1. Introduction
The rapid growth of scientific literature across various domains has created an overwhelming amount of information, making it increasingly challenging for researchers to extract relevant knowledge and insights from the vast amounts of available data. The ability to effectively extract information from multi-domain scientific documents is crucial for advancing research and facilitating knowledge discovery. Recent advances in natural language processing (NLP) and machine learning have paved the way for the development of automated information extraction methods. These methods can efficiently process large volumes of scientific texts and identify relevant information [
1].
Numerous studies have demonstrated the effectiveness of these methods in extracting information from scientific documents, including named entity recognition (NER) and relation extraction (RE) [
2]. However, the complexity and diversity of multi-domain scientific documents present considerable challenges to existing information extraction methods. This requires the development of more sophisticated and adaptable approaches [
3]. In contrast to the abundant availability of annotated corpora and text processing tools for English and Chinese, the public domain currently offers a very limited number of such resources for Kazakh. Moreover, the subject of information extraction from scientific texts has received scant attention from researchers, both in the Kazakh context and, to a lesser extent, in the Russian context. This underscores the scientific and practical significance of the present research.
While over 100 publicly available datasets exist for English in the field of NLP and Information Extraction (IE), including well-known benchmarks such as ACE2005 [
4], TACRED [
5], and SciERC [
6]-the situation is markedly different for other languages. For Russian, fewer than 10 scientific IE datasets are publicly available. Most of them are limited in scale or domain coverage, with resources like RuSERRC [
7] and NEREL [
8] representing some of the only large-scale efforts in this area.
In contrast, for Kazakh, only 2 to 3 general-purpose NER datasets are publicly available, most notably KazNERD [
9], which contains 112,702 sentences and 136,333 entity annotations. However, no open-access scientific corpora annotated for RE or domain-specific NER tasks currently exist for Kazakh. This lack of annotated scientific texts and tools makes research in Kazakh-language IE especially challenging and highlights the importance and novelty of developing resources and models for under-resourced languages.
The primary contributions of this study are as follows:
A new dataset of multi-domain scientific documents in Russian and Kazakh has been created, annotated with entities and relations.
A novel method for building a language model for relation extraction has been developed utilizing zero-shot learning techniques. This method performs effectively in the absence of substantial training data, making it particularly valuable for low-resource languages, such as Kazakh.
The remainder of this paper is organized as follows.
Section 2 reviews related work on the subject of scientific information extraction.
Section 3 provides a detailed description of the annotation process used in the analysis of the data.
Section 4 and
Section 5 present the methodology, experimental setup, and results for entity recognition.
Section 6 details the experimental setup and the results obtained from the application of RE methods. The subject of
Section 7 is the discussion of the insights and limitations. Finally,
Section 8 concludes the paper and outlines future work.
2. Related Work
IE from multi-domain scientific documents is a crucial area of research, enabling automated knowledge discovery, entity linking, and relation extraction across various disciplines. Numerous datasets have been developed to support these tasks, covering domains such as biomedical literature, artificial intelligence (AI), and machine learning (ML). These resources typically include annotations for NER, RE, coreference resolution, and event extraction.
One of the most prominent datasets in scientific text processing is SciERC, which focuses on identifying scientific entities, extracting inter-entity relations, and resolving coreference chains. The dataset consists of annotated scientific abstracts from AI, NLP, and ML research, and has become a benchmark for evaluating scientific IE models. Complementing this, SciER [
2] provides full-text publications in the ML and AI domains with manual annotations for entities and relations, supporting more granular and document-level information extraction.
While the development of English-language scientific IE datasets has been extensive, the situation for other languages is notably different. For Russian, the NEREL-BIO [
10] dataset provides annotations across general and biomedical domains. Baseline NER models evaluated on this dataset have demonstrated strong results, with F1-scores ranging from 70% to 83%, depending on entity type and model architecture.
The challenges are even more pronounced for Kazakh, where the scarcity of domain-specific datasets and pre-trained language models significantly hinders progress. KazNERD [
9] remains the most prominent resource, though no corpora for relation extraction or domain-specific NER tasks are currently available.
