedings of the 31st Conference on Neural Information Processing Systems (NeurIPS), Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Prokhorenkova, L.; Gusev, G.; Vorobev, A.; Dorogush, A.V.; Gulin, A. CatBoost: Unbiased Boosting with Categorical Features. In Proceedings of the 32nd Conference on Neural Information Processing Systems (NeurIPS), Montréal, QC, Canada, 3–8 December 2018. [Google Scholar]
- Bauer, E.; Kohavi, R. An Empirical Comparison of Voting Classification Algorithms: Bagging, Boosting, and Variants. Mach. Learn. 1999, 36, 105–139. [Google Scholar] [CrossRef]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning Representations by Back-Propagating Errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2980–2988. Available online: https://openaccess.thecvf.com/content_ICCV_2017/papers/Lin_Focal_Loss_for_ICCV_2017_paper.pdf (accessed on 15 May 2025).
- Arik, S.O.; Pfister, T. TabNet: Attentive Interpretable Tabular Learning. In Proceedings of the AAAI Conference on Artificial Intelligence, Virtual Event, 2–9 February 2021; Volume 35, pp. 6679–6687. Available online: https://arxiv.org/abs/1908.07442 (accessed on 18 July 2025).
- Bergstra, J.; Bengio, Y. Random Search for Hyper-Parameter Optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Rockenschaub, P.; Akay, E.M.; Carlisle, B.G.; Hilbert, A.; Wendland, J.; Meyer-Eschenbach, F.; Näher, A.F.; Frey, D.; Madai, V.I. External validation of AI-based scoring systems in the ICU: A systematic review and meta-analysis. BMC Med. Inform. Decis. Mak. 2025, 25, 5. [Google Scholar] [CrossRef]
- Guo, H.; Li, Y.; Shang, J.; Gu, M.; Hu, Y.; Gong, B. Learning from Class-Imbalanced Data: Review of Methods and Applications. Expert Syst. Appl. 2017, 73, 220–239. [Google Scholar] [CrossRef]
- van den Goorbergh, R.; van Smeden, M.; Timmerman, D.; Van Calster, B. The Harm of Class Imbalance Corrections for Risk Prediction Models. medRxiv 2022. [Google Scholar] [CrossRef]
- Lobo, J.M.; Jiménez-Valverde, A.; Real, R. AUC: A Misleading Measure of the Performance of Predictive Distribution Models. Glob. Ecol. Biogeogr. 2008, 17, 145–151. [Google Scholar] [CrossRef]
- Saito, T.; Rehmsmeier, M. The Precision-Recall Plot Is More Informative than the ROC Plot When Evaluating Binary Classifiers on Imbalanced Datasets. PLoS ONE 2015, 10, e0118432. [Google Scholar] [CrossRef]
- Sasaki, Y. The Truth of the F-Measure. Teach. Tutor. Mater. 2007, 1, 1–5. [Google Scholar]
- López, V.; Fernández, A.; García, S.; Palade, V.; Herrera, F. An Insight into Classification with Imbalanced Data: Empirical Results and Current Trends on Using Data Intrinsic Characteristics. Inf. Sci. 2013, 250, 113–141. [Google Scholar] [CrossRef]
- Kubat, M.; Matwin, S. Addressing the Curse of Imbalanced Training Sets: One-Sided Selection. In Proceedings of the 14th International Conference on Machine Learning (ICML), Nashville, TN, USA, 8–12 July 1997; pp. 179–186. [Google Scholar]
- Fawcett, T. An Introduction to ROC Analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Matthews, B.W. Comparison of the Predicted and Observed Secondary Structure of T4 Phage Lysozyme. Biochim. Biophys. Acta Protein Struct. 1975, 405, 442–451. [Google Scholar] [CrossRef]
- Powers, D.M.W. Evaluation: From Precision, Recall and F-Measure to ROC, Informedness, Markedness and Correlation. J. Mach. Learn. Technol. 2011, 2, 37–63. [Google Scholar]
- Tschoellitsch, T.; Seidl, P.; Böck, C.; Maletzky, A.; Moser, P.; Thumfart, S.; Giretzlehner, M.; Hochreiter, S.; Meier, J. Using Emergency Department Triage for Machine Learning-Based Admission and Mortality Prediction. Eur. J. Emerg. Med. 2023, 30, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Greco, S.; Ishizaka, A.; Tasiou, M.; Torrisi, G. On the Methodological Framework of Composite Indices: A Review of the Issues of Weighting, Aggregation, and Robustness. Soc. Indic. Res. 2019, 141, 61–94. [Google Scholar] [CrossRef]
- Altman, D.G.; Bland, J.M. Diagnostic Tests. 1: Sensitivity and Specificity. BMJ 1994, 308, 1552. [Google Scholar] [CrossRef] [PubMed]
- Shreffler, J.; Huecker, M.R. Type I and Type II Errors and Statistical Power. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, January 2025. Available online: https://www.ncbi.nlm.nih.gov/books/NBK557530/ (accessed on 20 May 2025).
- de Vos, J.; Visser, L.A.; de Beer, A.A.; Fornasa, M.; Thoral, P.J.; Elbers, P.W.G.; Cinà, G. The Potential Cost-Effectiveness of a Machine Learning Tool That Can Prevent Untimely Intensive Care Unit Discharge. Value Health 2022, 25, 359–367. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).