To address these gaps, recent research has explored deep learning and morphology-aware approaches tailored to the agglutinative nature of Kazakh. A hybrid neural model integrating word semantics, morphological embeddings, and graph attention networks has shown promising results in general-domain NER, achieving an F1-score of 88.04% [
11]. Similarly, the inclusion of root/entity embeddings and tensor layers in neural architectures has helped mitigate data sparsity and improve performance for morphologically rich languages like Kazakh [
12], with reported F1-scores exceeding 85% on KazNERD subsets.
A recent review of Kazakh morphological analysis methods highlights the differences between traditional machine learning models—such as Conditional Random Fields (CRFs) [
13]—and modern deep learning approaches, including RNNs, BERT [
14], and Transformer-based models. It emphasizes the need for Kazakh-specific adaptations in model architecture and training data [
15]. Studies comparing morphological parsers indicate that RNNs often outperform Transformers, achieving F1-scores between 85% and 89%, likely due to their compatibility with the language’s morphological complexity [
16].
Multi-task learning approaches have also been applied successfully in low-resource settings. For instance, integrating auxiliary tasks into NER training has yielded improved performance for Kazakh with limited annotated data, achieving F1-scores in the range of 84–87% [
17].
Despite these advances, scientific IE for both Russian and Kazakh remains fragmented and limited in scope. While Russian has begun to build domain-specific resources, Kazakh research remains focused largely on general-domain NER. The lack of annotated corpora, domain-specific datasets, and robust pre-trained models continues to be a major barrier, underscoring the urgent need to develop new resources and tools for these under-resourced languages.
3. Data Preparation
The methodology developed for data annotation for the tasks of entity and relation extraction consists of the following steps. Firstly, abstracts of scientific papers published in open-access journals from 2018 to 2024 were collected. These journals were recommended by the National Committee of the Republic of Kazakhstan to publish primary scientific research results. As the abstracts in such journals are written in Kazakh and Russian, both languages were collected.
The texts encompass four knowledge domains: Information Technology (Al-Farabi Kazakh National University. Journal of Mathematics, Mechanics and Computer Science. CC BY-NC 4.0 license.
https://bm.kaznu.kz/index.php/kaznu/issue/archive (accessed on 17 July 2025)), Linguistics (Al-Farabi Kazakh National University. KazNU Bulletin. Philology Series. CC BY-NC 4.0 license.
https://philart.kaznu.kz/index.php/1-FIL/issue/archive (accessed on 17 July 2025)), Medicine (Semey Medical University. Science and Healthcare. CC-BY 4.0 license.
https://newjournal.ssmu.kz/en/publication/releases/ (accessed on 17 July 2025)) and Psychology (Al-Farabi Kazakh National University. Journal of Psychology and Sociology. CC BY-NC 4.0 license.
https://bulletin-psysoc.kaznu.kz/index.php/1-psy/issue/archive (accessed on 17 July 2025)). The selection of these domains was guided by the goal of creating a diverse and representative dataset for evaluating performance in multi-domain settings. These domains were chosen to capture a broad spectrum of language use, conceptual structures, and knowledge representation styles, ensuring that the evaluation covers a wide range of challenges typically encountered in real-world applications of NLP and knowledge-based systems.
Information Technology reflects highly technical and structured content, characterized by frequent use of abbreviations and standardized terminologies. It represents domains with rapidly evolving vocabularies. Linguistics is a meta-domain concerned with the structure and function of language itself. It introduces specialized terminology that is conceptually abstract and often overlaps with other domains (e.g., cognitive science). The Medicine domain is representative of high-stakes, evidence-based fields where precision and disambiguation are critical. Medical texts are rich in domain-specific entities and hierarchical taxonomies, making them an essential test case for semantic understanding. The Psychology domain bridges the sciences and humanities. It includes both clinical terminology and theoretical constructs. It represents fields with complex conceptual relations and less standardized terminologies.
Secondly, our objective was to propose a relatively universal annotation scheme suitable for any domain of knowledge. The selection of entity and relation types was guided by principles of maximum expressiveness and non-redundancy. The annotation process was conducted in two stages. Initially, entities were annotated separately from relations.
3.1. Entity Annotation
The collected corpus was annotated for two categories of entities: scientific terms (TERM) and numerical values (VALUE). Terms are defined as words or phrases used in a specific domain to precisely denote particular concepts, phenomena, or objects. To illustrate this point, consider the field of Information Technology, where terms encompass the nomenclature of methods, architectures, models, programming languages, and related concepts. Similarly, in the domain of Medicine, terminology includes the nomenclature of diseases, symptoms, medications, diagnostic procedures, and other relevant concepts. It is important to note that abbreviations are also considered terms. Entities of the VALUE type are defined as numerical values accompanied by supplementary information, such as context or units of measurement. These quantitative or qualitative indicators are employed to describe specific data that can be measured or evaluated.
In the initial phase of the project, entities in the source texts were annotated using the “gpt-4o-mini” model [
18]. The methodology employed was predicated on cross-lingual transfer learning with a one-shot learning technique. The LLM prompt comprised an English-language instruction and a document with entity annotations in a specified format (see
Appendix A). Each entity is assigned a unique identifier. Entities are annotated in the following format: [Entity|Identifier|Label]. The label TERM indicates that the highlighted set of words is a term, while the label VALUE is used for numerical values. This markup format has been demonstrated to assist in the reduction of expenditure pertaining to API calls to LLMs.
The annotations were manually corrected, and each text was independently reviewed by two annotators using Label Studio. The moderator proceeded to address any residual ambiguities by referring to the annotation guidelines (
https://github.com/tvbat/sci-text-miner-scimdix (accessed on 17 July 2025)). An annotator instruction comprises a comprehensive description of various cases and numerous illustrative examples with explanatory notes for the domains of knowledge and languages under consideration. The consistency of the annotation was calculated using a standard statistical measure, namely Cohen’s kappa coefficient [
19]. The mean value obtained was 0.73, which is indicative of high annotation quality. The statistics for our dataset are presented in
Table 1 and
Table 2, respectively.
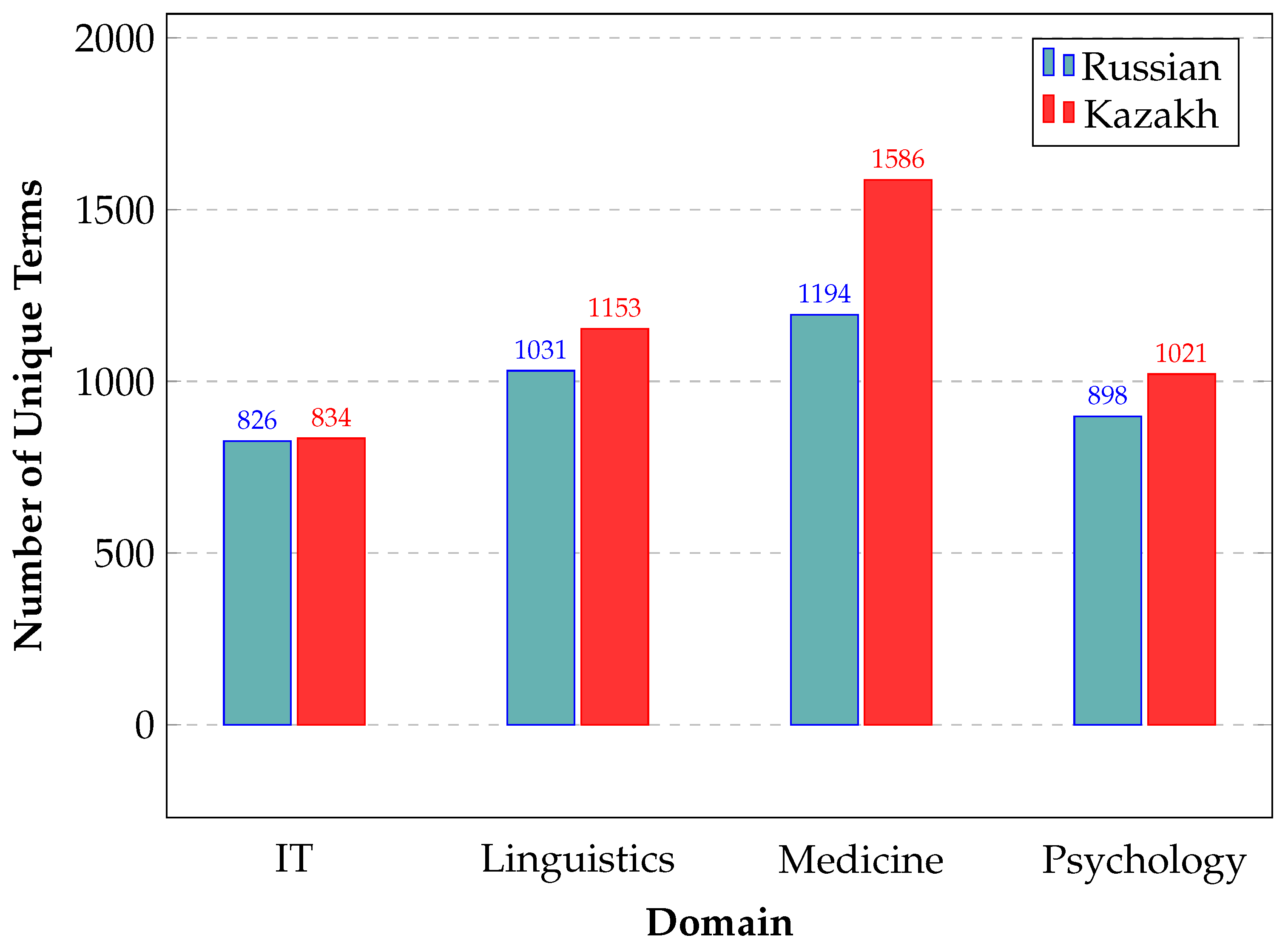
Specialized frequency dictionaries of terms for each domain have been compiled. The total number of unique terms is 3,949 for Russian and 4,594 for Kazakh.
Figure 1 presents detailed information organized by domain. As can be observed, the Medical domain shows a greater discrepancy in the number of unique terms between Kazakh and Russian. It may presumably be attributed to the linguistic features of the Kazakh language. For example, due to differences in word formation, a single medical term in Russian might correspond to multiple equivalents in Kazakh. Alternatively, because of semantic granularity, one term in Russian may cover a broad concept, whereas in Kazakh, distinct terms might be used for different aspects of that concept.
3.2. Relation Annotation
The initial step in the annotation process for relations between two entities within a text (document-level relations) is the identification of so-called morphological clusters. In the event of the same term appearing in different word forms (for example, in singular and plural forms or in different grammatical cases, which is relevant for Kazakh and Russian) or multiple times in the text, the identifiers of that term and its various word forms are merged into a single cluster. This stage of data preparation is referred to as deduplication. The execution of the process was conducted in an automated manner, employing the “gemini-2.0-flash” model. In the subsequent phase, the process of automatic relation annotation was executed, once more employing the “Gemini-2.0-Flash” model. For the purpose of this study, both languages were used, with the prompts for the LLM being written in English. In addition to the instructions, the prompt comprised a list of relation types accompanied by their respective descriptions, as well as exemplars of the required format for both input and output data. The prompt used for relation annotation is provided in the
Appendix B. The final stage of the data preparation process for relation extraction entailed the manual correction of the automatic annotations.
The annotation scheme of semantic relations includes six types: The following terms are to be noted: ’HAS_CHARACTERISTIC‘, ’HAS_PART‘, ’HAS_USE‘, ’HAS_VALUE‘, ’SUBCLASS_OF‘, and ’SYNONYM‘. It is important to note that a single entity can be involved in multiple relations concurrently. In the event of a sentence comprising multiple homogeneous entities that are semantically linked to another entity, it is essential that the relation is specified for each such pair. It is important to note that multiple relations may appear in a single sentence. The selection of relation classes was made in accordance with the following principles:
The relation should link terms within scientific texts.
The relation should have an unambiguous interpretation.
The relation types should be broadly universal to cover any knowledge domain.
Table 3 presents the relation types with descriptions and Wikidata properties. It is important to note that the HAS_CHARACTERISTIC relation differs from the HAS_PART relation in that HAS_PART (a meronymy relation) refers to components, not attributes. It is evident that ’HAS_CHARACTERISTIC’ exhibits a divergence from ’SUBCLASS_OF’ due to the nature of the latter, which is predicated on a hyponymy relation, signifying class membership rather than properties.
Table 4 and
Table 5 provide the distribution of relation types across knowledge domains for both languages.
4. Entity Recognition Methods
In this study, we have elected to utilize the following methods as baselines for the purpose of entity recognition: BERT, LLaMA, spaCy and GLiNER. To evaluate entity recognition models, the SciMDIX dataset was randomly split into 80% for training and 20% for testing.
4.1. BERT-Based Model
The present study adopted the method based on ruBERT [
20], obtaining the optimal results for the extraction of scientific terms from texts in Russian [
7]. The method combines a BERT model pre-trained on Russian texts with scientific term dictionaries collected in a semi-automatic manner, along with some heuristics. It is evident that these heuristics encompass a number of key principles. Firstly, it is evident that there is an absence of verbs and adverbs in terms. Secondly, prepositions are removed as the first token of a term. It should be noted that there are several other factors to be considered.
4.2. LLaMA-Based Model
The present study investigates the refinement of [
21] with 3B parameters, representing one of the most recent iterations of a publicly accessible large language model (LLM). In the training phase, the AdamW optimizer with 8-bit precision was employed in order to accelerate computations. Furthermore, the Low-Rank Adaptation (LoRA) technique [
22] was implemented. This method facilitates the optimization of LLMs through the reduction of the number of parameters necessary for model adaptation. This, in turn, minimizes the demands on memory and computational resources.
The model underwent training in four distinct domains: Information Technology, Linguistics, Medicine, and Psychology. Initially, each domain was trained separately, followed by joint training across both domains. The learning rate was set to , with a batch size of 2. The training was conducted for 2 epochs, with a maximum of 180 steps per epoch. The experiments were conducted using a single NVIDIA Tesla A100 GPU with 80 GB of memory.
4.3. GLiNER
Furthermore, experiments were conducted utilising GLiNER (Generalist Model for Named Entity Recognition using Bidirectional Transformer) [
23]. This model employs a smaller-scale bidirectional language encoder as opposed to the utilization of large autoregressive models. The proposed methodology treats NER as a matching task between entity type embeddings and textual span representations in latent space, rather than a generation task. This approach naturally resolves scalability issues and enables bidirectional context processing, leading to richer and more contextualized representations. The model displays remarkable cross-lingual robustness, thereby emphasizing its strong generalization capabilities.
4.4. SpaCy Model
As one of the NER baselines, we considered a CNN-based model with the spaCy tok2vec embedding layer [
24]. This model produces token-level vector representations that capture both meaning and context. Furthermore, it is characterized by its capacity for facile adaptation to diverse linguistic and knowledge domains.
The spaCy tok2vec model was evaluated on the prepared dataset through two different experiments. In the initial instance, the training and testing processes were executed in a manner that was distinct for each individual domain-specific dataset. In the second case, all domain datasets were combined for joint training and testing. The training parameters that were determined to be optimal for this model on the specified dataset are outlined below: learning rate = 0.001, batch size = 1000, number of epochs = 100, eval_frequency(steps) = 200, dropout = 0.1. During the training process, overfitting was observed at epoch 59 (corresponding to 11,800 iteration steps) on the Russian data and at epoch 43 (corresponding to 8,600 steps) on the Kazakh data. The Adam optimizer was utilized to optimize the parameters. Among the models, spaCy tok2vec demonstrated the best overall performance.
5. Results of Entity Recognition
A comprehensive analysis of model effectiveness, sensitivity to language, and the impact of domain characteristics was conducted by analyzing the results obtained during experiments on entity recognition across four models (BERT, LLaMA, GLiNER, and spaCy). These models were evaluated on two languages (Kazakh and Russian) and four domains (IT, Linguistics, Medicine, and Psychology).
The BERT-based model (see
Table 6) demonstrates a strong dependency on language resources. The model performs significantly better on Russian, with an average F1 score of 68.88% across all domains, compared to only 34.50% for Kazakh. This discrepancy likely reflects the availability of richer pretrained embeddings and annotated data for Russian. Among the individual domains, BERT achieves its highest performance in the Linguistics domain for Kazakh (F1 = 41.37%) and in the IT domain for Russian (F1 = 74.71%). However, the overall results for Kazakh remain modest, highlighting the limitations of BERT in low-resource language settings.
The LLaMA model (see
Table 7) provides a noticeable improvement over BERT, particularly for Kazakh. The average F1 score for Kazakh increases from 34.50% (BERT) to 53.36% (LLaMA), while Russian maintains a slightly higher average of 54.44%. Although the gap between languages persists, it is narrower than with BERT, suggesting that LLaMA is more adaptable to under-resourced contexts. These findings indicate that LLaMA can generalize better across domains and languages compared to BERT.
GLiNER results (see
Table 8) demonstrate a significant enhancement in performance, particularly with regard to the Kazakh language. The mean F1 score for Kazakh is 75.37%, while Russian achieves a mean score of 79.65%. GLiNER demonstrates particular efficacy in the domains of Linguistics and Medicine for the Russian language and in the domain of Linguistics for Kazakh, indicating its proficiency in semantically rich and structurally regular texts. Furthermore, the model maintains a satisfactory balance between precision and recall, indicating consistent entity recognition across domain-specific variations.
The spaCy model (see
Table 9) outperforms all other models by a large margin, achieving state-of-the-art results across both languages and all domains. For Kazakh, spaCy attains an average F1 score of 96.84%, while for Russian it reaches 97.36%. The model maintains high scores in both precision and recall, reflecting its robustness and reliability. Particularly noteworthy is its performance in the Linguistics+IT domain, where it achieves an F1 score of 97.94% for Kazakh and 98.30% for Russian. These results demonstrate spaCy’s strong ability to generalize across different languages and specialized domains, making it the most effective model in this study.
To sum up the findings, while BERT encounters challenges with low-resource languages such as Kazakh, LLaMA demonstrates moderate enhancements, and GLiNER exhibits substantial gains, particularly in domains characterized by linguistic richness. Notably, spaCy achieves near-perfect scores across various evaluation metrics, indicating minimal performance disparity between Kazakh and Russian. Furthermore, the study emphasizes the viability of spaCy and GLiNER for implementation in practical applications involving under-resourced languages.
6. Relation Extraction Methods
The extraction of relations is a key component in the process of structuring knowledge from scientific texts. In academic writing, semantic connections between entities such as concepts, terms, authors, methods, and findings are often implicitly expressed within varying contexts and across different levels of text granularity, ranging from individual sentences to whole paragraphs. This inherent complexity renders the extraction of such relations a particularly arduous task. The issue is further compounded in low-resource language settings, such as Kazakh, where there is a paucity of annotated corpora available. In order to address these challenges, a relation classification model based on zero-shot learning is employed, which allows for the extraction of meaningful relations even in the absence of training examples for target relation types, even in another language.
There are a number of approaches to the classification of relations between entities. The prevailing methodologies in this field depend on the utilization of supervised learning over fully annotated corpora, frequently employing transformer-based architectures. In contrast, alternative approaches involve the exploration of generative models that generate textual descriptions of relations given a contextual input. However, such techniques generally require substantial amounts of annotated data, which restricts their applicability in low-resource languages or niche scientific domains. Zero-shot learning constitutes an alternative strategy, whereby auxiliary semantic information is employed to facilitate the generalization of models to unseen classes or languages. In this particular instance, textual descriptions of relation types are utilized as the auxiliary semantic information. The latter approach is implemented in the model that was used in our study to transfer knowledge from Russian to Kazakh while working with scientific texts.
In order to evaluate the applicability of the proposed model for the classification of scientific relations, a series of experiments was conducted using SciMDIX, a newly introduced dataset in this paper containing Russian and Kazakh scientific documents annotated with entities and semantic relations.
6.1. Data Preprocessing
Transformer-based language models function as the fundamental encoding framework. These models, such as BERT [
14], E5 [
25] have achieved a high level of recognition for their capacity to generate context-sensitive representations that are able to account for the syntactic and semantic relations between words in a sentence. Vector representations are computed for each token in the sequence, from which three key components are extracted: firstly, the [CLS] token, which captures the global sentence context; secondly, the averaged embedding of all tokens belonging to the first entity; and thirdly, the averaged embedding for the second entity. This explicit bracketing serves to guide the attention mechanism of the model, focusing it on entity-relevant parts of the input. These three vectors are then concatenated and processed through fully connected neural layers with non-linear activations, resulting in a unified feature vector that encodes both global and entity-specific information.
6.2. Model Architecture
This section presents a novel relation extraction method distinguished by its integration of transformer-based encoding, explicit entity marking, and multimodal learning. This integration yields a model capable of effectively generalizing to unseen relation types, adapting to new domains, and facilitating cross-lingual transfer.
Unlike the study [
26], whose authors propose a Relation Contrastive Learning (RCL) method for zero-shot relation extraction, the method proposed in this study, in addition to RCL, uses multimodal feature spaces, including textual descriptions of relation classes. Both methods enhance the zero-shot capability of models but use different strategies: RCL focuses on separating classes in a learned high-dimensional space, while our method uses integrated semantic representations for improved relation extraction. The proposed method for building the RE model consists of the following main steps:
Constructing a unified feature vector representation leveraging entity embeddings and contextual information (see
Section 6.1).
Generating contextualized vector representations of textual descriptions of relations using a pre-trained Transformer model (such as BERT or E5).
Building a shared feature space for two modalities using cross-entropy minimization or triplet loss techniques. The selected strategy is based on a multimodal representation that integrates both textual context and class-level semantics to construct a unified feature space for relation classification. Specifically, it combines the distributed representations of the sentence context, the two entity mentions, and the textual definitions of possible relation classes. These elements are encoded in unison to form rich, transferable semantics that support zero-shot inference by projecting both seen and unseen relation types into a shared representation space.
6.3. Training and Evaluation Details
The RE model was evaluated in two learning settings: a standard multi-class classification scenario and a zero-shot evaluation setup. In the first scenario, the model was trained and tested on the Russian-language subset of SciMDIX, which was initially split into training and test sets with an 80/20 ratio, respectively. In the second scenario, the model was trained on the same training set (consisting of texts in Russian) and applied to a cross-lingual relation classification task in Kazakh. The presented results in
Table 10 demonstrate the performance of several baseline and advanced models in both fully supervised (Russian → Russian) and cross-lingual zero-shot (Russian → Kazakh) relation classification scenarios, measured by F1-score.
Across the supervised setting, the R-BERT [
27] model shows the highest F1-score (0.687), which is consistent with its design for fine-grained token-level relation extraction using explicit entity markers and full supervision. However, its performance significantly deteriorates in the cross-lingual scenario (0.564), highlighting a common limitation in traditional supervised models when applied to unseen languages.
By contrast, the proposed model shows highly balanced performance across both settings. It achieves 0.639 F1-score on the Russian test set (second-best overall) and reaches the highest F1-score (0.640) under the zero-shot setting. This finding indicates that the model generalizes well beyond language boundaries, likely due to its use of textual class descriptions and multimodal representation learning that aligns entity-context information with semantic type definitions in a shared embedding space.
Among alternative approaches, we observe that E5-based models perform surprisingly well in cross-lingual adaptation (0.637), outperforming their performance in the supervised setting and nearly matching the results of the proposed method. This suggests that encoder-only models pretrained with task-agnostic objectives may yield useful representations for transfer learning. However, the proposed model remains the only one showing consistently high values in both scenarios, without substantial performance degradation, making it a robust option for low-resource and cross-lingual scientific information extraction.
These results underscore the importance of model architecture and training strategy in designing systems for multilingual relation extraction. In particular, leveraging external semantic knowledge through textual descriptions and multimodal integration appears to be a key factor in achieving reliable generalization to unseen relation types and languages.
In addition to its primary function of relation classification, the proposed model demonstrates potential for use in automatic or semi-automatic annotation of relations within newly developed scientific corpora. Its zero-shot architecture, combined with the ability to interpret textual descriptions of relation types, enables the model to identify likely semantic connections between entities in previously unseen domains. This makes it particularly valuable in contexts involving domain adaptation and limited annotated data resources.
Although this annotation scenario has not yet been validated through empirical experimentation, the model’s design and zero-shot performance indicate its potential suitability for such applications. Investigating the use of the proposed model to provide weak or auxiliary supervision for relation classification represents a promising direction for future research in scientific information extraction.
7. Discussion
This study makes a valuable contribution to the information extraction from scientific documents, particularly in the context of multi-domain data and low-resource languages. The creation of a novel annotated dataset and the evaluation of entity recognition and relation extraction models offer significant insights into the challenges and opportunities in this area. However, a thorough review of the findings indicates numerous constraints that necessitate further deliberation.
Our findings indicate that spaCy, a relatively lightweight model, often surprisingly outperformed larger, more complex models as BERT and LLaMA. This counterintuitive result can be attributed to several factors. Firstly, SpaCy library provides full-fledged models and pipelines for several production-oriented NLP tasks (
https://spacy.io/models (accessed on 17 July 2025),
https://spacy.io/usage/processing-pipelines (accessed on 17 July 2025)). Secondly, we fine-tuned spaCy’s NER model for Russian and Kazakh languages to further optimize its performance for the domains considered in the study. SpaCy’s pre-trained models, though smaller, are often trained on large, general-purpose corpora, providing a strong foundation for transfer learning to scientific domains. In contrast, while LLaMA possesses extensive knowledge, its general-purpose nature may require substantial fine-tuning on domain-specific data to achieve optimal performance. The lack of sufficient domain-specific training data in our low-resource setting may have hindered LLaMA’s ability to adapt effectively. Furthermore, the computational cost of fine-tuning LLaMA is significantly higher than that of spaCy, making it less practical for many real-world applications. While GLiNER generally performs well, its comprehensiveness and accuracy may be limited for low-resource and non-Latin languages like Kazakh. To draw more reliable conclusions and determine whether the observed performance differences between models are significant, appropriate statistical testing should be conducted.
The annotation process itself introduces potential biases that could affect the quality of the dataset and the performance of the models trained on it. Annotators may have varying interpretations of the annotation guidelines, leading to inconsistencies in the annotations. One of the significant limitations of this study lies in the uneven distribution of entities and relations across the four domains in our dataset. The Medicine domain contains a higher number of entities compared to IT, Linguistics, and Psychology. Models trained and evaluated on this dataset may exhibit a preference for recognizing entities prevalent in the Medicine domain, leading to inflated performance metrics for this domain and potentially underestimating performance in other domains. To verify this, future work should address this issue by employing techniques such as data augmentation, re-sampling, or cost-sensitive learning to mitigate the impact of class imbalance.
While our RE model demonstrates promising zero-shot capabilities, its domain transferability remains a key concern. The model’s performance relies heavily on the quality and relevance of the textual definitions of relation classes used during inference. If these definitions are ambiguous, incomplete, or not representative of the target domain, the model’s performance could suffer significantly. Furthermore, its ability to generalize to unseen relations is limited by its reliance on pre-trained language models and the knowledge they encode. If a relation is not well-represented in the pre-training data, our model might struggle to accurately identify it. Future research should focus on enhancing the model’s ability to transfer across domains. This could involve integrating domain-specific knowledge into the model or designing approaches for automatically generating more informative and precise relation definitions.
8. Conclusions
In the present paper, a novel multi-domain dataset of scientific documents in Russian and Kazakh is presented, annotated with entities and relations. A comprehensive analysis of model effectiveness was conducted by analyzing the results obtained during experiments on entity recognition across four models (BERT, LLaMA, GLiNER and spaCy). These models were evaluated on two languages (Kazakh and Russian) and four domains (IT, Linguistics, Medicine, and Psychology).
In order to solve the relation classification task under zero-shot learning conditions, we proposed a new method for building a language model that leverages a multimodal representation of input data by combining the context of the sentence, mentions of the entities, and textual definitions of the relation classes. Exploring this method for generating weak or auxiliary supervision for relation classification signifies a promising avenue for future research in the domain of scientific information extraction.